Abstract
Brown adipose tissue (BAT) is specialized for heat generation and energy expenditure as a defense against cold and obesity; in both humans and mice increased amounts of BAT are associated with a lean phenotype and resistance to development of the metabolic syndrome and its complications. Here we summarize recent research showing that several BAT-expressed microRNAs miRNAs) play important roles in regulating differentiation and metabolism of brown and beige adipocytes; we discuss the key mRNA targets downregulated by these miRNAs and show how these miRNAs affect directly or indirectly transcription factors important for BAT development. We suggest that these miRNAs could be part of novel therapeutics to increase BAT in humans.
Keywords: Brown, beige adipocytes, microRNAs, BAT, adipogenesis
Types of adipose tissue
Mammals have two types of adipose tissue: brown and white, with opposing functions. White adipose tissue (WAT) consists of two main pools – subcutaneous (SAT) and visceral adipose tissue (VAT). WAT stores energy in the form of triglycerides (TGs), and is an important regulator of whole body homeostasis. It functions as endocrine organ and secretes a number of adipocyte-derived hormones, such as adiponectin, leptin and resistin (1–7), with well established and crucial functions regulating energy homeostasis and glucose and lipid metabolism, not only in adipose tissue but also in liver and muscle.
Brown adipose tissue (BAT) on the other hand, is a specialized form of adipose tissue found in humans, rodents and some hibernating animals. It differs from WAT in location, morphology and function. BAT is found in discrete locations, has a distinct brown color due to rich vascularization and a high density of mitochondria, and plays a major role in energy expenditure and non-shivering thermogenesis as it catabolizes lipids to generate heat. The BAT-specific mitochondrial uncoupling protein 1 (UCP1) short circuits the proton motive force across the inner mitochondrial membrane, generating heat rather than ATP (8, 9). BAT differentiation and full UCP1 mRNA expression can be induced by prolonged cold exposure, by beta-adrenergic stimulation leading to elevated intracellular cyclic AMP (cAMP) levels (10) (11) (12), and by thyroid hormones (13, 14). Until recently, cold-induced thermogenesis in BAT was considered of importance only in hibernators, rodents, and neonatal mammals. However, recent evidence strongly supports the existence of functional BAT in adult humans (15–18) (19).
Brown and beige fat cells
While disruption of normal WAT development causes ectopic lipid storage and severe pathology (lipodystrophy) in both humans and experimental animals, loss of BAT function is linked to obesity and metabolic diseases (20). Promotion of BAT development in animals, on the other hand, leads to increased energy expenditure without causing dysfunction of other tissues. Indeed, experimental increases of BAT in several rodent models have been associated with a lean and healthy phenotype (21–23), suggesting that manipulation of BAT levels and thus uncoupled respiration is a desirable therapeutic objective against obesity and its pathological consequences.
BAT is localized to distinct anatomical sites, including interscapular, perirenal, and axillary depots. Additional brown fat cells - known as “beige” cells - emerge in SAT in response to cold exposure (11, 21, 24) (25), a process commonly referred to as WAT browning. Following prolonged cold exposure, adult C57BL/6 wild type (WT) mice show an obvious transition from SAT into brown-like adipose tissue (brite) that contains a high number of UCP1-positive mitochondria, a characteristic feature of BAT (25). Although these beige cells (see Text Box 1) have a very low basal level of UCP1 gene expression, unlike a typical white fat cell they retain a remarkable ability to robustly activate UCP1 expression. Thus, both in rodents and humans beige adipocytes are a distinct type of thermogenic fat cells able to trigger a program of respiration and energy expenditure that is equivalent to that of classical brown fat cells (25). Indeed, expression profiling and other data has demonstrated the presence of UCP1-possitive cells in humans in the neck and upper chest region, and has indicated that these might be beige, rather than brown adipocytes (72).
Text BOX 1: Defining beige cells.
Beige adipocytes emerge within WAT following prolonged cold exposure (12, 80) and β-adrenergic stimulation (24). Unlike the “classical” BAT which originates from PAX7 / MYF5-possitive progenitors that also give rise to skeletal muscle (26), these inducible beige adipocytes originate form PAX7 / MYF5-negative precursors (27). It is not clear whether the beige adipocytes originate by “brown conversion” of white fat cells (11, 81), or whether this “transdifferentiation” is due to the presence of a distinct cell type with this predisposition. It is also not clear whether the beige adipocytes originate from the same MYF5-negative precursors as the white preadipocytes, or whether “beige-specific” preadipocytes exist that are committed to this cells type, as recently suggested (25). Classical brown adipocytes express high levels of UCP1 also in the absence of appropriate stimuli, and have constantly high thermogenic activity. Beige cells have a very low basal level of UCP1, but can robustly respond to cAMP to induce UCP1 and activate a thermogenic program to levels similar to those seen in the brown cell lines from the interscapular depot (82). The UCP1-possitive cells within the supraclavicular region of healthy human adults have a molecular phenotype that is much more similar to murine beige fat cells rather than brown fat cells (25). The beige adipocytes differ from classical BAT cells also in the expression of certain genes: they do not express the transcription factors ZIC1, LHX8, MEOX2; have low PRMD16 levels; and retain expression of HOXC9 as a WAT-specific gene (25). Recent reports however suggest that deep human neck brown adipocytes most closely resemble cells from the classical BAT lineage in the mouse (83), sharing similar capacity for high rates of energy expenditure seen in rodent iBAT. In this study, the following genes were used as markers: MPZL2, HOXC9, EBF3, FBXO31 and LEP (for the white adipocytes); TNFRSF9, TMEM26 and SHOX2 (for the beige adipocytes); and UCP1, LHX8 and ZIC1 (for the brown adipocytes).
Transcription factors important for BAT development
Brown fat cells can arise from both a myogenic MYF5 positive lineage and from MYF5 negative white fat precursors (26, 27). In vivo lineage tracing experiments in mice showed that bipotential MYF5 positive progenitor cells can give rise to two discrete cell types, myocytes and brown adipocytes in the interscapular fat depot, pointing to a similar developmental origin of these cells. In contrast, beige cells within white adipose depots are derived from a different linage and are MYF5 negative (26, 27). In both MYF5 positive and negative preadipocytes, brown differentiation is promoted by increased expression of the gene encoding the transcription factor PRD1-BF1-RIZ1 homologous domain containing 16 (PRDM16) (26, 27). In particular, PRDM16 expression inhibits myogenesis and promotes BAT differentiation from the common bipotential MYF5 positive progenitor. Thus, PRDM16 has emerged as a brown adipose determination factor that controls the switch between skeletal myoblasts and brown fat cells. PRDM16 also regulates the switch between white and beige adipocytes, thus regulating the development of both brown and beige adipocytes in classic BAT depots, and SAT, respectively (26, 27). Other well established key transcription factors that drive both brown and white fat cell differentiation include the peroxisome proliferator-activated receptor-γ (PPARγ) and CCAAT/enhancer-binding proteins (C/EBPs) (28–33). C/EBPβ is one of the crucial switches in brown fat cell fate determination and is a key transcriptional inducer of UCP1 expression and the thermogenic program (31, 34). Treatment of both mouse and human WAT with PPARγ activators also induces the expression of UCP1 as well as peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) and other nuclear proteins important for mitochondrial biogenesis (4, 16). The expression of PGC1α is elevated upon cold exposure (26) and is needed for BAT thermogenesis (28). PGC1α controls mitochondrial biogenesis and respiration through induction of uncoupling proteins and nuclear respiratory factors (27). cAMP responsive element binding protein (CREB) and PPARα are direct activators of PGC1α, while the interaction of PPARα with PRMD16 and cAMP-mediated pathways is necessary for full activation of PGC1α gene transcription (35). Another key regulator of adipose cell metabolism is the transcription factor forkhead-box-C2 (FOXC2) (36). Expression of FOXC2 in adipose tissue induced emergence of beige cells from WAT, which show elevated expression of thermogenic genes, including UCP1 and PGC1α (37). For a comprehensive review on the transcriptional control of brown fat development readers are referred to (38).
