Background: Fatty acid amide hydrolase (FAAH) belongs to the family of amidase signature proteins and is involved in N-acylethanolamine (NAE) metabolism.
Results: New synthetic phenoxyacyl-ethanolamide compounds increased the amidohydrolase activity of FAAH.
Conclusion: Phenoxyacyl-ethanolamide compounds increased the activity of the FAAH enzyme in plants and animals.
Significance: New properties of FAAH proteins were revealed with these phenoxyacyl-ethanolamide compounds, and the potential for their applications in vivo was demonstrated.
Keywords: Arabidopsis, Enzyme Kinetics, Lipids, Plant, Signaling, N-Acylethanolamines, Fatty Acid Amide Hydrolase, Synthetic Compounds
Abstract
N-Acylethanolamines (NAEs) are involved in numerous biological activities in plant and animal systems. The metabolism of these lipids by fatty acid amide hydrolase (FAAH) is a key regulatory point in NAE signaling activity. Several active site-directed inhibitors of FAAH have been identified, but few compounds have been described that enhance FAAH activity. Here we synthesized two sets of phenoxyacyl-ethanolamides from natural products, 3-n-pentadecylphenolethanolamide and cardanolethanolamide, with structural similarity to NAEs and characterized their effects on the hydrolytic activity of FAAH. Both compounds increased the apparent Vmax of recombinant FAAH proteins from both plant (Arabidopsis) and mammalian (Rattus) sources. These NAE-like compounds appeared to act by reducing the negative feedback regulation of FAAH activity by free ethanolamine. Both compounds added to seedlings relieved, in part, the negative growth effects of exogenous NAE12:0. Cardanolethanolamide reduced neuronal viability and exacerbated oxidative stress-mediated cell death in primary cultured neurons at nanomolar concentrations. This was reversed by FAAH inhibitors or exogenous NAE substrate. Collectively, our data suggest that these phenoxyacyl-ethanolamides act to enhance the activity of FAAH and may stimulate the turnover of NAEs in vivo. Hence, these compounds might be useful pharmacological tools for manipulating FAAH-mediated regulation of NAE signaling in plants or animals.
Introduction
Fatty acid amide hydrolase (FAAH)4 belongs to the superfamily of amidase signature proteins (1–3). FAAH enzymes are found in diverse groups of organisms, including both plants and animals. FAAH enzymes hydrolyze a broad range of N-acylethanolamines (NAEs) to corresponding free fatty acid and ethanolamine (4–6) and can also act on primary acyl amides as well as acyl esters (3). In plants, one type of FAAH has been characterized (7), whereas two distinct FAAHs have been described in animal systems (2, 8). The homology between plant (Arabidopsis) and mammalian (rat) FAAH proteins at the amino acid level is somewhat low over the full length of the proteins. However, the amidase signature sequences (with core catalytic residues) between plant and animal FAAH share up to 60% similarity at the amino acid level (1). The x-ray crystallography data of the rat protein provided new insights about the mode of action of this enzyme in NAE hydrolysis identifying a Ser-Ser-Lys catalytic triad (9, 10). Although no tertiary structure has been determined yet for plant FAAH, a structural homology model was developed for the amidase domain of the Arabidopsis FAAH protein, and conserved catalytic residues were identified experimentally (10, 11).
