Summary
Srs2 dismantles presynaptic Rad51 filaments and prevents its re-formation as an anti-recombinase. However, the molecular mechanism by which Srs2 accomplishes these tasks remains unclear. Here we report a single molecule fluorescence study of the dynamics of Rad51 filament formation and its disruption by Srs2. Rad51 forms filaments on ssDNA by sequential binding of primarily monomers and dimers in a 5’ to 3’ direction. One Rad51 molecule binds to 3 nucleotides and six monomers are required to achieve a stable nucleation cluster. Srs2 exhibits ATP-dependent repetitive scrunching of ssDNA and this activity prevents reformation of the Rad51 filament. The same activity of Srs2 cannot prevent RecA filament formation, indicating its specificity for Rad51. Srs2’s DNA unwinding activity is greatly suppressed when Rad51 filaments form on duplex DNA. Taken together, our results reveal an exquisite and highly specific mechanism by which Srs2 regulates Rad51 filament formation.
Introduction
DNA double strand breaks (DSBs) are the principle cytotoxic damage repaired by homologous recombination (HR), which is mediated by the Rad51 recombinase in eukaryotes (Paques and Haber, 1999; Symington, 2002). However, inappropriate or untimely HR leads to mutagenic and oncogenic consequences. Therefore, specific mechanisms have evolved to regulate HR and to minimize deleterious outcomes (Hu et al., 2007; Sung and Klein, 2006; Wu and Hickson, 2006). In the budding yeast Saccharomyces cerevisiae, Srs2 is one of the major HR regulators that functions in the early stages of the HR pathway. Srs2 is an ATP-dependent 3’-5’ DNA helicase and ssDNA translocase (Antony et al., 2009) that prevents HR by dismantling Rad51 filaments(Krejci et al., 2003; Le Breton et al., 2008; Veaute et al., 2003). Srs2 mutants that cannot bind or hydrolyze ATP fail to disrupt Rad51 filaments and display a hyper-recombination phenotype when subjected to genotoxic stress (Krejci et al., 2004). This anti-recombinase activity requires direct interaction of Srs2 with Rad51 and may require Srs2 translocation along ssDNA (Antony et al., 2009). Consistent with this view, Srs2 cannot remove a Rad51 mutant that lacks the Srs2 interaction domain (Colavito et al., 2009; Seong et al., 2009). This same mechanism may be preserved in the human helicase RECQ5 which also disrupts human Rad51 filaments via direct physical contact (Schwendener et al., 2010).
We used single molecule fluorescence to investigate the molecular mechanism controlling the balance between Rad51 filament formation and disruption by Srs2. Using single molecule FRET (Förster resonance energy transfer), we find the Rad51 site size to be three nucleotides and its nucleation cluster to be six monomers. We employed an alternative detection method, PIFE (protein induced fluorescence enhancement) (Fischer et al., 2004; Hwang et al., 2011; Myong et al., 2009) to determine that Rad51 loading on a ss/ds DNA occurs directionally from the 5’ duplex junction to the 3’ ssDNA end. We also report a novel activity for Srs2 – a repetitive scrunching mechanism that enables an Srs2 monomer to prevent reformation of Rad51 filaments. This activity occurs after the initial clearance of Rad51. Together, both activities contribute to its anti-recombinase function. Finally, the DNA unwinding activity of Srs2, which requires multimers of Srs2 (Antony & Lohman, unpublished data) was greatly diminished by Rad51 bound to duplex DNA, indicating a further balance between the helicase and anti-recombinase activity of Srs2. Our study reveals novel mechanistic details that govern the early stages of the HR DNA repair pathway.
Results
Rad51 monomers bind to a 3 nucleotides and six monomers form a nucleation cluster
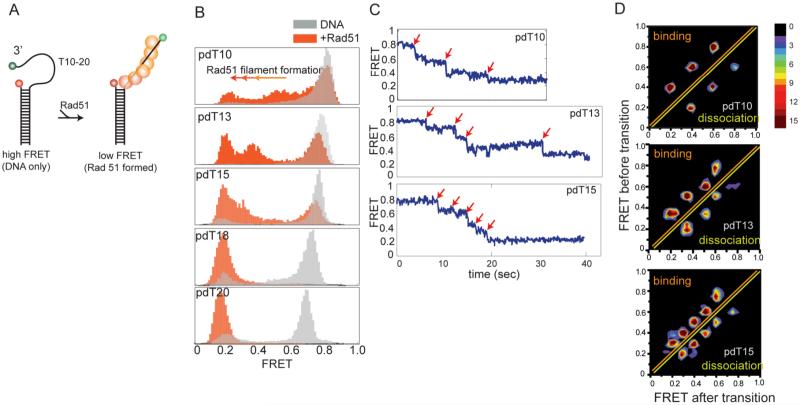
To visualize Rad51 filament formation, we prepared DNA substrates that mimic a resected DNA intermediate found during homologous recombination, which are the preferred substrates for Rad51 formation and strand invasion (Mazin et al., 2000). These are partial DNA duplexes with variable lengths of 3’ single stranded (ss) DNA composed of poly deoxythymidine, T10, 13, 15, 18 and 20, which we will refer to as pdT10 to pdT20 hereafter (Figure 1A). Each DNA possesses two fluorophores, Cy3 and Cy5 located at the 3' end of the ssDNA and at the duplex DNA end, respectively (Figure 1A), such that DNA alone exhibits high FRET due to the high flexibility of the ssDNA (Murphy et al., 2004) and a FRET decrease occurs upon Rad51 binding due to stretching of the ssDNA. The histograms of FRET values built from over 5,000 molecules of DNA show FRET peaks at around 0.85, 0.8, 0.75, 0.7, 0.65 for pdT10, 13, 15, 18 and 20, respectively (Figure 1B, gray histograms). Upon addition of Rad51 (1μM) with ATP (1mM) and MgCl2(10mM), the high FRET peaks disappear and another low FRET population arises within the first three minutes, indicating formation of Rad51 filaments on ssDNA (Figure 1B, orange histograms). The complete shift from high to low FRET observed for pdT18 and pdT20 indicates formation of stable Rad51 filaments whereas the partial shift seen for pdT15 and shorter ssDNA suggests a less stable filament.