General role of miRNAs in regulation of adipogenesis
MiRNAs) are a class of evolutionarily conserved 20 to 24 nucleotide RNAs that act as negative post-transcriptional regulators of target gene expression (39, 40). These have emerged as a fundamental mechanism controlling tissue development and function (41). The miRNA biosynthesis pathway includes processing of miRNAs precursors in the nucleus by the RNA type III endonuclease termed Drosha, and in the cytosol by the RNA type II endonuclease Dicer into ~22 nucleotide-long miRNAs (42) (43) (44) (see Text Box 2). MiRNAs silence transcripts by facilitating cleavage of target mRNAs via Argonaut proteins (catalytic components of the RNA-induced silencing complex), and/or by Argonaut acting as a physical barrier to translation, depending on the level of complementarity of the miRNA with their target mRNAs (45). Important for mRNA target recognition is the miRNA “seed” sequence that is centered on nucleotides 2–8 from the 5’ end. This sequence hybridizes with a sequence generally in the 3' untranslated region (3’UTR) of the mRNA (45). Computational and experimental analyses have revealed that almost half of the mammalian transcriptome is targeted by miRNAs. On average, a single miRNA family can regulate hundreds of targets (up to ~400), and the level of regulation on a single transcript tends to be mild, rarely exceeding 2 fold (46).
TEXT BOX 2: MicroRNAs, the basics.
MiRNAs are ~22 nucleotide long evolutionarily conserved RNA molecules that act at a posttranscriptional level to regulate gene expression in metazoans and plants. MiRNAs are transcribed by RNA polymerase II or III as primary transcripts, called pri-miRNAs. In the nucleus, pri-miRNAs are processed at the bottom of their stem loop into ~70 nucleotide long pre-miRNAs by the RNA type III endonuclease termed Drosha, together with its cofactor, the RNA binding protein DGCR8. Pre-miRNAs are then exported from the nucleus by the nuclear transport receptor Exportin 5 into the cytosol, where they are further cleaved by the RNA type II endonuclease Dicer into ~22 nucleotide-long miRNA duplexes. One strand of the mature miRNA is preferentially retained in the miRNA induced silencing complex (miRISC), consisting of several proteins of the Argonaute family, of which Argonaute-2 (AGO2) cleaves the target mRNA. MiRNAs in the miRISC mediate the mRNA target recognition by imperfect pairing, inducing translational repression and/or AGO2 mediated mRNA cleavage (42) (43) (44).
MiRNA expression is often developmental or tissue enriched. Recent data indicate that miRNAs play a key role in modulating cell differentiation and metabolism of many tissue types in vivo, including adipocytes (47) (48, 49). Conditional aP2-CRE Dicer knock-out (KO) mice revealed distinct requirements for Dicer in the formation of adipose cells and provided genetic evidence that miRNAs regulate adipogenesis. The Dicer KO mice, which express the CRE recombinase at high levels during white and brown adipocyte differentiation, are half the size of control mice, and exhibit a mild shivering phenotype (50). The mutants have a severe reduction of WAT and a 50% reduction in spleen weight, but no differences in the weight of various other tissues relative to overall body mass. Interestingly, these knock out animals showed massive decreases in the transcript levels of BAT genes that regulate thermogenesis, including UCP1, PGC1a, cyclooxygenase 1b (COX1B), and cell death-inducing DNA fragmentation factor alpha subunit-like effector A (CIDEA), as well as the expression of the PPARγ2 isoform and fatty acid synthase (FAS), indicating that miRNAs may affect the thermogenic functions of BAT. However, there was no reduction in BAT mass up to three weeks of age, when these mice typically died. It is important to note that the early mortality of the aP2-Dicer KO mice is a limitation in studies of the global importance of miRNAs in brown fat differentiation. Since aP2 (also known as fatty acid binding protein 4 - FABP4) is not adipose specific and is expressed in other tissues including macrophages, the phenotype of these mice might be influenced by changes in extra-adipose sites. It would be of interest to investigate whether these Dicer knockout mice, or other types of adipocyte miRNA-deficient mice, would show differences in BAT differentiation when challenged with cold exposure or a high fat diet. Undoubtedly, future studies will address the general role of miRNAs in brown fat differentiation at later stages using different CRE inducible knockouts of Dicer or other miRNA processing enzymes.
A number of miRNAs are known to direct early stages of white adipocyte differentiation, the fate determination step during which multipotent mesenchymal stem cells (MSCs) are induced to become adipocytes. Other miRNAs are essential for terminal differentiation, where cells acquire a mature adipocyte morphology and express mature adipocyte markers such as aP2, glucose transporter type 4 (GLUT4) and others (Reviewed in (51)). Here we summarize recent progress in understanding the role of individual miRNAs in brown adipogenesis.
MiR-193b-365, a brown fat enriched microRNA cluster essential for brown fat differentiation
A comparison between miRNAs expressed in skeletal muscle and white and brown adipose tissue identified 91 miRNAs that were differentially expressed between these tissues and potentially important for brown fat development (52). Among those, miR-193b and miR-365, which are co-transcribed as a single pri-miRNA, were strongly expressed in BAT, but not WAT or muscle. Using locked nucleic acid (LNA)-mediated technology to inhibit miR-193a/b and/or miR-365 in the stromal-vascular fraction (SVF) of brown fat or in purified brown preadipocytes from mice, there was a marked reduction in brown adipocyte differentiation, as monitored by decreased lipid accumulation and decreased mRNA expression of adipogenic markers common to brown and white fat, including adiponectin, C/EBPα, aP2, and PPARγ. Even more marked decreases were observed in the expression of brown fat-enriched markers including UCP1, PPARα, PGC1α, deiodinase 2 (DIO2), PRDM16 and CIDEA. In addition, and despite their low expression in white fat, both miR-193 and miR-365 were required for white adipocyte differentiation, indicating that they are general regulators of adipogenesis.