The activity of FAAH is a key regulatory feature of the NAE signaling pathway. The regulated accumulation of NAEs influences numerous functions in plants and animals. The functional activities and physiological effects of NAEs are mostly terminated by their degradation to FFA and ethanolamine. In plants, NAEs are involved in seedling establishment and growth (6). Exogenously applied N-lauroylethanolamine (NAE 12:0) arrests seedling growth, and this is evident through marked changes in root architecture and elongation (12, 13). This inhibitory effect of NAEs on seedling growth occurs in part through a complex interaction with ABA (abscisic acid) signaling machinery during the embryo-to-seedling transition that remains incompletely understood (14–16). On the other hand, reductions of endogenous seed NAE levels through the overexpression of FAAH in Arabidopsis results in enhanced seedling growth and increased size of roots, cotyledons, and other plant organs (13). Other physiological processes have been attributed to FAAH-mediated alteration of NAE levels in plants, such as flowering time, which is induced by the expression and translocation of the FT (flowering locus T) protein from leaves to the vegetative meristem. FAAH-overexpressing plants exhibited an early flowering phenotype in both inductive and non-inductive growth conditions, and this was associated with lower NAE levels and higher expression of FT and other key flowering genes (16). Still other work has attributed changes in host susceptibility to pathogens (17, 18) or changes in phytohormone signaling pathways (11, 14, 18) with altered FAAH expression.
In animals, FAAH-mediated NAE changes are part of the so-called “endocannabinoid signaling pathway,” and this pathway plays a central regulatory role in many physiological and behavioral processes (19). The most widely studied NAE in animal systems is the N-arachidonylethanolamine, known also as anandamide (NAE 20:4) (20), but other NAE species with overlapping or unique functions are known as well. As an example, N-linoleoylethanolamine (NAE 18:2) and N-palmitoylethanolamine (NAE 16:0) are involved in neuron protection in the retinal ganglion cell layer against excessive extracellular glutamate and against oxidative stress for the HT22 cells, respectively (21, 22). Anandamide was identified as the first endogenous ligand of the cannabinoid receptors (CB1 and CB2) and is involved in activating many of the important endocannabinoid pathways (20, 23, 24). Anandamide and other NAEs have been associated with different processes, such as pain modulation, memory, anxiety, appetite, etc. (25–27). Their levels are controlled largely through hydrolysis by FAAH. Thus, FAAH has become a major therapeutic target for many disorders that involve NAE signaling in situ.
Several approaches have been employed to increase the level of NAEs in plants or animals, including the direct application of NAEs or pharmacological reagents that inhibit NAE degradation. Utilization of general serine protease inhibitors and/or specific inhibitors of FAAH activity, such as phenylmethylsulfonyl fluoride (PMSF), 5Z,8Z,11Z,14Z-eicosatetraenyl-methyl ester phosphonofluoridic acid (MAFP), and 3′-(aminocarbonyl)[1,1′-biphenyl]-3-yl)-cyclohexylcarbamate (URB597), have been reported to elevate endogenous levels of NAE and to extend or amplify processes regulated by NAE signaling (28). Genetic approaches have been developed to reduce FAAH expression (FAAH knockouts) in mice and Arabidopsis (13, 29), although this approach has shown limited success, especially in plants where it appears that there are redundant pathways for NAE catabolism and where it has been difficult to raise endogenous NAE levels dramatically in vivo. On the other hand, it has been possible to overexpress FAAH in plants and reduce NAE levels to some extent to influence several physiological processes, including growth, defense, and flowering (13, 16, 18). However, there is limited information on chemical compounds that reduce the NAE content in plant and animal systems via enhanced FAAH activity.
Among the multitude of renewable resources, cashew nut shell liquid is an important by-product of the cashew nut industry that is currently used for green chemicals and technologies (30). More than 32% of the cashew shell is cashew nut shell liquid, the key constituent of cashew nut shell liquid being cardanol, a bio-based non-isoprene lipid, consisting of a rich mixture of phenolic lipids: 5% 3-(n-pentadecyl)-phenol (PDP), 50% 3-(8Z-pentadecenyl)phenol, 16% 3-(8Z,11Z-pentadecadienyl)phenol, and 29% 3-(8Z,11Z,14-pentadecatrienyl)phenol. Cardanol's unique properties stem from the varying degree of cis-double bonds and an odd number of hydrocarbons with easily accessible saturated and unsaturated hydrocarbon chains (31).