Figure 1.
Rad51 monomer binds 3 nucleotides and a stable nucleation requires 6 monomers (A) Schematic diagram of DNA constructs used in Rad51 binding experiment. Partially duplexed DNA of various poly-Thymidine tail lengths (T10-T20) contain donor (Cy3) at 3’ end and acceptor (Cy5) at duplex junction, FRET value decreases upon Rad51 formation. (B) FRET values collected over 5000 single molecules are built into a FRET histogram. DNA-only high FRET molecules (gray) transition to low FRET (orange) upon Rad51 formation. A complete shift to low FRET is observed in T18 and T20. (C) Single molecule FRET traces show up to three, four and five steps observed for T10, T13 and T15, reflecting a monomer binding steps. (D) Transition Density Plot (TDP) for each DNA built by collecting FRET values before (y-axis) and after (x-axis) transition from single molecules that show monomer binding steps.
Examples of single molecule FRET traces show that the addition of Rad51 induces a transition from high FRET to low FRET in a stepwise manner, likely representing the sequential binding of Rad51 molecules (Figure 1C). For pdT10, 13 and 15, a maximum of three, four and five steps of FRET decrease were observed, suggesting the binding of up to three, four and five Rad51 monomers. We collected FRET values from 40-50 single molecule traces that display clear steps and built a transition density plot (TDP) where the y- and x- axes represent FRET before and after the transition, respectively. As shown in Figure 1D, three, four and five peaks are observed in the TDP for T10, 13 and 15, above the diagonal line, reflecting that one Rad51 monomer binds to three nucleotides (nt). This is consistent with the observations in the crystal structure of the Rad51 filament (Conway et al., 2004) and the RecA-ssDNA filament (Chen et al., 2008; Joo et al., 2006) Due to the unstable binding of Rad51 to these shorter lengths of ssDNA, we obtained mirror image peaks below the diagonal line, reflecting dissociation of Rad51. This set of data suggests that a Rad51 monomer binds 3 nucleotides of ssDNA and that six monomers are required to form a stable nucleation cluster.
In addition to the FRET steps arising from monomer binding, we also observe single molecule traces with larger steps of FRET decrease and increase representing binding and dissociation, respectively, of larger Rad51 oligomers (Figure S1B). Based on the well-defined FRET states corresponding to monomer Rad51 binding, the observed FRET values (>800 molecules) were assigned as monomers to tetramers (Figure S1A-C). The analysis suggests that approximately 75% of Rad51 binds as either a monomer or dimer to pdT15 (Figure S1D). We note that binding of Rad51 oligomers larger than tetramers cannot be distinguished from photobleaching of the Cy5 dye, yet over 95% of the molecules exhibited smaller FRET changes corresponding to binding of Rad51 monomers, dimers or trimers.
Rad51 filaments form in the 5’ to 3’ direction from the duplex junction toward ssDNA
Next, we examined the directionality of Rad51 filament formation. Ensemble stopped-flow experiments suggest a 5’ to 3’ directional bias for Rad51 filament formation, however the substrate used was ssDNA without a duplex junction (Antony et al., 2009). The DNA substrates used in our experiments mimic a resected DNA, an intermediate found in early stage of homologous recombination. Although FRET is useful for visualizing monomer binding, it does not inform us whether filament growth occurs with directionality on ssDNA (e.g. growing from 3’ to 5’ or 5’ to 3’). We took advantage of an alternative fluorescence method, smPIFE whereby the intensity of a Cy3 fluorophore increases upon protein binding in its vicinity (Antony et al., 2009; Fischer et al., 2004; Hwang et al., 2011; Myong et al., 2009). The two DNA constructs used in the PIFE assay had a Cy3 fluorophore located at either the 3’ or near the 5’ end of the ssDNA (Figure 2A, C)(Modesti et al., 2007; Ristic et al., 2005). When Rad51 was added, the DNA with Cy3 at the 3’ end showed a multi-step Cy3 fluorescence increase (Figure 2B) whereas the one with Cy3 positioned at the duplex junction showed a single step increase in Cy3 fluorescence immediately after Rad51 addition (Figure 2D). This result indicates that the Rad51 filament extends from the 5’ toward the 3’ end of the ssDNA. We note that the intensity increase is greater for Cy3 at the 3' end because the dye is located at a conformationally more flexible position which is more sensitive to the PIFE effect. In contrast, the Cy3 near the duplex is more sterically restricted due to the neighboring DNA bases on both sides, resulting in a lower enhancement of the fluorescence (Sanborn et al., 2007; Sorokina et al., 2009). The same pattern is also observed when the intensities of single molecule traces are averaged. The intensity increase was normalized by its minimum value to allow a more direct comparison of the rates of increase at both positions (Figure 2E). The rate is approximately 3.3 fold higher for the 5’Cy3 than the 3’Cy3, indicating that the filament initiates preferentially from the 5’ end (duplex junction). Both sets of data are consistent with initiation of the Rad51 filament at the duplex junction and assembly/extension towards the 3’ end of the ssDNA. Rad51 can also form a filament on double stranded (ds) DNA (Krogh and Symington, 2004; Modesti et al., 2007; Ristic et al., 2005), but the rate of filament assembly on dsDNA is approximately 6 fold lower (0.05 s−1) than the rate observed for assembly on ssDNA (0.28 s−1; Figure S2A-E). Based on these results, Rad51 binding to dsDNA does not influence the directionality of filament formation on the ssDNA substrates tested here.
