Background: CDK12 mutations occur in ovarian cancer.
Results: These mutations impaired CDK12 kinase activity. Additionally, disabling CDK12 in ovarian cancer cells reduced BRCA1 levels and disrupted homologous recombination repair.
Conclusion: CDK12 mutations that impair kinase activity likely disrupt homologous recombination.
Significance: Defects in homologous recombination caused by CDK12 mutations may predict sensitivity to chemotherapy agents, including poly(ADP-ribose) polymerase inhibitors.
Keywords: BRCA1, CDK (Cyclin-dependent Kinase), DNA Damage, Homologous Recombination, Ovarian Cancer, CDK12, Cisplatin, Poly(ADP)-ribose Polymerase, RAD51, Veliparib
Abstract
Mutations in the tumor suppressors BRCA1 and BRCA2, which encode proteins that are key participants in homologous recombination (HR) repair, occur in ∼20% of high grade serous ovarian cancers. Although only 20% of these tumors have mutations in BRCA1 and BRCA2, nearly 50% of these tumors have defects in HR. Notably, however, the underlying genetic defects that give rise to HR defects in the absence of BRCA1 and BRCA2 mutations have not been fully elucidated. Here we show that the recurrent somatic CDK12 mutations identified in ovarian cancers impair the catalytic activity of this kinase, which is involved in the transcription of a subset of genes, including BRCA1 and other DNA repair genes. Furthermore, we show that disabling CDK12 function in ovarian cancer cells reduces BRCA1 levels, disrupts HR repair, and sensitizes these cells to the cross-linking agents melphalan and cisplatin and to the poly(ADP-ribose) polymerase (PARP) inhibitor veliparib (ABT-888). Taken together, these findings suggest that many CDK12 mutations are an unrecognized cause of HR defects in ovarian cancers.
Introduction
BRCA1 and BRCA2 mutations are found in ∼20% of high grade epithelial ovarian cancers, the histotype that accounts for two-thirds of ovarian cancer deaths (1–3). These tumor suppressors maintain genomic stability by facilitating homologous recombination (HR),2 a process that repairs double-stranded DNA breaks and restores stalled replication forks (4). During HR repair, BRCA1 coordinates the early steps of this repair pathway by interacting with proteins that process DNA into single-stranded DNA tracts, which are then coated with RAD51, in a process facilitated by BRCA2 (5). These RAD51-single-stranded DNA filaments then serve as substrates during the search for homologous sequences to complete the HR repair process. Correspondingly, cells with defects in BRCA1 or BRCA2 exhibit impaired HR repair (2, 6), which causes sensitivity to DNA-damaging agents that inflict lesions repaired by HR. One such lesion is an interstrand DNA cross-link, which is induced by cross-linkers such as cisplatin and other platinum salts that are the backbone of ovarian cancer therapies (2). Similarly, cells exhibiting HR defects are also selectively sensitive to poly(ADP-ribose) polymerase (PARP) inhibitors (7–9). Consistent with these findings, PARP inhibitors have shown impressive activity in BRCA1/2-mutant ovarian cancers (10–13).
Although only 20% of ovarian cancers harbor BRCA1 or BRCA2 mutations, nearly 50% of ovarian cancers show defects in HR repair (1). This phenomenon has been attributed to loss of BRCA1 expression (due to BRCA1 promoter methylation) and rare mutations in other genes that directly participate in HR (3, 6, 14). Despite this progress, it remains unclear whether the full spectrum of genetic alterations that cause HR defects has been identified.
A high throughput exome sequencing effort of 316 high grade serous ovarian cancers by The Cancer Genome Atlas Research Network uncovered a series of genes that are recurrently mutated (3). One of the most frequently altered genes was CDK12, which was mutated in ∼3% of these tumors, a prevalence similar to that observed for somatic mutations in BRCA1 (3.5%) and BRCA2 (3.2%) in this series. Interestingly, although CDK12 is not known to directly participate in HR, a recent study showed that depleting CDK12 reduced levels of BRCA1 and other proteins (e.g. ATR, FANCI, and FANCD2) involved in responses to DNA damage (15). In this study, the mechanistic basis for reduced BRCA1 expression was ascribed to the ability of CDK12 to phosphorylate Ser-2 in the heptapeptide repeat (Tyr-Ser-Pro-Thr-Ser-Pro-Ser) located in the C-terminal domain (CTD) of RNA polymerase II, an event that regulates mRNA elongation (15, 16). Although Ser-2 CTD phosphorylation is generally attributed to CDK9 (17), a kinase that is closely related to CDK12, these studies firmly established that 1) CDK12 also phosphorylates this site and 2) CDK12 regulates a small subset of genes, including BRCA1.
Notably, however, the functional effects of the CDK12 mutations identified in ovarian cancers have not been evaluated. Also, it remains unclear whether CDK12 mutations affect HR in ovarian cancer cells and whether disabling CDK12 sensitizes cells to DNA-damaging agents and PARP inhibitors. Here we show that 1) nearly all ovarian tumor-associated CDK12 kinase domain mutations severely impede catalytic activity, 2) cells expressing catalytically inactive CDK12 have defects in HR, and 3) disabling CDK12 in ovarian cancer cells not only disrupts HR but also robustly sensitizes cells to DNA cross-linking agents and PARP inhibitors. Taken together, these observations suggest that ovarian cancer-associated CDK12 mutations cause HR defects, a finding that may help identify tumors that will respond to specific therapeutic interventions, including PARP inhibitors.