Here, we synthesized a new set of phenoxyacyl-ethanolamides from this cashew nut shell liquid renewable resource with structural similarity to NAEs to investigate their effects on FAAH activity and to explore the potential utility of natural mixtures in cardanol ethanolamide compared with the pure, saturated analog pentadecylphenoxy ethanolamide. These phenoxyacyl-ethanolamides were not substrates for FAAH; however, we measured a positive effect of these compounds on NAE hydrolysis by recombinant FAAH. The increase in enzyme turnover rate is probably through relief from product inhibition by ethanolamine, a property not previously appreciated for either plant or mammalian FAAH enzymes. It is possible that compounds like these phenoxyacyl-ethanolamides might prove useful in manipulating NAE levels in vivo through their actions on FAAH.
EXPERIMENTAL PROCEDURES
Materials
[1-14C]Lauric acid was from Amersham Biosciences, [1-14C]palmitic acid and [1-14C]arachidonic acid were purchased from PerkinElmer Life Sciences. Ethanolamine, anandamide, isopropyl-β-d-thiogalactopyranoside, and Triton X-100 were from Sigma. N-Dodecyl-β-d-maltoside (DDM) was from Calbiochem. Sprague-Dawley rat embryonic day 18 cortical neurons, NeuroPapain, NeuroPrep medium, and NeuroPure plating medium were obtained from Genlantis (San Diego, CA). Neurobasal media, B27 supplement, B27 antioxidant-free supplement, GlutaMAX I, and calcein-AM were purchased from Invitrogen. BD PureCoatTM amine plates were obtained from BD Biosciences. DMSO and 2-hydroxypropyl-β-cyclodextrin were purchased from Sigma. tert-Butyl hydroperoxide (tBHP) was obtained from Acros Organics (part of Thermo Fisher Scientific). PBS and penicillin/streptomycin were purchased from Lonza (Walkersville, MD). Palmitoylethanolamide (NAE 16:0) was from Best West Laboratories (Salt Lake City, UT), MAFP was from Tocris Biosciences, and URB597 was from EMD Millipore. Silica Gel G (60 A)-coated glass plates for thin layer chromatography (10 × 20 cm or 20 × 20 cm, 0.25-mm thickness) were from Whatman. Different species of N-[1-14C]acylethanolamines (and non-radiolabeled NAEs) were synthesized from ethanolamine and corresponding 1-14C-labeled fatty acids (and non-radiolabeled FFAs) by first producing the fatty acid chloride (32) and purifying by thin layer chromatography (TLC) as described elsewhere (33). The chemical compounds 3-n-pentadecylphenol-ethanolamide and cardanolethanolamide (cardanol-EA) were produced as described below.
Synthesis of PDP-EA and the More General Mixed Species Cardanol-EA
For the cardanol-EA, in a round bottom flask fixed with a magnetic stirrer, cardanol-methyl ester (3.74 g, 10 mmol) was added, followed by dichloromethane (25 ml) and triethylamine (1.4 ml, 10 mmol). The reaction mixture was stirred for 2 min, followed by the dropwise addition of ethanolamine (0.66 ml, 11 mmol) in an ice bath with constant stirring. The resultant mixture was stirred at room temperature for about 6–8 h. After completion of the reaction, as identified using TLC, ice-cold water was added, and the lipids were extracted with ethyl acetate. The organic phase was separated and dried over anhydrous sodium sulfate, filtered, and concentrated. Pure product as a colorless liquid was obtained by column chromatographic purification. At 5 °C, the viscous liquid solidifies into a pale yellow solid. Yield = 78%. 