Figure 2.
Rad51 binds in 5’ to 3’ direction (A) Schematic of 3’ Cy3 labeled DNA used in PIFE assay. (B) Single molecule trace displaying a gradual and stepwise increase in Cy3 signal. (C) Schematic of 5’ Cy3 labeled DNA used in PIFE assay. (D) Single molecule trace showing an abrupt increase in Cy3 signal. (E) Normalized average intensity plot for both DNA constructs showing slower increase in 3’ Cy3 DNA compared to 5’ Cy3 DNA. Errors in fit results are in standard error of mean (SEM). (F) Schematic summary of Rad51 binding in 5’ to 3’ direction.
Srs2 monomer displays repetitive movement on DNA
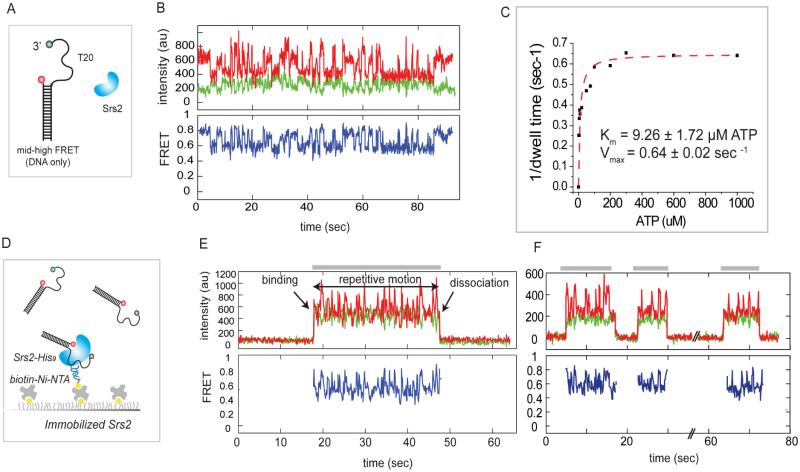
To probe the translocation activity of Srs2 which is believed to be necessary for its anti-recombinase function (Antony et al., 2009), we examined its ATP stimulated movement on pdT20 (Figure 3A). We examined a well characterized C-terminal deletion mutant, Srs2CΔ276 which exhibits ATP-driven translocation and anti-recombinase activity (Antony et al., 2009). When Srs2 was added with ATP, we obtained single molecule traces of highly repetitive FRET fluctuations reflecting repeated motion of Srs2 on ssDNA which induces repetitive distance changes between the two ends of the ssDNA (Figure 3B). These fluctuations require ATP hydrolysis since they are not seen in the absence of ATP or presence of ATPγS (Figure S3A). Furthermore, the periodicity measured by collecting dwell times (δt) from over three hundred events shows a dependence on ATP concentration (Figure S3B, C). A fit of these data to the Michaelis-Menten equation yields Km = 9.26±1.72 μM ATP and Vmax = 0.64±0.02/sec (Figure 3C). To test if the FRET fluctuations arise from the action of a monomer of Srs2 protein or successive binding of multiple monomers, we tethered a histidine tagged Srs2 (0.5-1nM) to a surface treated with biotinylated Ni-NTA (Figure 3D) and added Cy3-Cy5 labeled DNA substrate (non-biotinylated). Here, we monitored FRET fluctuations, sometimes in bursts representing the repetitive motion of a Srs2 monomer on a single DNA (Figure 3E-F)). The DNA binding is highly specific to the presence of surface tethered Srs2 as we do not see any fluorescence spots in the absence of Ni-NTA or Srs2. This tethered Srs2 assay enables us to clearly distinguish the moment of DNA binding as an abrupt appearance of FRET, duration of repetitive motion as a continuous FRET fluctuation (gray bars) and the DNA dissociation as the disappearance of FRET (Figure 3E, F). This provides direct evidence that a monomer of Srs2 is sufficient for the repetitive motion on the resected DNA. We note that this activity is not the initial directional translocation of Srs2 on ssDNA, which occurs at a rate of 300 nts/sec (Antony et al., 2009), which is beyond our time resolution of 30-100 ms, but what occurs after the fast translocation.
Figure 3.
Srs2 exhibits repetitive motion on ssDNA (A) Schematic diagram of FRET DNA used in Srs2 assay. (B) Single molecule traces displaying Srs2 induced FRET fluctuation. (C) The inverse of FRET peak to peak dwell time was fitted to Michaelis-Menten equation. Errors in fit results are in SEM. (D) Schematic diagram of tethered protein assay. Histidine (9x) tagged Srs2 was immobilized to surface and non-biotinylated FRET-DNA substrate was applied. (E) Single molecule traces show that the repetitive motion arises from a single monomer of Srs2. DNA binding, Srs2 mediated repetitive motion and dissociation from Srs2 are shown as an appearance of FRET, FRET fluctuation and disappearance of FRET, respectively. (F) Single molecule trace shows multiple bursts of repetitive events arising from single Srs2 monomer.