EXPERIMENTAL PROCEDURES
Materials
Antibodies and suppliers were as follows: RAD51 (Calbiochem, PC130), BRCA1 (Cell Signaling Technology, 9010), CDK12 (Sigma, WH0051755M2), HSP90 (D. Toft, Mayo Clinic, H9010), S-peptide (18), hemagglutinin (HA, 12CA5, Covance), fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G, tetramethylrhodamine isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch), and horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (Cell Signaling Technology). Other reagents were obtained as follows: cisplatin (Teva Pharmaceuticals), veliparib (ABT-888) (Selleck Chemicals or ChemieTek), streptavidin-agarose (Millipore), and [γ-32P]ATP (MP Biomedicals, 38101X). Glutathione S-transferase (GST)-CTD was expressed in Escherichia coli and purified using standard methods. siRNAs (Dharmacon) used were: luciferase, 5′-CTTACGCUGAGUACUUCGA-3′ (19); BRCA1-1, 5′-GUGGGUGUUGGACAGUGUA-3′ (20); CDK12-1, 5′-GAAAGAAGACAAACAGAAA-3′; CDK12-2, 5′-GAGACUAGACAAUGAGAAA-3′; CDK12-3, 5′-GAAGAACAGTATTGACATA-3′; and CDK12-5, 5′-GTAGATACAAGTAGAGAAT-3′. All other reagents were from Sigma-Aldrich.
Cell Lines, Cell Culture, and Transfections
OVCAR-3 and K562 cells were from the American Type Culture Collection (Manassas, VA). OVCAR-5 and OVCAR-8 were gifts from D. Scudierio (NCI, National Institutes of Health), and OVCAR-8-DR-GFP, which has a genomically integrated DR-GFP substrate for HR repair assays, was described previously (21). OVCAR-3 cells were cultured in RPMI 1640 medium supplemented with 8% fetal bovine serum (Atlanta Biologicals or Sigma) and 10 μg/ml insulin. All other cells were cultured in the same medium but without insulin. Cells were expanded upon receipt, cryopreserved, and reinitiated from cryopreserved stocks every 3–6 months. Transfection of siRNAs and plasmids was as described (21).
DR-GFP HR Assays, Clonogenic Assays, Cell Cycle Analysis, and Immunoblotting
Analysis of OVCAR-8-DR-GFP cells transfected with pCβASceI was described previously (21). Clonogenic assays, cell cycle analysis, and immunoblotting were performed as described (22, 23).
Plasmids
Human CDK12 cDNA (BC140854, Open Biosystems) and CDK9 cDNA (Addgene plasmid 28102 (24)) were subcloned into the pSFB vector, which appends in-frame, N-terminal S-peptide, FLAG, and streptavidin-binding peptide tags (25). Rat cyclin K cDNA (BC129121, Open Biosystems) was subcloned into pcDNA3 (Invitrogen) containing tandem C-terminal HA tags. RNA polymerase II CTD was amplified from mouse genomic DNA and subcloned into pGEX-KG to create an in-frame fusion to GST. CDK12 mutations were generated using QuikChange site-directed mutagenesis (Agilent). All plasmids generated were confirmed by Sanger sequencing.
CDK12 in Vitro Kinase Assay
K562 cells (1.5 × 107 cells) transfected with the indicated plasmids were cultured for 16–24 h and lysed in 30 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 0.5% Triton X-100, 1 mm Na3VO4, 10 mm 2-glycerophosphate, 10 μg/ml leupeptin, 5 μg/ml aprotinin, 5 μg/ml pepstatin, 20 nm microcystin-LR. The cleared lysate was incubated with streptavidin-agarose beads with rotation for 2–3 h at 4 °C. Beads were washed three times with lysis buffer supplemented with 500 mm NaCl, 1% Triton X-100, and 0.1% SDS (without leupeptin, aprotinin, pepstatin, and microcystin-LR); washed twice with 40 mm HEPES, pH 7.6, 0.5 mm EDTA, 0.5 mm EGTA, 1 mm 2-glycerophosphate, 1 mm NaF, 2 mm dithiothreitol, and 2 mm magnesium acetate; and incubated with 20 μl of kinase reaction mix containing 4.8 μg of GST-CTD, 20 μCi of [γ-32P]ATP, and 50 μm ATP. The reaction was incubated for 10 min at 30 °C, stopped with SDS-PAGE sample buffer, heated to 95 °C for 5 min, separated by SDS-PAGE, and transferred to Immobilon-P. Membrane-bound radioactivity was detected and quantitated by phosphorimaging (Storm, GE Healthcare) before immunoblotting to detect tagged proteins.