1H NMR (CDCl3, 300 MHz): δ 7.17 (s, 2H), 6.68–6.83 (m, 3H), 5.36 (d, 3H), 4.45 (s, 2H), 3.72 (s, 2H), 3.49 (s, 3H), 2.78 (s, 1H), 2.56 (d, 2H), 2.14 (s, 1H), 2.01 (s, 2H), 0.86–1.57 (m, 20H); 13C NMR (CDCl3, 75 MHz): δ 169.91, 157.33, 145.25, 130.28, 130.15, 130.00, 129.68, 122.58, 122.43, 115.18, 114.99, 111.88, 111.69, 67.36, 61.85, 42.04, 36.15, 31.99, 31.59, 29.93, 29.45, 29.41, 22.89, 14.33. High resolution MS analysis showed [M + H]+, m/z 404.3163 (for the principal ethanolamide species in the cardanol-EA preparation) compared with the calculated mass for C25H41NO3, [M + H]+ of m/z 404.3165. To synthesize and purify the PDP-EA (N-(2-hydroxyethyl)-2-(3-n-pentadecylphenoxy) acetamide), a similar scheme was used as for cardanol-EA, except pure PDP methyl ester was used to generate the acylethanolamide. Purity of crude product was greater than 90%. Pure PDP-EA product, as colorless crystals, was obtained by column chromatographic purification. Yield = 92%. 1H NMR (CDCl3, 300 MHz): δ 7.18 (s, 2H), 6.84 (s, 1H), 6.73 (s, 2H), 4.77 (s, 1H), 4.47 (s, 2H), 4.11 (s, 1H), 3.72 (s, 2H), 3.45 (s, 2H), 2.56 (s, 2H), 2.02 (s, 1H), 1.57 (s, 2H), 1.25 (s, 22H), 0.87 (s, 3H); 13C NMR (CDCl3, 75 MHz): δ 169.76, 157.29, 145.35, 129.72, 122.66, 115.20, 114.91, 111.89, 67.34, 62.18, 42.12, 36.16, 32.13, 31.61, 29.89, 22.91, 14.48. High resolution MS analysis of the PDP-EA matched exactly with the calculated mass for C25H43NO3, [M + H]+ of m/z 406.3321.
Plasmid Constructs
The recombinant plasmid, rat FAAH1-pTrcHis2 (NCBI accession number NP_077046), was provided by Dr. Benjamin Cravatt's laboratory (34), and the plasmid At-FAAH-pTrcHis2 (At5g64440, UniProt number Q7XJJ7) was constructed as described previously (7). The expression constructs were introduced into chemically competent Escherichia coli TOP10 cells as host as described in the manufacturer's instructions.
Protein Expression and Solubilization for Enzymatic Assays from Different Bacteria Cultures (E. coli)
The different cell lines were grown in 250 ml of LB medium with 100 μg·ml−1 filtered ampicillin to an A600 of 0.6 and induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 4 h at 37 °C. Each culture was centrifuged at 5000 rpm for 10 min at 4 °C in a Beckman tabletop centrifuge (rotor, GH 3.7). The pelleted cells expressing rat FAAH1 or At-FAAH1 were resuspended in 10 ml of lysis buffer A (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% Triton X-100) or 10 ml of lysis buffer B (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.2 mm DDM). After incubation on ice for 30 min, resuspended cells were sonicated on ice with 10 30-s bursts at 50% intensity with 30-s cooling (ice) periods between bursts. Each crude lysate was centrifuged at 13,000 × g for 20 min at 4 °C in a Sorvall RC 5C model ultracentrifuge (Sorvall rotor, SS-34). The supernatant was applied to a QiQexpress® nickel-nitrilotriacetic acid fast start (Qiagen®) column, and the proteins were purified according to the manufacturer's instructions. The purified fractions (2 ml) were concentrated, and imidazole was removed with buffer C (50 mm BisTris propane-HCl, pH 9.0, 0.2 mm DDM) by filtration-centrifugation using Centricon YM-30 (Millipore, Bedford, MA) devices. The protein concentration was estimated by Bradford reagent (Sigma) against a BSA standard curve, and the purity of the proteins was evaluated by SDS-PAGE and Western blotting. The rat or Arabidopsis thaliana FAAH (At-FAAH) proteins were aliquoted (20 μl) and stored at −80 °C for up to several months and thawed once for use.