One mechanism employed by miR-193a/b to regulate adipogenesis is direct negative regulation of Runt-related transcription factor 1; translocated to 1 RUNX1t1), an inhibitor of both white and brown adipogenesis. Among other conserved miR-193b targets are Cdon (also named Cdo) and insulin-like growth factor-binding protein 5 (IGFBP5), both previously implicated as pro-myogenic factors (53–56). CDON is a cell surface receptor that stimulates post-translational activation of myogenic bHLH factors and E proteins, and increases transcription of muscle-specific genes. IGFBP5 is required for myogenesis and knocking it down also impairs myogenic differentiation of C2C12 myoblasts and primary skeletal muscle cells in vitro.
Ectopic expression of miR-193b in C2C12 myoblasts repressed myogenesis and decreased the mRNA levels of myogenic markers including the paired box gene 3PAX3 and MYOD. It also promoted a significant upregulation in the mRNAs encoding selective brown fat markers, such as UCP1, CIDEA, PRDM16 and PPARα, as well as adipogenic markers including PPARγ, CEBPα and aP2. More strikingly, ectopic expression of miR-193b in these myoblasts, when cultured in a proadipogenic medium, induced formation of brown adipocytes that exhibited UCP1 - mediated thermogenesis.
Forced expression of PRDM16 in primary white preadipocytes or C2C12 myoblasts, on the other hand, induced mir193b–365 expression, at least in part by inducing expression of PPARα, suggesting that this miRNA cluster is indirectly activated PRDM16. Thus, a feed-forward loop might exist, whereby overexpression of miR-193b promotes PRDM16 expression, and its inhibition results in decreased PRDM16 expression, thus ensuring differentiation of brown adipocytes from bipotential brown adipocyte/myocyte progenitors (Figure 1).
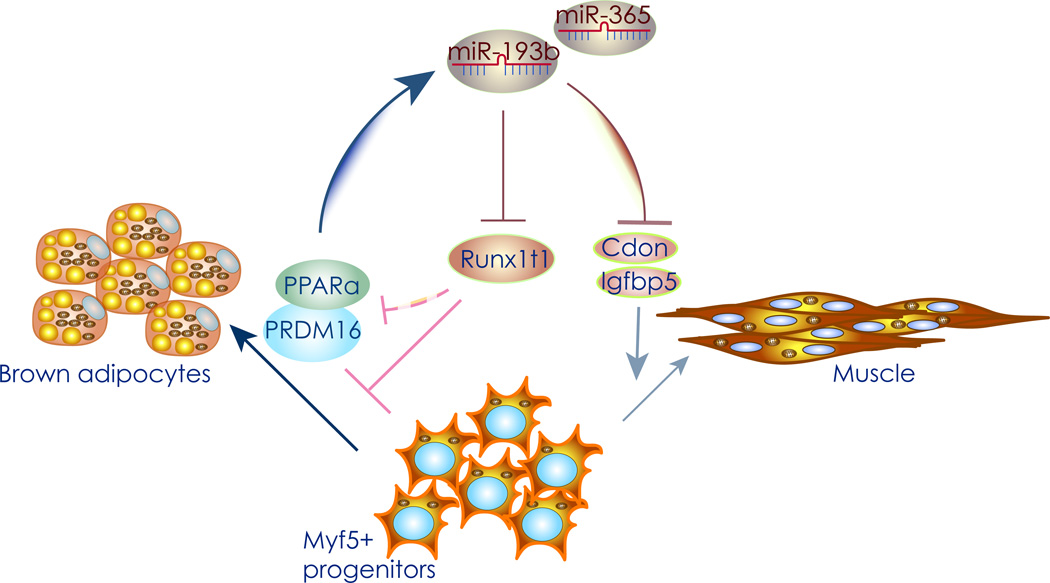
Figure 1. MiR-193b stimulates brown adipogenesis.
Graphical representation of the miR-196b/365 pathway. Runx1t1 is an inhibitor of brown adipogenesis, while Cdon and Igfbp5 are pro-myogenic factors. Increased miR-193b expression during adipogenesis directly inhibits CDON, IGFBP5, and Runx1t1 expression, thus increasing the levels of PRDM16 and PPARα. The resulting increase in the levels of these two transcription factors promotes brown adipogenesis, and further up-regulates miR-193/365 expression as part of a positive feedback loop.
Collectively, these results demonstrate that the miR-193b-365 cluster contributes to the regulation of brown fat versus muscle lineage determination, by directly repressing myogenesis.
Decreased miR-133 expression during cold exposure promotes brown adipogenesis
Several other miRNAs were found also to be differentially expressed between muscle, white and brown fat (52). MiR-1 and -133a/b, derived from the same miRNA polycistron and transcribed together, have distinct roles in modulating skeletal muscle cell proliferation and differentiation, and were recently implicated in brown adipocyte differentiation (51). Specifically, miR-133a and b, previously thought to be exclusively expressed in muscle, is strongly expressed in brown fat (57), although to a lower extent than in muscle (52). MiR-133a and b are involved in cell specification of mouse and human embryonic stem (ES) cells (58), differentiation of mouse myotubes (59), and mouse development (60). MiR-133a and b are downregulated in mouse and human models of cardiac hypertrophy, and may play a role in the underlying pathogenesis of the disease (61, 62).
A comparative analysis of miRNA microarrays collected from BAT of C57Bl/6N mice held either at room temperature or after cold exposure identified a set of miRNAs that regulate BAT differentiation and whose expression changes in response to cold (63). MiR-133 was one of the most downregulated miRNAs after cold exposure. Interestingly, miR-133 contains a highly evolutionarily conserved octamer seed motif within its 5’ segment complementary to a conserved sequence in the 3′UTR of PRMD16 mRNA. Indeed miR-133 directly binds to and suppresses the mRNA encoding PRDM16, thus inhibiting brown adipogenesis from both brown and white progenitor cells (63). Similarly, miR-133 inhibition using antimiRs in MYF5 positive brown precursors, or MYF5 negative preadipocytes derived from SAT of wt mice, led to a dramatic increase of UCP1 levels during brown differentiation. This was accomplished by increased PRDM16 expression, and accompanied by an increase other brown and general adipocyte differentiation markers, ultimately increasing the number of differentiated brown adipocytes. Further, short-hairpin knockdown of PRDM16 abrogated the increase in brown differentiation seen after miR-133a/b inhibition, confirming PRDM16 mRNA as the critical molecular effector of miR-133a/b. MiR-133 overexpression, on the other hand, antagonized brown fat differentiation in both progenitor cell types.
The mechanism underlying miR-133 mediated inhibition of brown adipocyte differentiation involves myocyte-specific enhancer factor 2C (MEF2C), a protein known to regulate myogenesis and vascular development. Indeed, miR-133 is under the transcriptional control of MEF2C, in both brown fat and muscle (63) (64). Adrenergic stimulation and the subsequent increase in cAMP levels that also occurs after cold exposure, leads to decreased MEF2 expression, resulting in marked down-regulation of miR-133 (Figure 2). This triggers de-repression of PRDM16, leading to a marked increase in brown differentiation from both MYF5 positive and negative precursor cells.