RAD51 Foci Assay
To assess the impact of BRCA1 and CDK12 depletion on RAD51 foci formation, OVCAR-8 cells were transfected twice with siRNAs. To determine how CDK12 mutants affect RAD51 foci formation, OVCAR-8 cells were transfected with CDK12 siRNAs (CDK12-3 and CDK12-5) that target the 3′-untranslated region of endogenous CDK12, and 24 h later they were transfected with the same siRNAs plus empty vector or plasmids that encode wild-type or mutant SFB-tagged CDK12 (which also encodes enhanced green fluorescent protein to identify transfected cells). For both experimental approaches, 40–48 h after the second transfection, cells were plated in 8-well chamber slides (Lab-Tek II), allowed to attach for 4–6 h, irradiated with 10 grays of ionizing radiation (RS-2000, RadSource), incubated at 37 °C for 18–22 h, fixed in 4% paraformaldehyde in PBS for 10 min, and permeabilized in 0.25% Triton X-100 in PBS. Slides were blocked in PBS containing 3% bovine serum albumin and 0.05% Triton X-100, incubated with 1:250 dilution anti-RAD51 antibody (Calbiochem) for 2 h at 20 °C, and stained with 1:200 dilution fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G (for cells transfected with siRNAs only) or 1:200 dilution tetramethylrhodamine isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (for cells transfected with siRNAs plus plasmids). Cells were then counterstained with 1 μg/ml Hoechst 33342. For cells transfected with siRNAs only, 100 cells were counted. For cells transfected with siRNAs plus plasmids, 100 enhanced green fluorescent protein-positive cells were counted. For both assays, cells with more than five foci were considered positive for RAD51 foci formation (26, 27). Images were acquired with a Zeiss LSM 510 microscope and processed with the manufacturer's software.
RESULTS
Tumor-associated CDK12 Mutations Abrogate Catalytic Activity
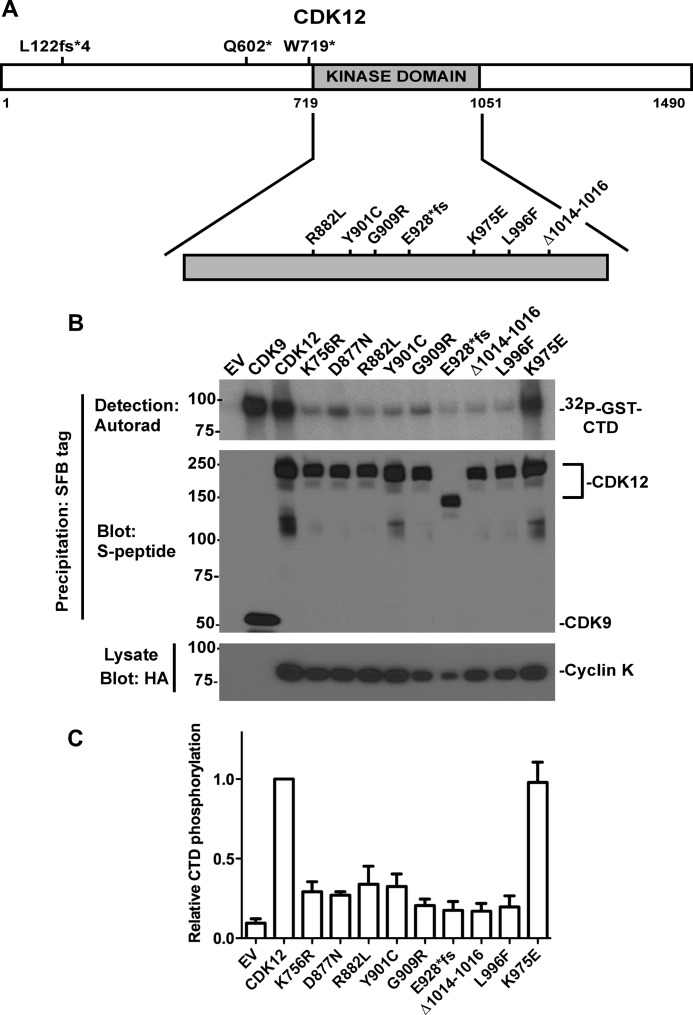
The ovarian cancer-associated CDK12 mutations identified by The Cancer Genome Atlas Network exome sequencing effort (3) can be roughly divided into two groups. In one group, frameshift or point mutations introduce premature stop codons that precede the kinase domain such that the predicted protein would not have kinase activity (Fig. 1A). In the second group, missense, frameshift, or deletion mutations occur in the kinase domain itself (amino acids 719–1051). Notably, however, the effects of the kinase domain mutations on CDK12 catalytic activity have not been characterized. Therefore, we compared the kinase activities of wild-type CDK12 with the kinase domain mutants using RNA polymerase II CTD as a substrate. Wild-type and mutant SFB-tagged CDK12s were transiently expressed with HA-tagged cyclin K, the cyclin partner for CDK12 (28, 29), and the CDK12 complexes were purified by streptavidin-agarose bead precipitation. As controls for these experiments, we transfected cells with SFB-tagged CDK9 (which also phosphorylates RNA polymerase CTD) and with CDK12 mutants (K756R and D877N), which disrupt the conserved catalytic lysine and the conserved DFG motif, respectively, and are predicted to inactivate the kinase. We observed that wild-type CDK12 phosphorylated the CTD substrate nearly as effectively as CDK9 (Fig. 1B), and that the K756R (catalytic lysine) and D877N (conserved DFG motif) had severely impaired kinase activity, thus validating the assay. Importantly, all but one of the kinase domain mutations (K975E) severely disrupted kinase activity (Fig. 1, B and C). Taken together, these results show that many, but not all, CDK12 kinase domain mutations found in ovarian cancers inactivate its catalytic activity.