Srs2 displays repetitive scrunching of ssDNA at ss/ds junction
The FRET traces obtained above suggest that Srs2 repeatedly brings the 3’ end of the ssDNA close to the duplex junction. To probe the mechanistic basis of this repetitive action, we varied the lengths of the ssDNA from pdT13 to pdT70 (Figure 4A). We observe similar FRET fluctuations when the tail length is between 13 and 20, with similar amplitude changes as depicted both in single molecule traces (Figure 4B) and FRET histograms taken from over one hundred events per given tail length (Figure 4C). As the tail length is increased to 25 and 30 nts, FRET fluctuations occur at a lower FRET range with less pronounced FRET peaks. Such FRET patterns completely disappear for the pdT70 DNA (Figure 4B, C). Dwell time analyses performed on all of the tail lengths (T13-30) suggest that the fluctuations are not length dependent beyond 20 nts, indicating that the repetitive movement is limited to ~18-20 nts of ssDNA length (Figure 4D, E). Both the FRET pattern and dwell time analysis point to a mechanism whereby Srs2, while bound at the junction periodically reels in a finite segment of ssDNA of approximately 18-20 nts, rather than the entire length of ssDNA. To test this hypothesis further, we prepared three substrates with tail lengths of pdT30, pdT40 and pdT50, but with each containing two dyes separated by a fixed contour length of 15 nts (Figure S4A). Consistent with the previous experiment, we observed the same FRET fluctuation with similar periodicity for all three DNA substrates (Figure S4B-D). Based on these data, we propose a mechanism in which Srs2 after the initial translocation on ssDNA, situates itself near the duplex junction while pulling in a finite length of ssDNA in a repetitive manner. Experiments with fluorescently labeled Srs2 further support this model of junction anchored Srs2 repetitively scrunching the 3’ tail as FRET fluctuations are only seen on a short tail with a dye at 3’ end of DNA (Figure S4E-L). It is interesting to note that this activity involves an apparent 5’ to 3’ directed movement, opposite of its 3’ to 5’ directional translocation (Antony et al., 2009). We refer to the repetitive Srs2 activity as a "scrunching" of ssDNA. Such scrunching activity for a protein was first defined in the context of RNA polymerase (RNAP) whereby the single stranded DNA template strand is accumulated in the active site pocket of RNAP in the transcription initiation phase (Brieba and Sousa, 2001; Cheetham and Steitz, 1999; Garcia-Diaz et al., 2009; Kapanidis et al., 2006; Revyakin et al., 2006; Tang et al., 2009; Tang et al., 2008). Here, we describe a similar scrunching activity of a helicase that functions in a DNA repair pathway. It really does not seem to be the same phenomenon. It might be better to call it something else.
Figure 4. Srs2 repetitively scrunches short segment of ssDNA.
(A) Schematic of FRET-DNA constructs with various tail lengths (T13-T70). (B) Single molecule traces show similar range of FRET fluctuation in T13-T20, but much less prominent and lower FRET range fluctuation in T25-T30 and no fluctuation in T70. (C) FRET histograms of single molecule traces of Srs2 on various ssDNA tail lengths. (D) FRET peak to peak dwell time δt. (E) Dwell times collected from various tail lengths. Error bars denote SEM.
Srs2 anti-recombinase activity
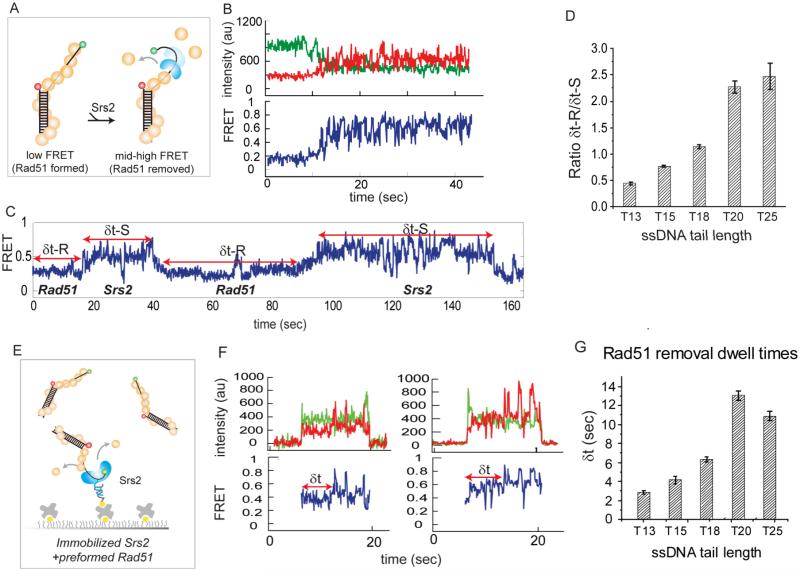
Next, we tested if the scrunching activity of Srs2 contributes to its anti-recombinase activity. We formed a Rad51 filament on Cy3/Cy5 labeled pdT20 DNA (Figure 5A) and added Srs2 (20-200nM) together with Rad51 to maintain the same Rad51 concentration. The representative single molecule trace shows a steady low FRET transitioning to a high FRET followed by FRET fluctuations, suggesting a complete clearance of the Rad51 filament (Figure 5B). The initial FRET increase corresponds to the dismantling of Rad51 by Srs2 translocation (Antony et al., 2009) whereas the FRET fluctuation reflects the repetitive scrunching of Srs2 that occurs after the removal process. This result suggests that after Srs2 removes Rad51 filament, it stays on the DNA and prevents re-binding of Rad51 by repeatedly scrunching the ssDNA. This can also be advantageous for subsequent binding of RPA to reinforce the clearance of the Rad51 filament.