SDS-PAGE and Western Blotting of Purified FAAH Proteins
Each aliquot (rat or At-FAAH protein) was separated by SDS-PAGE (10% resolving gels) as described previously (7). The proteins were visualized in gels by Coomassie Blue staining, or proteins were electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes (0.2 μm; Bio-Rad) according to the protocol described elsewhere (10). The recombinant proteins expressing the His tag at the C terminus were detected by chemiluminescence using a 1:2000 dilution of mouse monoclonal anti-His antibodies (ABGENT, San Diego, CA) and a solution of 1:4000 dilution of goat anti-mouse IgG conjugated to a peroxidase (Bio-Rad).
FAAH Assays on Purified Proteins from Different E. coli Cell Lines
The NAE amidohydrolase assays were conducted as described previously (7, 10, 33) with few modifications. The reactions were conducted for 30 min at 30°C, 120 rpm, in 150 μl of BTP buffer (50 mm BisTris propane-HCl, pH 9.0) containing different concentrations of radiolabeled NAEs, the new NAE-like compounds, and different concentrations of purified protein (see figure legends for more details of the composition of each reaction mixture). Enzyme reactions were terminated by the addition of hot isopropyl alcohol (70 °C). The lipids were extracted, and the distribution of the radioactivity was evaluated by radiometric scanning of TLC plates as described elsewhere (10).
Ethanolamine Inhibition Assays on Purified FAAH Enzymes
Assays containing 0.3 μg of purified protein were first incubated with 100 μm PDP-EA or cardanol-EA and then with different concentrations of ethanolamine (0–100 mm) (Vf = 150 μl of buffer C). Reactions were initiated by adding 100 μm radiolabeled NAE and terminated as described above. The lipids were extracted, and the total distribution of the radioactivity was calculated as above.
Plant Material and Cultures
Ten mg of A. thaliana (ecotype Col-0) seeds were surface-sterilized and then stratified in the dark for 2 days at 4 °C prior to sowing in liquid (75 ml) or solid Murashige and Skoog medium (14). Growth of seedlings was in a 16-h light/8-h dark cycle (60 μmol·m−2·s−1) for 11 days at 20 °C.
Plant Protein Extraction
After 11 days in liquid culture, the plant material was rinsed with milliQ water and then blotted dry. With a mortar and pestle, the plant material (500 mg) was ground with liquid nitrogen and then with 2 ml of plant protein solubilization solution (0.1 m potassium phosphate buffer, pH 7.2, 400 mm sucrose, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 1 mm MgCl2) with 0.2 mm DDM. The crude extract was centrifuged at 2000 rpm at 4 °C for 10 min in a Beckman (rotor, GH 3.7). The supernatant, containing the total solubilized proteins, was stored at 4 °C for up to 2 days.
Plant Amidohydrolase Assays
The reactions were conducted at 30 °C for 2 h in BTP buffer (50 mm BisTris propane-HCl, pH 9.0) with 300 μm PDP-EA or cardanol-EA and 200 μm radiolabeled NAE 12:0. Reactions were initiated by adding 5 μg of total protein and terminated by the addition of hot isopropyl alcohol. Lipids were extracted and analyzed as above.