FIGURE 1.
CDK12 mutations identified in ovarian cancer impair kinase activity. A, CDK12 mutations identified in ovarian cancers. fs*, frameshift that causes premature truncation. B, detergent-soluble proteins from K562 cells transfected with empty vector (EV) or transiently expressing SFB-tagged CDK9, wild-type CDK12, or CDK12 mutants were immunoblotted for cyclin K-HA expression (bottom panel) or were precipitated with streptavidin-agarose (precipitates SFB tag), washed, and subjected to kinase assays with GST-CTD and [γ-32P]ATP as substrates. Kinase reactions were separated by SDS-PAGE and transferred to Immobilon P membrane, and 32P-labeled GST-CTD products were detected by autoradiography (Autorad, top panel). The membrane was then immunoblotted with anti-S peptide antibody, which detects the SFB tag (middle panel). C, phosphorimaging quantitation of [32P]CTD from four independent experiments, mean ± S.D.
CDK12 Depletion Decreases Homologous Recombination
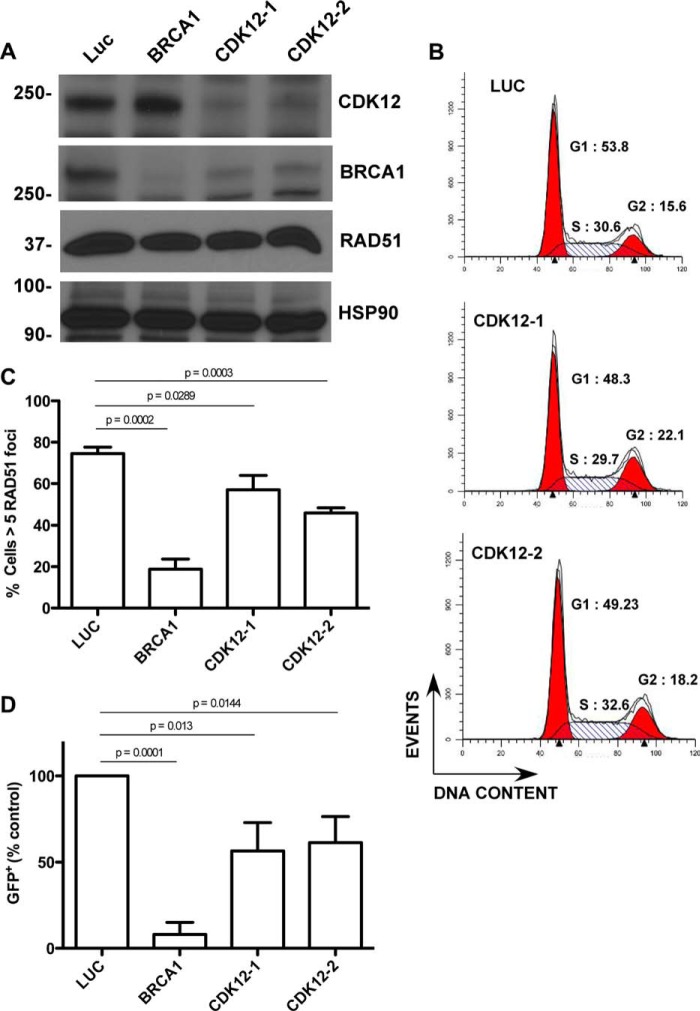
To test the effect of disabling CDK12 in ovarian cancer cells, we transfected OVCAR-8 cells, which contain wild-type BRCA1 (30) and CDK12 (31), with two independent siRNAs that target CDK12. As shown in Fig. 2A, CDK12 depletion markedly reduced BRCA1 levels in these cells, consistent with results previously reported in HeLa cells (15), but did not appreciably alter the cell cycle (Fig. 2B). To determine whether CDK12 depletion affected HR, we next examined the assembly of RAD51 foci at sites of double-strand breaks induced by ionizing radiation and the ability of the cells to carry out HR repair of a genomically integrated DR-GFP substrate, which is composed of tandem inactive green fluorescent protein (GFP) fragments that flank an I-SceI restriction enzyme site (32). Depletion of BRCA1 blocked ionizing radiation-induced RAD51 foci formation (Fig. 2C) and HR repair of the DR-GFP substrate following I-SceI expression (Fig. 2D). Similarly, CDK12-depleted cells also formed fewer RAD51 foci (Fig. 2C) and showed reduced recombination repair of DR-GFP (Fig. 2D). Collectively, these results show that disabling CDK12 reduces BRCA1 levels and disrupts HR repair in ovarian cancer cells.
FIGURE 2.