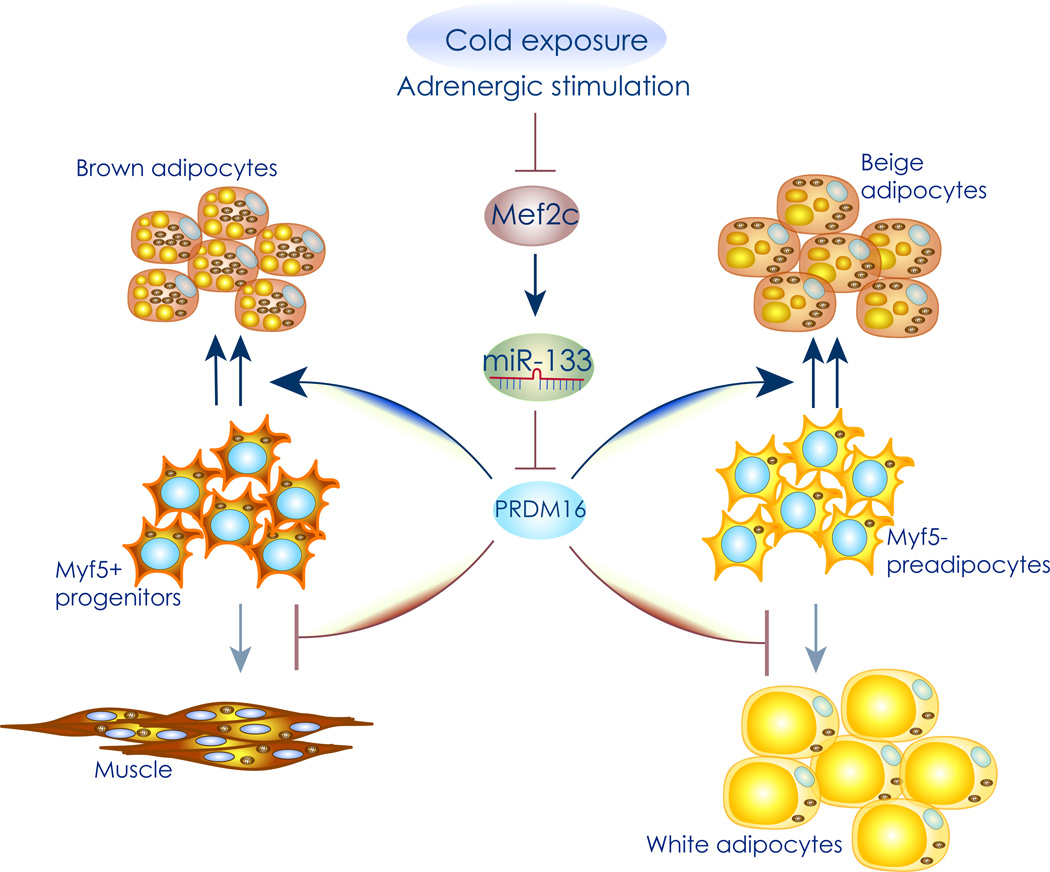
Figure 2. Decreased miR-133 expression during cold exposure promotes brown adipogenesis.
Graphical representation of the miR-133 pathway. Cold exposure and beta adrenergic stimulation increase cyclic AMP levels, leading to a decrease in MEF2C expression and resulting in marked downregulation of miR-133. Since miR-133 represses expression of the master brown adipocyte differentiation factor PRMD16 in both MYF5 positive, and MYF5 negative progenitor cells, this leads to increased brown and beige adipocyte differentiation, respectively.
Brown adipocytes that have differentiated in the absence of miR-133 are fully functional, as shown by increased levels of mitochondrial activity and increased responsiveness to beta adrenergic stimulation (63). Since MEF2C and miR-133 downregulation lie upstream of PRDM16, which as noted above induces miR-193b/365 expression through PPARα, it will be of interest to investigate a possible link between the expression of these miRNAs. It is tempting to speculate that upon cold exposure, miR-133 downregulation represents a triggering event that leads to increased PRDM16 expression, which then stimulates an increase in miR-193b levels further promoting brown adipogenesis.
Interestingly, while cold exposure up to 48 hrs caused a marked downregulation of miR-133 in BAT and SAT (63), it failed to induce changes in the expression of this miRNA in muscle or induction of BAT cells from muscle progenitors. This emphasizes the essential role of BAT and SC beige cells as the first line of defense to cold. However, it was recently reported that cold exposure for up to one week decreases muscle miR-133 levels, leading to increased brown adipogenesis from satellite cells within regenerating muscle tissue (65), an event mechanistically linked to PRDM16 de-repression. While we do not yet understand the nature of this temporal difference, it will be interesting to determine whether repression of miR-133 and MEF2C expression in muscle in vivo is under direct beta adrenergic regulation, or whether additional activation pathways, presumably secondary to the initial steps towards brown adipogenesis in BAT and SAT, are also relevant to the conversion of satellite cells into beige cells.
During chondrocyte maturation, stimulation with parathyroid hormone (PTH)-related peptide or forskolin leads to dephosphorylation of histone deacetylase 4 (HDAC4) on serine 246 by protein phosphatase 2A (PP2A), which induces nuclear translocation of the deacetylase and interaction with and repression of MEF2 activity (66). In addition, in muscle cells miR-1 binds to and downregulates the mRNA encoding the HDAC4 protein that in turn represses transcription (59), and both the myomiRs, miR-1 and -206 are downergulated after cold exposure in BAT (63). Although inhibition of miR-1 does not seem to cause a significant effect on BAT differentiation (52), it will be interesting to determine whether dual miR-1/206 silencing ultimately affects miR-133 expression through HDAC4 and MEF2C, providing an alternative pathway of activating the miR-133/miR-193 cascade in either or both MYF5 positive and negative progenitor cells.
Role of MiR-196a in beige cell formation
Members of the Hox family of homeobox genes (Hox genes), proteins that confer segmental identity during embryonic development, have distinct expression patterns between BAT and WAT, indicating that they might play a role in the determination of these two adipocyte cells types. Among clustered HOX genes HOXC8 gene had the highest expression levels in human WAT-progenitor cells, where it acts by repressing the expression of brown fat genes crucial for differentiation. HoxC8 was also shown to be down-regulated during brown adipogenesis (67).