Primary Neuronal Culture
Commercially obtained Sprague-Dawley rat embryonic day 18 cortical tissue was allowed to settle for 5–10 min at room temperature, the shipping medium was removed (reserved at 37 °C), and the tissue was enzymatically treated with NeuroPapain dissolved in NeuroPrep medium (2 mg·ml−1, 2.5 ml) for 28 min at 37 °C with gentle swirling every 7 min. Treated tissue was centrifuged for 1.5 min at 200 × g, and the cells were dissociated by gentle trituration in 1 ml of the reserved shipping medium. Cells were collected by centrifugation as above and resuspended by gentle trituration in 1 ml of NeuroPure plating medium. Viable cells were counted (Nexcelom Auto T4; Nexcelom Bioscience LLC, Lawrence, MA), resuspended in 9 ml of NeuroPure plating medium plus enough neurobasal medium (supplemented with B27; GlutaMAX I (2 mm), penicillin (50 units/ml), and streptomycin (50 μg·ml−1)) to achieve a density of 250,000 cells/ml, and seeded in BD PureCoatTM black-walled amine-coated 96-well plates in a 100-μl volume. Cultures were maintained at 37 °C, 5% CO2, 95% humidity for 7 days prior to experiments with a 50% medium exchange on day 3.
Effects of Cardanol-EA on Cultured Primary Neurons
One hundred millimolar cardanol-EA in DMSO was diluted 1:5 in warm 40% 2-hydroxypropyl-β-cyclodextrin dissolved in DMSO and incubated for 10 min at 50 °C prior to serial dilution in warm neurobasal medium containing antioxidant-free B27 supplement, GlutaMAX I (2 mm), penicillin (50 units/ml), and streptomycin (50 μg·ml−1). Growth medium was exchanged for 100 μl of antioxidant-free medium containing serial dilutions of cardanol-EA or vehicle (0.6% DMSO, 0.2% 2-hydroxypropyl-β-cyclodextrin), and plates were incubated as described above for 1–2 h. Oxidative stress was induced by the addition of concentrated tBHP to achieve a final concentration of 7.5 μm. Controls were treated with an equivalent volume of PBS. After 16–18 h, the cell culture medium was replaced with 100 μl of prewarmed PBS containing 5 μg·ml−1 calcein-AM, and plates were returned to the incubator for 30 min. Cell viability was determined by measuring calcein fluorescence on a FlexStation3 plate reader (Molecular Devices, Sunnyvale, CA) at 485/525 nm excitation/emission with a 515 nm emission cut-off, subtracting the background, and normalizing to the vehicle control. Three separate experiments obtained from different cultures (different animals) of primary neurons were performed. For each condition, six replicate wells were measured, and the mean value was used for statistical analyses. Data were analyzed and plotted using Prism version 5.0 (GraphPad Software Inc., La Jolla, CA).
Treatment of Cultured Primary Neurons with FAAH Inhibitors or Substrate
The FAAH substrate, NAE 16:0 (N-palmitoylethanolamine), was emulsified in 40% 2-hydroxypropyl-β-cyclodextrin dissolved in DMSO by sonication at 50 °C to a final concentration of 50 mm. The FAAH inhibitors MAFP and URB597 were dissolved in DMSO at a concentration of 100 mm. Growth medium was exchanged for 100 μl of antioxidant-free medium or antioxidant-free medium containing vehicle or serial dilutions of substrate or inhibitor, and plates were incubated as above. After 1 h, 5 μl of 2 × 10−8 m cardanol-EA in antioxidant-free medium prediluted as above was added to the conditions indicated, and plates were gently mixed and returned to the incubator. After another 1 h, oxidative stress was induced as described above. Viability assay, replicates, and analyses were performed as described above.