CDK12 depletion causes HR defects in ovarian cancer cell lines. A–C, OVCAR-8 cells were transfected twice with control luciferase (Luc), BRCA1-1, CDK12-1, or CDK12-2 siRNAs. 48 h after transfection, the cells were analyzed for CDK12, BRCA1, and RAD51 expression (A), analyzed for DNA content (B), or irradiated with 10 grays of ionizing radiation and stained to detect RAD51 foci (C). Mean ± S.D., n = 4; p values by paired t test. D, OVCAR-8-DR-GFP cells transfected with pCβASceI plasmid plus control luciferase (Luc), BRCA1, CDK12-1, or CDK12-2 siRNAs were analyzed by flow cytometry for GFP fluorescence 72 h after transfection. Data in D were normalized to luciferase-transfected cells for each experiment. Mean ± S.D., n = 4; p values by paired t test.
Tumor-associated CDK12 Mutant Decreases Homologous Recombination in Ovarian Cancer Cells
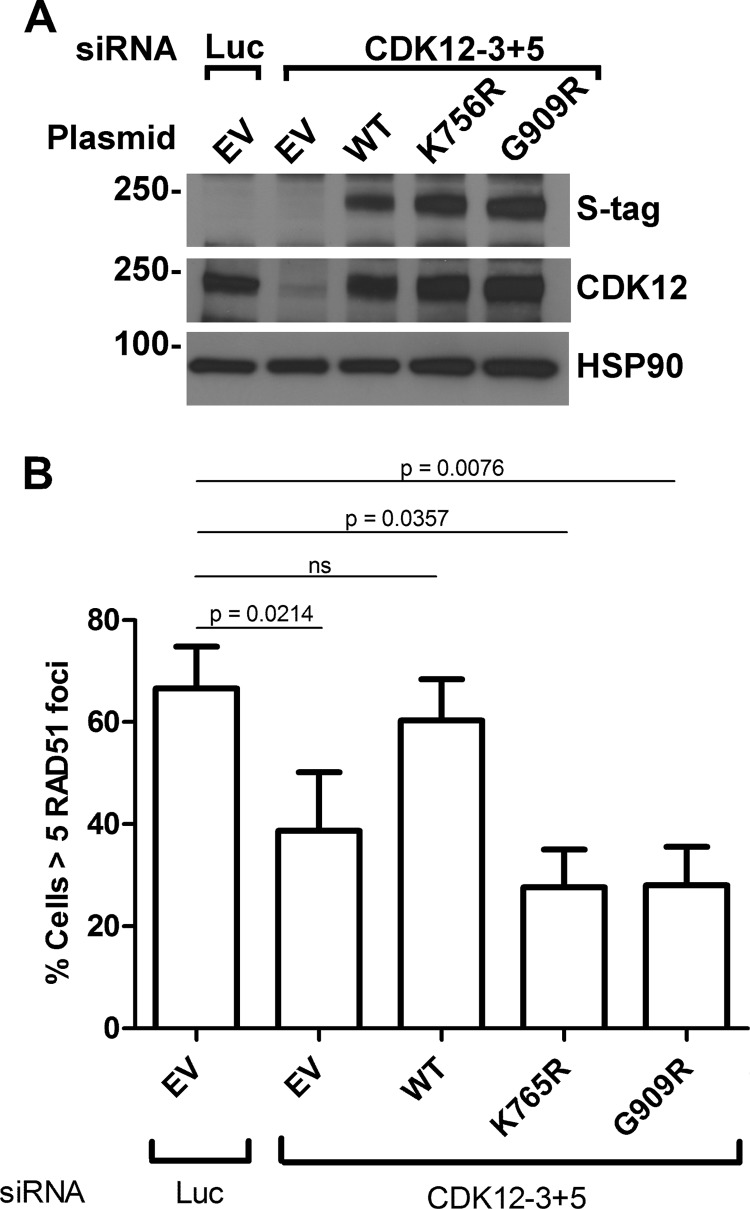
To determine whether a catalytically inactive CDK12 mutant causes HR defects in ovarian cancer cells, we depleted OVCAR-8 cells of endogenous CDK12 by cotransfecting two siRNAs that target the 3′-untranslated region. These cells were then additionally transfected with empty vector or plasmids that encode wild-type CDK12, kinase-dead CDK12 (K756R), or a tumor-associated CDK12 (G909R) mutant (all of which lack the 3′-untranslated region). CDK12-depleted OVCAR-8 cells reconstituted with the wild-type CDK12 restored CDK12 levels (Fig. 3A) and the formation of RAD51 foci (Fig. 3B). In contrast, expression of either the kinase-dead (K756R) or tumor-associated CDK12 mutant (G909R) at similar levels (Fig. 3A) did not restore RAD51 foci (Fig. 3B). Accordingly, these results indicate that catalytically inactive tumor-associated CDK12 mutants disrupt HR in ovarian cancer cells.
FIGURE 3.
Kinase-dead and tumor-associated CDK12 mutations cause HR defects in ovarian cancer cells. OVCAR-8 cells were transfected with luciferase (Luc) or CDK12-3 plus CDK12-5 siRNAs. 24 h later the cells were transfected with the same siRNAs plus empty vector (EV) or plasmids encoding SFB-tagged wild-type CDK12, kinase-dead CDK12 (K756R), or the tumor-associated mutant CDK12 (G909R). 40–48 h after the second transfection, cells were analyzed for SFB-CDK12 expression (S-tag) and endogenous CDK12 expression (A) or were irradiated with 10 grays of ionizing radiation and stained to detect RAD51 foci (B). Mean ± S.D., n = 3; p values by paired t test.