The miR-196a gene is located near the HoxC8 gene. miR-196a has extensive, evolutionarily conserved, sequence complementarity to HOXC8 mRNA, and downregulates HOXC8 expression during several stages of vertebrate development (68). In particular, miR-196a is sharply upregulated during brown fat differentiation of WAT precursors, as well as after cold exposure or beta adrenergic stimuli in vivo. This suggests a direct link between miR-196a upregulation, repression of HOXC8, and conversion of WAT cells into beige adipocytes. Indeed, fat-specific forced expression of miR-196a in mice induced generation of brown adipocyte-like cells in WAT (67). Insights into the mechanism by which HOXC8 represses brown adipogenesis came from chromatin immunoprecipitation (ChIP) assays that identified an enrichment of Hoxc8 bound to the C/EBPβ locus in the mouse genome. By luciferase reporter assays using homeodomain Hoxc8 mutants (HDm) lacking DNA-binding capacity it was demonstrated that Hoxc8 targets and represses C/EBPβ expression, in cooperation with HDAC3, through a C/EBPβ 3' regulatory sequence (Figure 3). Thus, an increase in miR-196a expression during beige adipogenesis suppresses Hoxc8 expression, leading to de-repression of C/EBPβ and promotion of differentiation of beige adipocytes. MiR-196a transgenic mice exhibited enhanced energy expenditure and resistance to obesity, suggesting that the induced beige adipocyte-like cells differentiated in the presence of miR-196a are metabolically functional.
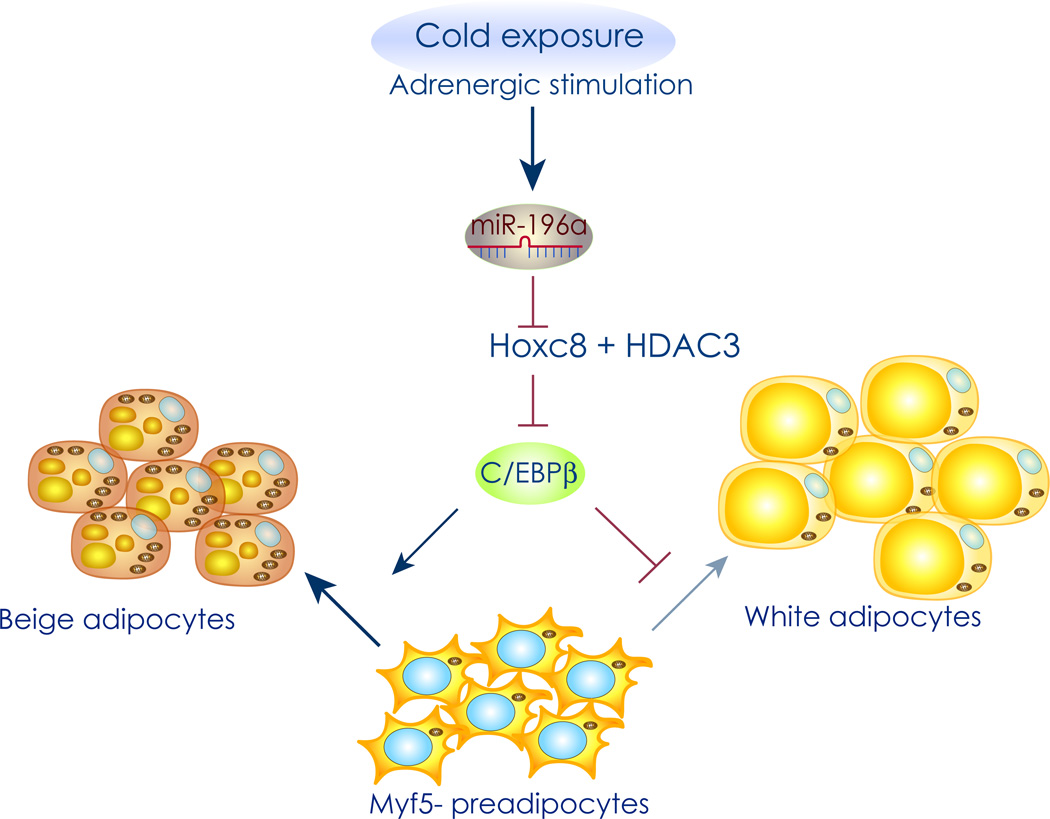
Figure 3. MiR-196a upregulation induces beige cell formation.
Graphical representation of the miR-196a pathway. Hoxc8 targets and represses C/EBPβ expression in cooperation with histone deacetylase 3 (HDAC3) through a C/EBPβ 3' regulatory sequence. Cold exposure and adrenergic stimulation lead to an increase in miR-196a expression, which directly suppresses Hoxc8 expression and thereby de-represses C/EBPβ expression. This leads to induction of beige adipogenesis exclusively in MYF5 negative progenitor cells.
Thus, the mir-196a/HOXC8 axis and the miR-133/PRDM16/193b axis seem to regulate different molecular pathways while both activating brown adipogenesis. It will be interesting to study whether simultaneous activation of both pathways will additively increase brown adipogenesis, possibly as a new way of treating metabolic diseases, primarily dyslipidemia, diabetes, and obesity.
Role of MiR-155 in brown and beige adipocyte differentiation
Comparison by deep sequencing of miRNA expression profiles of preadipocytes isolated from mouse BAT SVF with that of mature brown adipocytes identified miR-155 as being decreased during brown adipogenesis (69). MiR-155 is downregulated by transforming growth factor β1 (TGF β1) in epithelial (70), and brown fat cells (69), and TGF β1 potently inhibits adipogenesis in 3T3-L1 cells (71). In addition, overexpression of C/EBPβ leads to inhibition of miR-155 promoter activity in HIB-1B brown preadipocytes, while siRNA-mediated knockdown of C/EBPβ causes an increase in miR-155 expression in BAT-derived preadipocytes. Chromatin immunoprecipitation assays identified a distal site in the miR-155 promoter as the critical binding site for C/EBPβ, thus demonstrating that miR-155 is also regulated by this major brown fat transcriptional regulator (69). Interestingly, C/EBPβ is a miR-155 target gene in inflammatory processes as well as in in vitro models of white adipogenesis (72–75). MiR-155 directly downregulates C/EBPβ also in BAT and SAT preadipocytes, thus leading to suppression of brown or beige adipogenesis respectively. Thus miR-155 and C/EBPβ constitute a double-negative feedback loop (69) (Figure 4). Accordingly, BAT specific miR-155 transgenic animals show reduced BAT mass, altered BAT morphology, and reduced UCP1 expression. Conversely, miR-155 KO mice show resistance to cold induced drop of body temperature, higher levels of glycerol release and cellular respiration, and a decrease of the number and size of lipid droplets in BAT. Ultimately, UCP1 and PGC1α levels are increased in BAT and SAT in the miR-155 KO mice, demonstrating increased brown/beige fat activity in absence of miR-155 also in vivo.