Figure 5. Srs2 repetitive scrunching prevents Rad51 formation.
(A) Schematic of FRET DNA used in Rad51 removal assay. (B) Single molecule trace showing low to high FRET followed by FRET fluctuation. (C) Single molecule trace displaying dynamic exchange between Rad51 filament formation and Srs2 clearance/scrunching. (D) Ratio between filament formed duration and Srs2 cleared duration tested on varying tail. Error bars denote SEM. (E) Schematic of immobilized Srs2 assay to test if clearance and scrunching activity arises from Srs2 monomer. (F) Single molecule traces showing monomer Srs2 removing Rad51 filament and preventing reformation. (G) Dwell times corresponding to removal duration for different tail length. Error bars denote SEM.
While the repetitive scrunching by Srs2 serves to maintain the ssDNA clear of Rad51, this activity is transient, as more than 60% of single molecule traces display a dynamic exchange between the steady low FRET and the FRET fluctuations, representing reformation of Rad51 filaments and subsequent clearance by Srs2, respectively (Figure 5C). To test if the stability of the Rad51 filament influences its clearance by Srs2, we measured the dwell times corresponding to the duration of the Rad51 filament formation (δt-R, low FRET) and Srs2 induced clearance (δt-S, FRET fluctuation) (Figure 5C) for varying lengths of the ssDNA tail. We measured the two dwell times from over 50 traces for each tail length and calculated the ratio between δt-R and δt-S. This ratio provides a measure of the propensity for Rad51 filament formation over clearance (Figure 5D). The result shows that the longest ssDNA (T25) is five times more likely to be occupied by a Rad51 filament than the shortest ssDNA (T13). In other words, the T13 ssDNA is five times more likely to remain cleared of Rad51 by Srs2 than the longer T25 ssDNA. The ratio appears to be the same for T20 and T25, reflecting the similar stability of a Rad51 nucleation above T20. When Srs2 was added to the DNA prior to Rad51, the duration of Srs2 scrunching before Rad51 formation was substantially longer (Figure S5A-D) than when Srs2 was added post Rad51 formation, indicating an enhanced ability of Srs2 to prevent Rad51 binding. Nevertheless, Rad51 filaments eventually reform even under these circumstances and a dynamic exchange between Rad51 filament formation and removal by Srs2 follows as before (Figure 5C).
To test if Rad51 removal can be accomplished by an Srs2 monomer, we used the protein tethered assay where DNA pre-bound with a Rad51 filament was applied to the surface tethered Srs2 (Figure 5E). We observed a brief low FRET binding followed by high FRET and FRET fluctuations, indicating the binding of a Rad51-ssDNA filament to Srs2, followed by a quick clearance of Rad51 and Srs2 mediated scrunching of ssDNA respectively (Figure 5F). This assay offers two unique advantages. First, it enables a direct monitoring of a Srs2 monomer to clear Rad51. Second, it allows for precise measurement of the time required for a Srs2 monomer to clear a Rad51 filament, δt (Figure 5F). We collected the δt for the different ssDNA tail lengths (pdT13-pdT25). Consistent with the stability of Rad51 filaments shown above (Figure 5D), the removal time to clear the longest (T20, T25) was 4-5 fold higher than the shortest Rad51 filament (T13). The similar fold difference obtained in Figure 5D and 5G indicates that the overall filament stability of Rad51 on T20/25 is approximately five fold higher than on T13.
Srs2 cannot disassemble a RecA filament
To examine the specificity of Srs2 in clearing Rad51, we tested its ability to remove RecA filaments. We formed a stable RecA filament (Joo et al., 2006) and added Srs2 (200nM). In this case, less than 5% of the molecules showed FRET fluctuations induced by Srs2, indicating negligible removal of RecA (Figure S5E). Based on this result, we confirm that Srs2’s scrunching activity is specific for dismantling Rad51 filaments as the anti-recombinase activity of Srs2 requires a direct physical contact with Rad51 (Antony et al., 2009; Colavito et al., 2009).