CDK12 Depletion Sensitizes Ovarian Cancer Cells to DNA Cross-linking Agents and PARP Inhibition
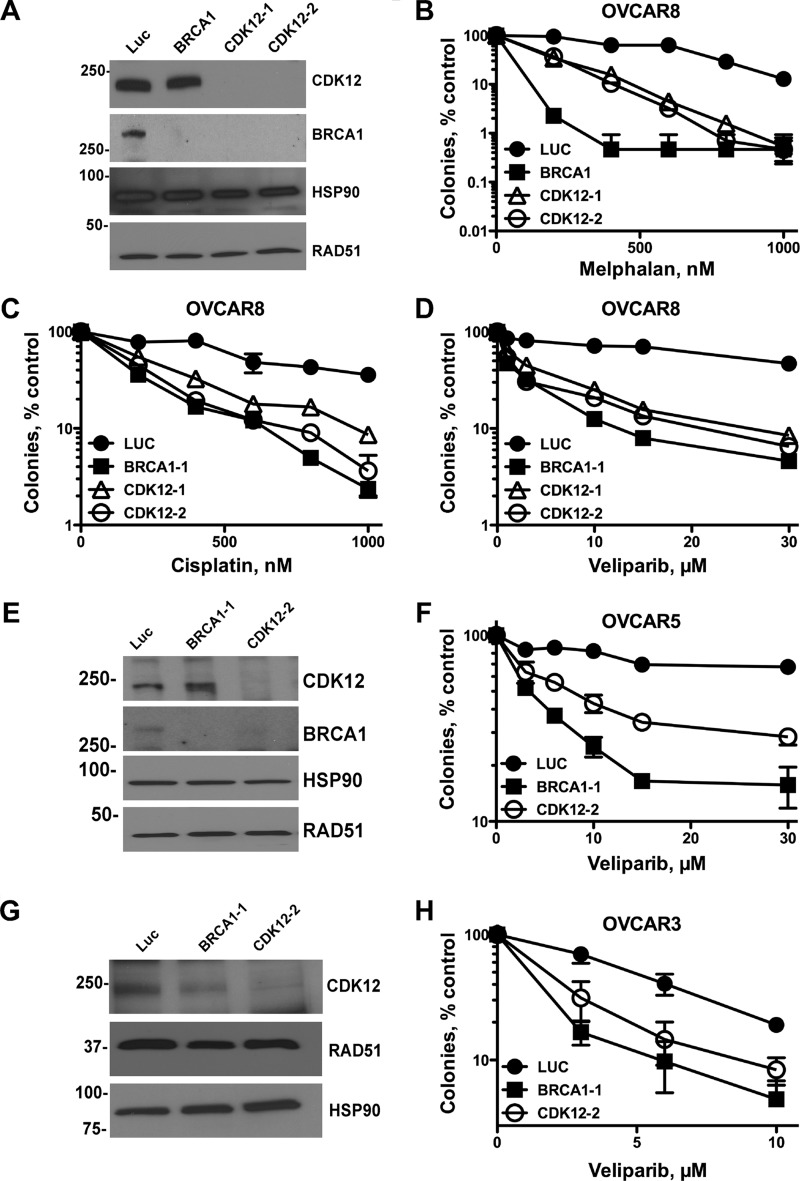
Defects in HR sensitize cells to DNA-damaging agents that inflict lesions repaired by this pathway, such as interstrand DNA cross-links. Furthermore, cells harboring defects in BRCA1, BRCA2, and other HR components are hypersensitive to PARP inhibitors when compared with cells with functional HR repair. To determine whether CDK12 sufficiently disrupts HR repair to sensitize to these agents, we treated cells with two cross-linking agents, melphalan, a bifunctional alkylating agent, and cisplatin, which is used to treat ovarian cancer, as well as the PARP inhibitor veliparib (ABT-888). Consistent with the defects in RAD51 foci formation and HR repair, CDK12- and BRCA1-depleted OVCAR-8 cells (Fig. 4A) were sensitized to melphalan (Fig. 4B), cisplatin (Fig. 4C), and the PARP inhibitor veliparib. Similarly, depletion of CDK12 and BRCA1 (Fig. 4E) from OVCAR-5 cells, which have wild-type BRCA1 (30) but harbor a mutation of unknown significance outside the kinase domain of CDK12(P1266L) (31), sensitized these cells to veliparib (Fig. 4F). Notably, even in OVCAR-3 cells, which have wild-type BRCA1 (30) and CDK12 (31), but in which BRCA1 levels are below our detection limit (data not shown), both BRCA1 and CDK12 depletion (Fig. 4G) sensitized cells to veliparib (Fig. 4H).
FIGURE 4.