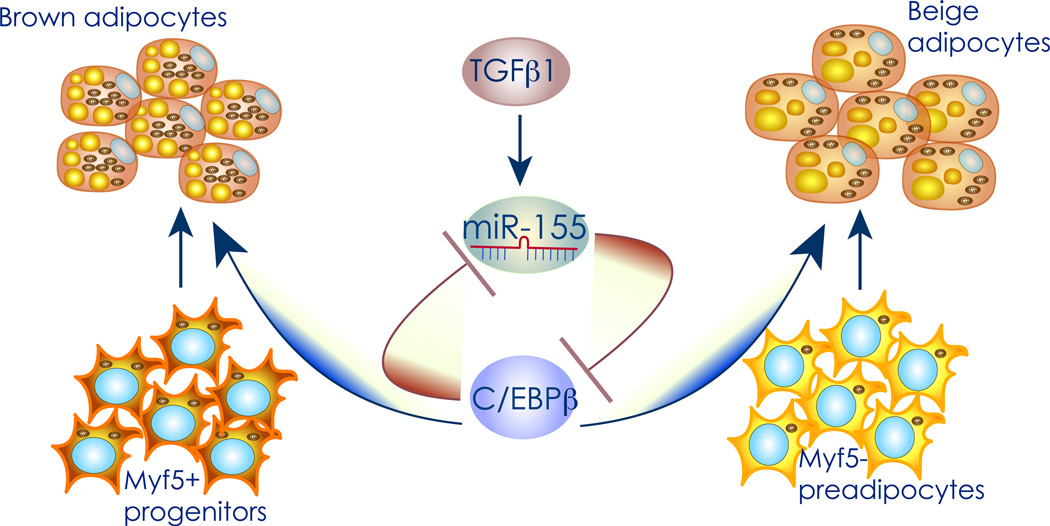
Figure 4. miR-155 controls the development of brown and beige cells.
Graphical representation of the miR-155 pathway. MiR-155 expression is positively regulated by TGFβ1, and suppresses the translation of C/EBPβ as it’s direct target. C/EBPβ induction during brown and beige adipogenesis inhibits transcription of the miR-155 gene, thereby forming a bistable loop for the regulation of adipogenesis either maintaining preadipocytes in an undifferentiated precursor state or initiating the brown adipogenic program.
Therapeutic potential of miRNA
Currently, the most effective approach of enhancing miRNA function is through adenovirus vectors (76). On the one hand, accumulating evidence indicates miRNA silencing as an emerging and novel way to treat several diseases (77) (76), further outlining the therapeutic potential of BAT relevant inhibitory miRNAs. These approaches rely on antimiRs with chemical modifications aiming to increase the binding affinity to the miRNA, enable nuclease resistance, and facilitate cellular uptake. AntimiRs currently in development as therapeutic tools use non- conjugated phosphorothioate antisense molecules incorporating various additional high affinity 2’ sugar modifications such as 2’-O-methoxyethyl (2’-MOE) and 2’-fluoro (2’-F) substituents. Alternatively, locked nucleic acid (LNA) and LNA-like conformationally restricted nucleotides can be used. Initial results of tongoing phase II clinical trials indicate that LNA-modified antimiR-122 is well tolerated and provides continuous and prolonged antiviral activity in patients infected with hepatitis C virus (HCV) (clinicaltrials.gov). Pre-clinical data on 2’-F-2’-MOE-modified antimiR-33 demonstrates a key role of this miRNA in cholesterol homeostasis and fatty acid metabolism in non-human primates. (78). Several other antimiR therapies are undergoing preclinical trials: downregulating miR-21 in hepatocellular carcinoma and kidney fibrosis; and miR-10b in glioblastoma. Since miR-193b/365, miR-133, miR-196a, and miR-155 potently regulate brown adipogenesis, and in the case of miR-133, -196a, and -155 differentiation of SAT preadipocytes into beige cells, modulating the levels of these miRNAs could lead to novel therapeutic strategies for promoting brown adipogenesis and thus for treatment of a variety of metabolic disorders.
Concluding remarks
Here we focused on five BAT miRNAs and discussed their importance in the differentiation of brown and beige adipocytes. Increased expression of miR-193b, miR-196a, or downregulation of miR-133 and miR-155, is sufficient to cause an increase in brown fat differentiation and function. Deregulated expressions of miRNAs are associated with various disorders, including obesity and diabetes. Recent studies show that in obese rodents there is altered expression of miR-143 and miR-103/107 in insulin-sensitive tissues and in the case of miR-103/107 also in humans. These might contribute to impaired insulin sensitivity and glucose homeostasis (47) (48) (49, 79). Interestingly, miR-103/107 inhibition promotes increased energy expenditure in the resting state (49), a phenotype associated with smaller adipocytes. It will be interesting to investigate whether this effect is, at least in part, due to effects of miR-103/107 inhibition on certain BAT functions. As many other miRNAs are essential for development of white adipose cells (51), it remains to be determined whether any of these are also important for the BAT formation.
Thus far most work on BAT-important miRNAs has involved cell cultures or transgenic mice; total body or lineage specific knockout mice will greatly inform the functions of these miRNAs in vivo. One example is miR-155 knock out mice, which show increased brown/beige fat activity as well as improved resistance to cold. Yet in some cases generating knockout mice will be challenging. For instance both miR-193 and miR-365 exist as two alleles: mir-193a-mir-365-2 and mir-193b-mir-365-1. Both alleles are transcribed as a single RNA from which the two microRNAs are generated (52). The miR-365 produced by both alleles is identical in sequence. MiR-193a and miR-193b differ in only three nucleotides but the seed sequences – the 8 nucleotides at the 5’ ends - of the two are identical. Since miR-193a and miR-193b share similar functions and are expected to downregulate common mRNA targets, knocking out both alleles will be essential. This is also the case for miR-133, as there are three known loci in the genome: miR-133a-1, miR-133a-2 and miR-133b, found on chromosomes 18, 20 and 6, respectively (46). This can be overcome in part by generating in vivo miRNA inhibitors that can promiscuously inhibit the distinct miRNA families. It is tempting to propose that miRNA based formulations aiming at increasing miR-193b and miR-196a, or inhibiting miR-155 global or mir-133 in BAT/SAT could represent novel therapeutic approach for combating metabolic diseases.
Table 1.
MiRNAs that regulate brown adipogenesis
| Name | Species found |
Expression profile |
Role in MYF5 positive cells |
Role in MYF5 negative cells |
Target genes |
REFS |
|---|---|---|---|---|---|---|
| MiR-193b/365 | Mouse Human | BAT, WAT, other | Promotes brown adipogenesis | Promotes white adipogenesis | CDON, IGFBP5, RUNX1t1 | 52 |
| MiR-133a/b | Mouse Human | Muscle, BAT, SAT | Inhibits brown adipogenesis | Inhibits beige adipogenesis | PRDM16 | 63, 65 |
| MiR-196a | Mouse Human | BAT, SAT | No effect | Promotes beige adipogenesis | HOXC8 | 67 |
| MiR-155 | Mouse Human | BAT, WAT | Inhibits brown adipogenesis | Inhibits beige adipogenesis | C/EBPβ | 69 |
Acknowledgments
We thank Drs. Markus Stoffel, Lei Sun, Maria Gustafsson Trajkovska, and Marko Knoll for insightful comments on the manuscript. MT is grateful to Prof. Gabriel Waksman for support and advices. This work is supported in part by a UCL starting grant to MT, and NIH grants DK047618, DK 068348, and 5P01 HL066105 to HFL.