Srs2 unwinding of DNA is suppressed by Rad51
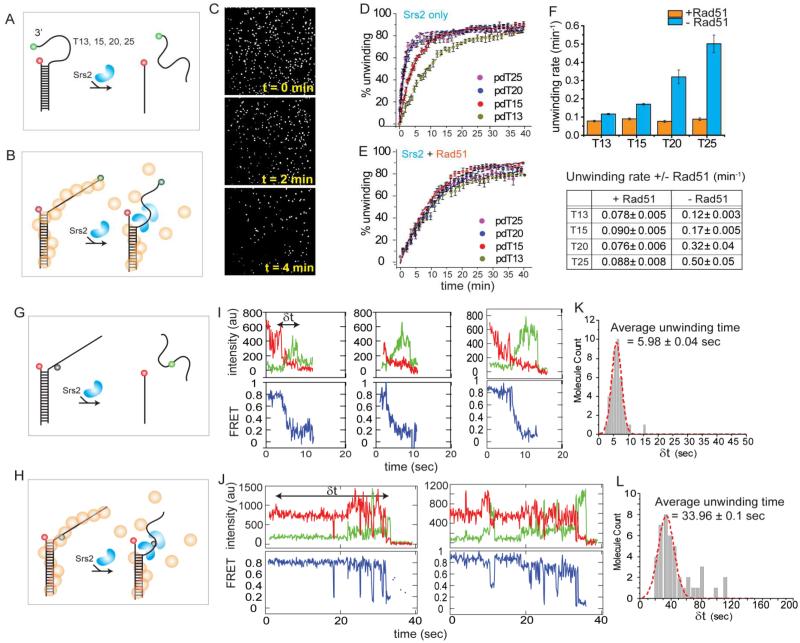
As a helicase, Srs2 can unwind double stranded (ds)DNA with a flanking 3’ ssDNA tail longer than 12 nucleotides (Krejci et al., 2004). We asked if this unwinding activity is modulated by the presence of a Rad51 filament. We confirmed Rad51 formation on dsDNA as shown before (Figure S2A-C). Complete unwinding is expected to result in the loss of the Cy3 (green) labeled strand because only the other strand (red) is immobilized to the surface (Figure 6A, B). Therefore, the unwinding kinetics was measured by the disappearance rate of the Cy3 (green) signal over time on all DNA constructs (Figure 6C). We counted molecules every 5-10 seconds after the addition of Srs2 on different regions of the microscope slide rather than continuous monitoring of one area to minimize photobleaching. Unwinding in the presence of Rad51 was performed by first forming the Rad51 filament and subsequently adding Srs2 and Rad51 together. Consistent with the previous finding (Krejci et al., 2004), the rate of unwinding is dependent on the length of the 3’ tail (Figure 6D, F). In the presence of Rad51, however, the unwinding rate was reduced to the same degree, independent of the tail length (Figure 6E, F). This agrees with results obtained in an ensemble study (Antony & Lohman, unpublished data). If the stability of the Rad51 filament on ssDNA were the main contributing factor, the unwinding rate is expected to show a tail length dependence. Our observation of a uniformly low unwinding rate regardless of tail length (Figure 6F) suggests that Rad51 formed on the duplex DNA is responsible for the inhibitory effect.
Figure 6. Rad51 suppresses Srs2 induced unwinding.
(A, B) FRET-DNA constructs with varying tail lengths (T13-T25) without and with Rad51. (C) Fluorescence spots left on surface can be counted for unwinding rate analysis. (D-F) Unwinding rate of Srs2 is substantially lower in the presence of Rad51. Error bars denote SEM. (G, H) Schematic of unwinding DNA constructs. Cy3 and Cy5 are located near duplex junction such that unwinding will result in FRET decrease. (I, J) Unwinding in the presence of Rad51 is delayed as indicated by slower FRET decrease. (K, L) Histograms and a Gaussian distribution fit show that the Srs2 unwinding duration is about 5-6 times longer in the presence of Rad51. Errors in fit results are in SEM.
To examine this effect more directly, we prepared a DNA substrate which has two dyes located near the duplex junction such that unwinding can be monitored as a decrease in FRET (Figure 6G, H). In the absence of Rad51, Srs2 (>20nM) induces a rapid FRET decrease as seen in a representative trace (Figure 6I). The dwell time corresponding to the unwinding duration (δt) collected from over 100 events is approximately 6 seconds (Figure 6K). In the presence of a Rad51 filament, however, unwinding is inhibited as seen by the long delay in the FRET decrease (Figure 6J), yielding an average unwinding time of 34 seconds (Figure 6L).
DNA unwinding measured using ensemble methods indicate that multiple Srs2 monomers or Srs2 oligomers are required for DNA unwinding (Antony & Lohman, unpublished data). Hence, we employed the Srs2 monomer tethered assay to test if monomer can unwind (Figure S6A). Over 95% of the binding events showed a stable high FRET (Figure S6B), indicating no DNA unwinding activity. Therefore, we conclude that Srs2 monomers cannot unwind DNA efficiently and that the unwinding seen with immobilized DNA is likely due to higher oligomeric forms of Srs2 or multimers of Srs2 binding to one DNA, consistent with ensemble studies (Antony & Lohman, unpublished data).
Discussion
Rad51 site size, nucleation cluster
We have used single molecule FRET to detect the formation of Rad51 filaments on ssDNA with lengths in the 10-20 nucleotide range where the distance change induced by Rad51 binding is detectable by FRET (Ha, 2001). We observed that Rad51 formed a stable filament on ssDNA as short as 18 nucleotides (Figure 1B). Based on the detection of three, four and five steps for Rad51 binding to pdT10, 13 and 15, we deduced a site size of three nucleotides per Rad51 monomer. Combining the site size with the minimum ssDNA length required for forming a stable Rad51 filament (pdT18), we infer a nucleation cluster size of six Rad51 monomers. These numbers are similar to those inferred for RecA (Joo et al., 2006) and consistent with the structural resemblance between the two proteins (Chen et al., 2008; Conway et al., 2004). Previous studies with human Rad51 (hRad51) reported that nucleoprotein filaments grow in multiples of trimers as a stable nucleation cluster (Hilario et al., 2009). However, this assay was performed on purely dsDNA (λ DNA) on which a Rad51 filament may have a different stability. Similar to our finding that the majority of Rad51 binds as monomers and dimers, a recent study by Bell et al demonstrated that RecA filament formation on ssDNA involves primarily monomer and dimer units and the net growth favors the 5’ to 3’ direction (Bell et al., 2012).