CDK12 depletion sensitizes ovarian cancer cell lines to DNA cross-linking agents and PARP inhibitors. OVCAR-8 (A–D), OVCAR-5 (E and F), or OVCAR-3 (G and H) cells were transfected with control luciferase (Luc), BRCA1, CDK12-1, or CDK12-2 siRNAs. 48 h after transfection, cells were analyzed for BRCA1 and CDK12 by immunoblotting (A, E, and G) and used in clonogenic assays. For clonogenic assays, cells were plated (OVCAR-8 and OVCAR-5, 300 cells per well; OVCAR-3, 2000 cells per well), allowed to adhere for 4–6 h, treated with cisplatin (C) for 24 h, washed, and re-fed. Alternatively, cells were treated with melphalan (B) or veliparib (D, F, and H) continuously and allowed to form colonies for 9–14 days. Colonies (>50 cells) were stained with Coomassie Blue and manually counted. Results shown are representative of four (OVCAR-8) and three (OVCAR-5) independent experiments.
DISCUSSION
A recent reanalysis of the large scale exome sequencing data of ovarian tumors found that CDK12 is frequently biallelically mutated in these tumors (33), raising the possibility that CDK12 is a tumor suppressor. Notably, however, the effects of these mutations on CDK12 kinase activity have not been previously assessed, thus limiting our understanding of how they may impact ovarian cancer biology. Here we show that the vast majority of kinase domain mutations abrogate catalytic activity. This finding, taken in conjunction with the finding that CDK12 mutations occur in both alleles (33), suggests that ovarian tumors harboring inactivating CDK12 mutations have severely impaired CDK12 function.
Our studies to address the consequences of disabling CDK12 in ovarian cancer cells confirmed that CDK12 depletion diminished BRCA1 levels in ovarian cancer cells. We note, however, that it is unlikely that BRCA1 is the only CDK12 target that contributes to effective HR. For example, depletion of ATR, which is also reduced by CDK12 knockdown (15), also disrupts HR (21). Similarly, CDK12 depletion also reduced the levels of FANCI and FANCD2 (15), two proteins that, when down-regulated, cause sensitivity to PARP inhibitors (8), a hallmark of defective HR.
Our further analyses of CDK12 revealed that depleting CDK12 reduced RAD51 foci formation and HR repair, two phenotypes that are associated with HR defects. Additionally, the kinase-dead CDK12 K756R mutant and the tumor-associated CDK12 G909R mutant did not restore impaired RAD51 foci formation, thus demonstrating that CDK12 catalytic activity is required for this function and showing that the catalytically impaired, tumor-associated CDK12 mutants also disrupt HR. Consistent with these findings, we then showed that depleting CDK12 sensitized ovarian cancer cells to the DNA cross-linking agent cisplatin, a representative of a drug class that is the backbone of ovarian cancer treatments. Finally, we showed that CDK12 depletion sensitized multiple ovarian cancer cell lines to PARP inhibition, an exciting therapeutic approach that relies on the synthetic lethality of PARP inhibition in cells with defects in HR repair.
While this manuscript was under revision, another study found that CDK12 depletion sensitized cells to PARP inhibition in a genome-wide synthetic lethality screen (34). Our results extend these observations by showing that the majority of tumor-associated CDK12 kinase domain mutations actually inactivate the catalytic activity of CDK12 and demonstrating that CDK12 catalytic activity is required for optimal homologous recombination.
Acknowledgments
We thank the Mayo Clinic Optical Morphology Laboratory and Advanced Genomics Technology Center.
This work was supported in part by the Mayo Clinic Ovarian Cancer SPORE Grant P50 CA136393 (through the National Institutes of Health) and Mayo Clinic Pobanz Family Predoctoral Research Fellowship (to P. M. J.).
- HR
- homologous recombination
- PARP
- poly(ADP-ribose) polymerase
- CTD
- C-terminal domain
- DR-GFP
- direct repeats of green fluorescent protein
- SFB
- S-peptide, FLAG, streptavidin-binding peptide.
REFERENCES
- 1. Hennessy B. T., Timms K. M., Carey M. S., Gutin A., Meyer L. A., Flake D. D., 2nd, Abkevich V., Potter J., Pruss D., Glenn P., Li Y., Li J., Gonzalez-Angulo A. M., McCune K. S., Markman M., Broaddus R. R., Lanchbury J. S., Lu K. H., Mills G. B. (2010) Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 28, 3570–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bast R. C., Jr., Mills G. B. (2010) Personalizing therapy for ovarian cancer: BRCAness and beyond. J. Clin. Oncol. 28, 3545–3548 [DOI] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roy R., Chun J., Powell S. N. (2012) BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holloman W. K. (2011) Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 18, 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turner N., Tutt A., Ashworth A. (2004) Hallmarks of 'BRCAness' in sporadic cancers. Nat. Rev. Cancer 4, 814–819 [DOI] [PubMed] [Google Scholar]
- 7. Martin S. A., Lord C. J., Ashworth A. (2008) DNA repair deficiency as a therapeutic target in cancer. Curr. Opin. Genet. Dev. 18, 80–86 [DOI] [PubMed] [Google Scholar]
- 8. McCabe N., Turner N. C., Lord C. J., Kluzek K., Bialkowska A., Swift S., Giavara S., O'Connor M. J., Tutt A. N., Zdzienicka M. Z., Smith G. C., Ashworth A. (2006) Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115 [DOI] [PubMed] [Google Scholar]