References
- 1.Zhang Y, et al. Positional cloning of the mouse obese gene and its human homologue (Translated from eng) Nature. 1994;372(6505):425–432. doi: 10.1038/372425a0. (in eng). [DOI] [PubMed] [Google Scholar]
- 2.Maffei M, et al. Increased expression in adipocytes of ob RNA in mice with lesions of the hypothalamus and with mutations at the db locus (Translated from eng) Proc Natl Acad Sci U S A. 1995;92(15):6957–6960. doi: 10.1073/pnas.92.15.6957. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelleymounter MA, et al. Effects of the obese gene product on body weight regulation in ob/ob mice (Translated from eng) Science. 1995;269(5223):540–543. doi: 10.1126/science.7624776. (in eng). [DOI] [PubMed] [Google Scholar]
- 4.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes (Translated from eng) J Biol Chem. 1995;270(45):26746–26749. doi: 10.1074/jbc.270.45.26746. (in eng). [DOI] [PubMed] [Google Scholar]
- 5.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity (Translated from eng) J Biol Chem. 1996;271(18):10697–10703. doi: 10.1074/jbc.271.18.10697. (in eng). [DOI] [PubMed] [Google Scholar]
- 6.Montague CT, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans (Translated from eng) Nature. 1997;387(6636):903–908. doi: 10.1038/43185. (in eng). [DOI] [PubMed] [Google Scholar]
- 7.Steppan CM, et al. The hormone resistin links obesity to diabetes (Translated from eng) Nature. 2001;409(6818):307–312. doi: 10.1038/35053000. (in eng). [DOI] [PubMed] [Google Scholar]
- 8.Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues (Translated from eng) Nat Rev Mol Cell Biol. 2005;6(3):248–261. doi: 10.1038/nrm1592. (in eng). [DOI] [PubMed] [Google Scholar]
- 9.Brand MD. The efficiency and plasticity of mitochondrial energy transduction (Translated from eng) Biochem Soc Trans. 2005;33(Pt 5):897–904. doi: 10.1042/BST0330897. (in eng). [DOI] [PubMed] [Google Scholar]
- 10.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance (Translated from eng) Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. (in eng). [DOI] [PubMed] [Google Scholar]
- 11.Himms-Hagen J, et al. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes (Translated from eng) Am J Physiol Cell Physiol. 2000;279(3):C670–C681. doi: 10.1152/ajpcell.2000.279.3.C670. (in eng). [DOI] [PubMed] [Google Scholar]
- 12.Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse (Translated from eng) FEBS Lett. 1984;167(1):10–14. doi: 10.1016/0014-5793(84)80822-4. (in eng). [DOI] [PubMed] [Google Scholar]
- 13.Bianco AC, Silva JE. Intracellular conversion of thyroxine to triiodothyronine is required for the optimal thermogenic function of brown adipose tissue (Translated from eng) J Clin Invest. 1987;79(1):295–300. doi: 10.1172/JCI112798. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva JE. Full expression of uncoupling protein gene requires the concurrence of norepinephrine and triiodothyronine (Translated from eng) Mol Endocrinol. 1988;2(8):706–713. doi: 10.1210/mend-2-8-706. (in eng). [DOI] [PubMed] [Google Scholar]
- 15.Hany TF, et al. Brown adipose tissue: a factor to consider in symmetrical tracer uptake in the neck and upper chest region (Translated from eng) Eur J Nucl Med Mol Imaging. 2002;29(10):1393–1398. doi: 10.1007/s00259-002-0902-6. (in eng). [DOI] [PubMed] [Google Scholar]
- 16.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans (Translated from eng) Am J Physiol Endocrinol Metab. 2007;293(2):E444–E452. doi: 10.1152/ajpendo.00691.2006. (in eng). [DOI] [PubMed] [Google Scholar]
- 17.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans (Translated from eng) N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virtanen KA, et al. Functional brown adipose tissue in healthy adults (Translated from eng) N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. (in eng). [DOI] [PubMed] [Google Scholar]
- 19.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men (Translated from eng) N Engl J Med. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. (in eng). [DOI] [PubMed] [Google Scholar]
- 20.Lowell BB, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue (Translated from eng) Nature. 1993;366(6457):740–742. doi: 10.1038/366740a0. (in eng). [DOI] [PubMed] [Google Scholar]
- 21.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity (Translated from eng) J Clin Invest. 1998;102(2):412–420. doi: 10.1172/JCI3155. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghorbani M, Claus TH, Himms-Hagen J. Hypertrophy of brown adipocytes in brown and white adipose tissues and reversal of diet-induced obesity in rats treated with a beta3-adrenoceptor agonist (Translated from eng) Biochem Pharmacol. 1997;54(1):121–131. doi: 10.1016/s0006-2952(97)00162-7. (in eng). [DOI] [PubMed] [Google Scholar]
- 23.Kopecky J, Clarke G, Enerback S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity (Translated from eng) J Clin Invest. 1995;96(6):2914–2923. doi: 10.1172/JCI118363. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cousin B, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization (Translated from eng) J Cell Sci. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. (in eng). [DOI] [PubMed] [Google Scholar]
- 25.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human (Translated from eng) Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch (Translated from eng) Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice (Translated from eng) J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development (Translated from eng) Mol Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. (in eng). [DOI] [PubMed] [Google Scholar]
- 29.Rosen ED, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway (Translated from eng) Genes Dev. 2002;16(1):22–26. doi: 10.1101/gad.948702. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen ED, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro (Translated from eng) Mol Cell. 1999;4(4):611–617. doi: 10.1016/s1097-2765(00)80211-7. (in eng). [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene (Translated from eng) EMBO J. 1997;16(24):7432–7443. doi: 10.1093/emboj/16.24.7432. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang ND, et al. Impaired energy homeostasis in C/EBP alpha knockout mice (Translated from eng) Science. 1995;269(5227):1108–1112. doi: 10.1126/science.7652557. (in eng). [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity (Translated from eng) Mol Cell. 1999;3(2):151–158. doi: 10.1016/s1097-2765(00)80306-8. (in eng). [DOI] [PubMed] [Google Scholar]
- 34.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex (Translated from eng) Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hondares E, et al. Peroxisome proliferator-activated receptor alpha (PPARalpha) induces PPARgamma coactivator 1alpha (PGC-1alpha) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16 (Translated from eng) J Biol Chem. 2011;286(50):43112–43122. doi: 10.1074/jbc.M111.252775. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cederberg A, et al. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance (Translated from eng) Cell. 2001;106(5):563–573. doi: 10.1016/s0092-8674(01)00474-3. (in eng). [DOI] [PubMed] [Google Scholar]
- 37.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis (Translated from eng) Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. (in eng). [DOI] [PubMed] [Google Scholar]
- 38.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development (Translated from eng) Cell Metab. 2010;11(4):257–262. doi: 10.1016/j.cmet.2010.03.005. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels (Translated from eng) Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs (Translated from eng) Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. (in eng). [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease (Translated from eng) Development. 2005;132(21):4653–4662. doi: 10.1242/dev.02073. (in eng). [DOI] [PubMed] [Google Scholar]