Rad51-ssDNA filaments grow directionally from 5’ to 3’
Although Rad51 binding is expected to be similar to RecA (Register and Griffith, 1985), Rad51 binding directionality has never been demonstrated on a resected DNA mimic with high resolution. In humans, Rad51 binding to resected DNA is controlled by BRCA2 where BRCA2 positions itself at the duplex junction and initiates nucleation of Rad51 in a 5’ to 3’ direction (Carreira and Kowalczykowski, 2011). In agreement with our finding, the overall net growth directionality for RecA, hRad51 and yeast Rad51 was shown to be 5’ to 3’ on both ssDNA and dsDNA although bi-directionality was also demonstrated (Bell et al., 2012; Galletto et al., 2006; Hilario et al., 2009; Joo et al., 2006). To directly monitor the directional bias of yeast Rad51 filament formation, we utilized a recently developed single molecule assay, smPIFE (Hwang et al., 2011; Myong et al., 2009). The 5’ to 3’ assembly was demonstrated in two ways; the difference in the rate of overall signal increase and the shape of the intensity increase (Figure 2B, D). In addition, the larger PIFE effect seen for the 3’ Cy3 is in agreement with our previous finding in which RecA binding to a similar substrate resulted in 3-4 fold increase in the fluorescence signal (Hwang et al., 2011).
Repetitive scrunching contributes to anti-recombinase activity
Previous single molecule studies have identified numerous helicases that translocate or unwind repetitively (Myong et al., 2007; Myong et al., 2009; Myong et al., 2005; Park et al., 2010; Tomko et al., 2010; Yodh et al., 2009). Two bacterial homologs of Srs2, Rep (Myong et al., 2005) and PcrA (Park et al., 2010) display repetitive translocation that resembles that of Srs2 in two aspects. First, all three translocases exhibit repetitive activity on single stranded DNA and second, they all involve looping of ssDNA although the mechanistic details differ. One clear difference is that Srs2 only acts on a small segment of ssDNA rather than the entire ssDNA length. Based on our results, we propose a scrunching model whereby Srs2 stations itself near the junction of ssDNA and dsDNA and repetitively reels in a short segment of ssDNA (Figure S4M). We hypothesize that this scrunching involves ATP stimulated conformational changes within Srs2 which results in a periodic tugging of a finite length of ssDNA.
Srs2 is a functionally unique helicase in yeast in that its recruitment is highly specific for the removal of Rad51 filaments (Antony et al., 2009; Krejci et al., 2003; Liu et al., 2011). As a control, we showed that Srs2 is unable to remove a RecA filament from ssDNA (Figure S5E). The repetitive scrunching by Srs2 we demonstrate here is not the dismantling of a Rad51 filament, but what is likely to occur after disruption of the filament. Hence our findings expand on the previously known role of Srs2’s anti-recombinase function (Antony et al., 2009; Krejci et al., 2003; Le Breton et al., 2008; Veaute et al., 2003). This repetitive mechanism at the site of a duplex junction can offer two advantages. First, the repetitive nature of scrunching can effectively maintain ssDNA clear of Rad51 binding. Second, the scrunching at the duplex junction enables efficient sequestering of ~18-20 nts of ssDNA, which coincides with the ~18 nt length required for formation of a stable Rad51 nucleation cluster. Taken together, the Srs2 mechanism we present here (Figure 7) represents a highly specific activity of an anti-recombinase both in terms of motion and position. It is important to note that the anti-recombinase activity of Srs2 does not prevent Rad51 formation permanently, but only acts to transiently counterbalance Rad51 filament formation.
Figure 7.
Schematic summary of Srs2 scrunching as an anti-recombinase mechanism for Rad51 clearance. Srs2 first translocates in the 3’ to 5’ direction along ssDNA to displace Rad51, then remains bound near the duplex junction and repetitively scrunches a short segment of ssDNA to prevent Rad51 reformation.
Suppressed unwinding activity of Srs2 in the presence of Rad51
The helicase activity of Srs2 is not correlated with its anti-recombinase function (Krejci et al., 2003; Prakash et al., 2009; Van Komen et al., 2003) whereas the ATP fueled translocase activity is required, but not sufficient for Rad51 filament removal (Antony et al., 2009; Krejci et al., 2004). Furthermore, it has been shown for other SF1 helicases that their translocation and unwinding activities are functionally separable in that enzyme oligomerization is often required for DNA unwinding activity (Lohman et al., 2008). We find that monomeric Srs2 can dislodge Rad51 filaments on ssDNA, but cannot unwind dsDNA, in agreement with ensemble studies (Antony & Lohman, unpublished data). Hence, Srs2 monomers are capable of removing Rad51 filaments from ssDNA without unwinding the duplex DNA. This behavior is thus similar to what has been observed and proposed for E. coli UvrD in its role as an anti-recombinase to remove RecA filaments (Tomko et al., 2010). Previous studies revealed that Rad51 plays a dual role; it targets Srs2 to homologous recombination intermediates and triggers ATP hydrolysis within the Rad51 filament, inducing Rad51 dissociation from the DNA (Antony et al., 2009). Here, we demonstrate yet another role of Rad51 in preventing DNA unwinding by Srs2. In contrast, one study reported that Rad51 enhances Srs2 unwinding (Dupaigne et al., 2008). However, those studies were performed uder very different conditions including the presence of RPA, use of up to a 25 fold higher Srs2 concentration (500nM) and a DNA with a much longer ssDNA tail (209 nts), all of which may favor the DNA unwinding reaction. Due to these differences, a direct comparison cannot be made. Our more direct real time assay shows that in the absence of RPA Rad51 inhibits the ability of Srs2 to unwind DNA.