- 9. Ratner E. S., Sartorelli A. C., Lin Z. P. (2012) Poly (ADP-ribose) polymerase inhibitors: on the horizon of tailored and personalized therapies for epithelial ovarian cancer. Curr. Opin. Oncol. 24, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M. J., Ashworth A., Carmichael J., Kaye S. B., Schellens J. H., de Bono J. S. (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 11. Fong P. C., Yap T. A., Boss D. S., Carden C. P., Mergui-Roelvink M., Gourley C., De Greve J., Lubinski J., Shanley S., Messiou C., A'Hern R., Tutt A., Ashworth A., Stone J., Carmichael J., Schellens J. H., de Bono J. S., Kaye S. B. (2010) Poly(ADP)-ribose polymerase inhibition: Frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 28, 2512–2519 [DOI] [PubMed] [Google Scholar]
- 12. Audeh M. W., Carmichael J., Penson R. T., Friedlander M., Powell B., Bell-McGuinn K. M., Scott C., Weitzel J. N., Oaknin A., Loman N., Lu K., Schmutzler R. K., Matulonis U., Wickens M., Tutt A. (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 376, 245–251 [DOI] [PubMed] [Google Scholar]
- 13. Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T., Matei D., Macpherson E., Watkins C., Carmichael J., Matulonis U. (2012) Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 366, 1382–1392 [DOI] [PubMed] [Google Scholar]
- 14. Banerjee S., Kaye S. B., Ashworth A. (2010) Making the best of PARP inhibitors in ovarian cancer. Nat. Rev. Clin. Oncol. 7, 508–519 [DOI] [PubMed] [Google Scholar]
- 15. Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B. M. (2011) The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25, 2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L. (2010) CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24, 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brès V., Yoh S. M., Jones K. A. (2008) The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20, 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hackbarth J. S., Lee S. H., Meng X. W., Vroman B. T., Kaufmann S. H., Karnitz L. M. (2004) S-peptide epitope tagging for protein purification, expression monitoring, and localization in mammalian cells. BioTechniques 37, 835–839 [PubMed] [Google Scholar]
- 19. Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- 20. Huehls A. M., Wagner J. M., Huntoon C. J., Karnitz L. M. (2012) Identification of DNA repair pathways that affect the survival of ovarian cancer cells treated with a poly(ADP-ribose) polymerase inhibitor in a novel drug combination. Mol. Pharmacol. 82, 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huntoon C. J., Flatten K. S., Wahner Hendrickson A. E., Huehls A. M., Sutor S. L., Kaufmann S. H., Karnitz L. M. (2013) ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 73, 3683–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huntoon C. J., Nye M. D., Geng L., Peterson K. L., Flatten K. S., Haluska P., Kaufmann S. H., Karnitz L. M. (2010) Heat shock protein 90 inhibition depletes LATS1 and LATS2, two regulators of the mammalian Hippo tumor suppressor pathway. Cancer Res. 70, 8642–8650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wagner J. M., Karnitz L. M. (2009) Cisplatin-induced DNA damage activates replication checkpoint signaling components that differentially affect tumor cell survival. Mol. Pharmacol. 76, 208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y., Dow E. C., Liang Y. Y., Ramakrishnan R., Liu H., Sung T. L., Lin X., Rice A. P. (2008) Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J. Biol. Chem. 283, 33578–33584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chini C. C., Chen J. (2006) Repeated phosphopeptide motifs in human Claspin are phosphorylated by Chk1 and mediate Claspin function. J. Biol. Chem. 281, 33276–33282 [DOI] [PubMed] [Google Scholar]
- 26. Tarsounas M., Davies D., West S. C. (2003) BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 27. Nakada S., Yonamine R. M., Matsuo K. (2012) RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res. 72, 4974–4983 [DOI] [PubMed] [Google Scholar]
- 28. Blazek D. (2012) The cyclin K/Cdk12 complex: an emerging new player in the maintenance of genome stability. Cell Cycle 11, 1049–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng S. W., Kuzyk M. A., Moradian A., Ichu T. A., Chang V. C., Tien J. F., Vollett S. E., Griffith M., Marra M. A., Morin G. B. (2012) Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol. Cell. Biol, 32, 4691–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stordal B., Timms K., Farrelly A., Gallagher D., Busschots S., Renaud M., Thery J., Williams D., Potter J., Tran T., Korpanty G., Cremona M., Carey M., Li J., Li Y., Aslan O., O'Leary J. J., Mills G. B., Hennessy B. T. (2013) BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 7, 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abaan O. D., Polley E. C., Davis S. R., Zhu Y. J., Bilke S., Walker R. L., Pineda M., Gindin Y., Jiang Y., Reinhold W. C., Holbeck S. L., Simon R. M., Doroshow J. H., Pommier Y., Meltzer P. S. (2013) The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 73, 4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pierce A. J., Johnson R. D., Thompson L. H., Jasin M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carter S. L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P. W., Onofrio R. C., Winckler W., Weir B. A., Beroukhim R., Pellman D., Levine D. A., Lander E. S., Meyerson M., Getz G. (2012) Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bajrami I., Frankum J. R., Konde A., Miller R. E., Rehman F. L., Brough R., Campbell J., Sims D., Rafiq R., Hooper S., Chen L., Kozarewa I., Assiotis I., Fenwick K., Natrajan R., Lord C. J., Ashworth A. (2014) Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 74, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]