- 42.Poy MN, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism (Translated from eng) Diabetes Obes Metab. 2007;9(Suppl 2):67–73. doi: 10.1111/j.1463-1326.2007.00775.x. (in eng). [DOI] [PubMed] [Google Scholar]
- 43.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay (Translated from eng) Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. (in eng). [DOI] [PubMed] [Google Scholar]
- 44.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay (Translated from eng) Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. (in eng). [DOI] [PubMed] [Google Scholar]
- 45.Bartel DP. MicroRNAs: target recognition and regulatory functions (Translated from eng) Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data (Translated from eng) Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esau C, et al. MicroRNA-143 regulates adipocyte differentiation (Translated from eng) J Biol Chem. 2004;279(50):52361–52365. doi: 10.1074/jbc.C400438200. (in eng). [DOI] [PubMed] [Google Scholar]
- 48.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity (Translated from eng) Diabetes. 2009;58(5):1050–1057. doi: 10.2337/db08-1299. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity (Translated from eng) Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. (in eng). [DOI] [PubMed] [Google Scholar]
- 50.Mudhasani R, et al. Dicer is required for the formation of white but not brown adipose tissue (Translated from eng) J Cell Physiol. 2011;226(5):1399–1406. doi: 10.1002/jcp.22475. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexander R, Lodish H, Sun L. MicroRNAs in adipogenesis and as therapeutic targets for obesity (Translated from eng) Expert Opin Ther Targets. 2011;15(5):623–636. doi: 10.1517/14728222.2011.561317. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L, et al. Mir193b-365 is essential for brown fat differentiation (Translated from eng) Nat Cell Biol. 2011 doi: 10.1038/ncb2286. (in Eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang JS, Mulieri PJ, Miller C, Sassoon DA, Krauss RS. CDO, a robo-related cell surface protein that mediates myogenic differentiation (Translated from eng) J Cell Biol. 1998;143(2):403–413. doi: 10.1083/jcb.143.2.403. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole F, Zhang W, Geyra A, Kang JS, Krauss RS. Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO (Translated from eng) Dev Cell. 2004;7(6):843–854. doi: 10.1016/j.devcel.2004.10.009. (in eng). [DOI] [PubMed] [Google Scholar]
- 55.Kang JS, et al. A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38alpha/beta MAPK activity and myogenic differentiation (Translated from eng) J Cell Biol. 2008;182(3):497–507. doi: 10.1083/jcb.200801119. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren H, Yin P, Duan C. IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop (Translated from eng) J Cell Biol. 2008;182(5):979–991. doi: 10.1083/jcb.200712110. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes (Translated from eng) J Cell Physiol. 2009;218(2):444–449. doi: 10.1002/jcp.21621. (in eng). [DOI] [PubMed] [Google Scholar]
- 58.Ivey KN, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells (Translated from eng) Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation (Translated from eng) Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart (Translated from eng) Genes Dev. 2008;22(23):3242–3254. doi: 10.1101/gad.1738708. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure (Translated from eng) Proc Natl Acad Sci U S A. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Care A, et al. MicroRNA-133 controls cardiac hypertrophy (Translated from eng) Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. (in eng). [DOI] [PubMed] [Google Scholar]
- 63.Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16 (Translated from eng) Nat Cell Biol. 2012;14(12):1330–1335. doi: 10.1038/ncb2612. (in eng). [DOI] [PubMed] [Google Scholar]
- 64.Liu N, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133 (Translated from eng) Proc Natl Acad Sci U S A. 2007;104(52):20844–20849. doi: 10.1073/pnas.0710558105. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin H, et al. MicroRNA-133 Controls Brown Adipose Determination in Skeletal Muscle Satellite Cells by Targeting Prdm16 (Translated from eng) Cell Metab. 2013;17(2):210–224. doi: 10.1016/j.cmet.2013.01.004. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozhemyakina E, Cohen T, Yao TP, Lassar AB. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway (Translated from eng) Mol Cell Biol. 2009;29(21):5751–5762. doi: 10.1128/MCB.00415-09. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mori M, Nakagami H, Rodriguez-Araujo G, Nimura K, Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells (Translated from eng) PLoS Biol. 2012;10(4):e1001314. doi: 10.1371/journal.pbio.1001314. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA (Translated from eng) Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. (in eng). [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit (Translated from eng) Nat Commun. 2013;4:1769. doi: 10.1038/ncomms2742. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA (Translated from eng) Mol Cell Biol. 2008;28(22):6773–6784. doi: 10.1128/MCB.00941-08. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ignotz RA, Massague J. Type beta transforming growth factor controls the adipogenic differentiation of 3T3 fibroblasts (Translated from eng) Proc Natl Acad Sci U S A. 1985;82(24):8530–8534. doi: 10.1073/pnas.82.24.8530. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He M, Xu Z, Ding T, Kuang DM, Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta (Translated from eng) Cell Mol Immunol. 2009;6(5):343–352. doi: 10.1038/cmi.2009.45. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S, Yang Y, Wu J. TNFalpha-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors (Translated from eng) Biochem Biophys Res Commun. 2011;414(3):618–624. doi: 10.1016/j.bbrc.2011.09.131. (in eng). [DOI] [PubMed] [Google Scholar]
- 74.Skarn M, et al. Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222 (Translated from eng) Stem Cells Dev. 2012;21(6):873–883. doi: 10.1089/scd.2010.0503. (in eng). [DOI] [PubMed] [Google Scholar]
- 75.Worm J, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF (Translated from eng) Nucleic Acids Res. 2009;37(17):5784–5792. doi: 10.1093/nar/gkp577. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles (Translated from eng) Nat Rev Drug Discov. 2012;11(11):860–872. doi: 10.1038/nrd3864. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Esteller M. Non-coding RNAs in human disease (Translated from eng) Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. (in eng). [DOI] [PubMed] [Google Scholar]
- 78.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides (Translated from eng) Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jordan SD, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism (Translated from eng) Nat Cell Biol. 2011;13(4):434–446. doi: 10.1038/ncb2211. (in eng). [DOI] [PubMed] [Google Scholar]
- 80.Loncar D, et al. The effect of intermittent cold treatment on the adipose tissue of the cat. Apparent transformation from white to brown adipose tissue (Translated from eng) J Ultrastruct Mol Struct Res. 1986;97(1–3):119–129. doi: 10.1016/s0889-1605(86)80012-x. (in eng). [DOI] [PubMed] [Google Scholar]
- 81.Cinti S. Adipocyte differentiation and transdifferentiation: plasticity of the adipose organ (Translated from eng) J Endocrinol Invest. 2002;25(10):823–835. doi: 10.1007/BF03344046. (in eng). [DOI] [PubMed] [Google Scholar]
- 82.Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: Is beige the new brown? (Translated from eng) Genes Dev. 2013;27(3):234–250. doi: 10.1101/gad.211649.112. (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat (Translated from eng) Nat Med. 2013 doi: 10.1038/nm.3112. (in Eng). [DOI] [PMC free article] [PubMed] [Google Scholar]