In conclusion, we present a comprehensive step-by-step process of the roles of Rad51 and Srs2 in the early stages of homologous recombination. Our results reveal an exquisite mechanism that controls the balance between the recombinase, Rad51 and the anti-recombinase, Srs2.
Experimental Procedures
Oligonucleotides
Unlabeled, Cy3-labeled and Cy5-labeled Oligonucleotides were purchased from Integrated DNA technologies (Coralville, IA). Internally labeled oligos contain amino modifier dT subsequently labeled using Cy3 monofunctional NHS esters (GE Healthcare, Princeton, NJ) as described (Joo and Ha, 2008).
Proteins
The wildtype Rad51 protein and Srs2 truncation mutant Srs21-898 (Srs2CΔ276) were overexpressed and purified as described (Antony et al., 2009). The bacterial wildtype RecA protein was purchased from New England Biolabs (Ipswich, MA).
Reaction Conditions for Rad51, RecA and Srs2
Standard Rad51 reaction buffer was 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2; standard RecA reaction buffer was 100 mM NaAc, 25 mM Tris-HCl (pH 7.5), 10 mM MgAc, with an oxygen scavenging system containing 0.8% v/v dextrose, 1 mg/ml glucose oxidase, 0.03 mg/ml catalase (Joo and Ha, 2008), and 2-mercaptoethanol (1% v/v), all items were purchased from Sigma Aldrich (St. Louis, MO). The measurements were performed at room temperature (21°C ± 1°C). 1 mM ATP was used in all experiments unless otherwise specified.
Data Analysis
Single molecule traces were analyzed using codes written in Matlab. FRET efficiency values were calculated as a ratio between acceptor intensity and total donor and acceptor intensity.
Supplementary Material
Highlights.
Rad51 has a site size of 3 nts and requires 6 monomers for a stable nucleation.
Srs2 translocates to near ss/ds DNA junction and scrunches in finite lengths of ssDNA.
Repetitive scrunching of Srs2 prevents reformation of Rad51 filament.
Srs2 unwinding is impeded by Rad51 filament formed on double stranded DNA.
Acknowledgment
The authors thank members of the Myong laboratory for helpful discussions. We thank Paul Hergenrother and Robert Tampé for providing biotin-tris-NTA used for Srs2 tethering and NTA-Atto 647 used for Srs2 labeling, respectively. Support for this work was provided by American Cancer Society (Research Scholar Grant; RSG-12-066-01-DMC), NIH Director’s New Innovator Award (1DP2GM105453), the Human Frontier Science Program (RGP0007/2012), the U.S. National Science Foundation Physics Frontiers Center Program (0822613) through the Center for the Physics of Living Cells for S.M. and NIH GM045948 for T.M.L.
Reference
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieba LG, Sousa R. T7 promoter release mediated by DNA scrunching. EMBO J. 2001;20:6826–6835. doi: 10.1093/emboj/20.23.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, Krejci L, Sung P. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009;37:6754–6764. doi: 10.1093/nar/gkp748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Larrea AA, Havener JM, Perera L, Krahn JM, Pedersen LC, Ramsden DA, Kunkel TA. Template strand scrunching during DNA gap repair synthesis by human polymerase lambda. Nat Struct Mol Biol. 2009;16:967–972. doi: 10.1038/nsmb.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci U S A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. In: Selvin PR, Ha T, editors. In Single-molecule techniques : a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2008. pp. 3–35. [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Le Breton C, Dupaigne P, Robert T, Le Cam E, Gangloff S, Fabre F, Veaute X. Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res. 2008;36:4964–4974. doi: 10.1093/nar/gkn441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Register JC, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. (3rd) 1985;260:12308–12312. [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn ME, Connolly BK, Gurunathan K, Levitus M. Fluorescence properties and photophysics of the sulfoindocyanine Cy3 linked covalently to DNA. J Phys Chem B. 2007;111:11064–11074. doi: 10.1021/jp072912u. [DOI] [PubMed] [Google Scholar]
- Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong C, Colavito S, Kwon Y, Sung P, Krejci L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J Biol Chem. 2009;284:24363–24371. doi: 10.1074/jbc.M109.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina M, Koh HR, Patel SS, Ha T. Fluorescent lifetime trajectories of a single fluorophore reveal reaction intermediates during transcription initiation. Journal of the American Chemical Society. 2009;131:9630–9631. doi: 10.1021/ja902861f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Roy R, Bandwar RP, Ha T, Patel SS. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:22175–22180. doi: 10.1073/pnas.0906979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Roy R, Ha T, Patel SS. Transcription initiation in a single-subunit RNA polymerase proceeds through DNA scrunching and rotation of the N-terminal subdomains. Mol Cell. 2008;30:567–577. doi: 10.1016/j.molcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM. 5'-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Reddy MS, Krejci L, Klein H, Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J Biol Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.