RESULTS
Phenoxyacyl-Ethanolamide Synthesis
We developed a simple method that proceeds by refluxing the mixture of phenolic lipids 3-PDP (Fig. 1, 1) or cardanol (2) and methylbromoacetate in the presence of K2CO3 as a base and 2-butanone as a solvent to generate the desired PDP-methylester (3) or cardanol-methylester (4), (35, 36). Amide bond formation of methylester by chemoselective reaction with ethanolamine as N-nucleophile in dry dichloromethane and triethylamine as base yielded the desired phenoxyacyl ethanolamides (5 and 6) in good yield and purity. The products were separated by column chromatography and characterized by both NMR spectroscopy (1H and 13C NMR) and mass spectrometric analysis. 1H NMR spectra of compounds 5 and 6 (Fig. 1) showed signals at δ 7.1 ppm for -NH proton and δ 2.7 ppm for -OH proton, respectively. The exchangeable nature of these protons was identified using D2O exchange studies. High resolution mass spectral analysis of PDP-EA and cardanol-EA showed a molecular ion peak (M + H+) at m/z 406.3321 and m/z 404.3163, respectively, which exactly matches with the theoretically calculated value (PDP-EA: [M + H]+, m/z 406.3321; cardanol-EA: [M + H]+, m/z 404.3165). These new NAE-like compounds, named 3-n-pentadecylphenolethanolamide (PDP-EA, 406.3 g·mol−1) and cardanol-ethanolamide (cardanol-EA, 404.3 g·mol−1), were dissolved as 10 mm stock solutions in DMSO for assays.
FIGURE 1.
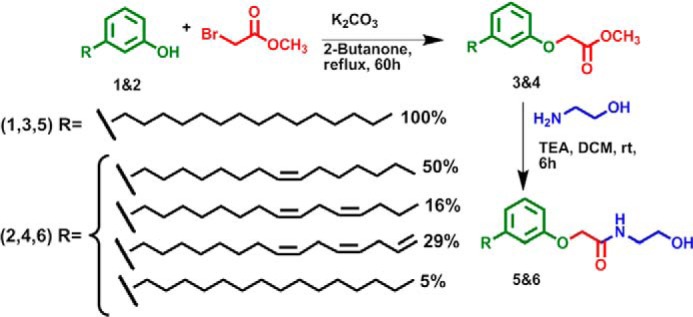
Chemical synthesis and structure of new NAE-like compounds. 1, PDP; 2, cardanol; 3, PDP-methylester; 4, cardanol-methylester; 5, PDP-EA (N-(2-hydroxyethyl)-2-(3-n-pentadecylphenoxy)acetamide); 6, cardanol-EA (mixture of phenoxyacyl-ethanolamides); R, acyl chain.
Protein Purification and Amidohydrolase Assays
Enzymatic assays were performed with two different purified recombinant proteins: At-FAAH (UniProt number Q7XJJ7) and rat FAAH (NCB accession number NP_077046) (34). Expression and purification of these proteins were monitored by SDS-PAGE and Western blotting (Fig. 2). Bands observed in the SDS-polyacrylamide gel are consistent with the molecular masses calculated for each protein plus the His tag (C terminus of the protein), which are 70 kDa for At-FAAH and 66 kDa for rat FAAH. Both proteins were also detected by Western blot using an anti-His tag monoclonal antibody with some unavoidable proteolytic degradation evident (Fig. 2, lane 5). The inclusion of serine protease inhibitors was avoided so as not to influence FAAH activity.
FIGURE 2.
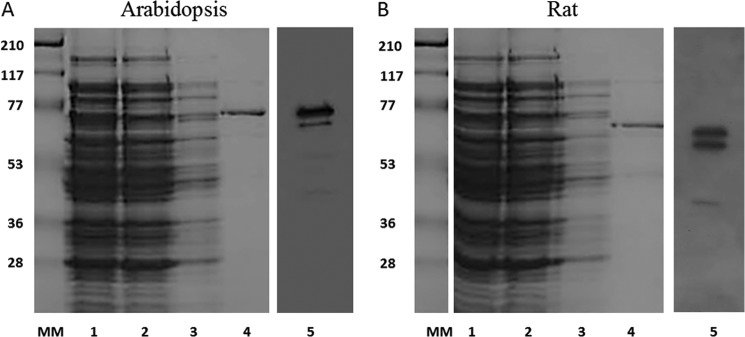
SDS-PAGE and Western blot of the enriched At-FAAH1 or rat FAAH from E. coli on a 12.5% polyacrylamide gel. A, SDS-PAGE and Western blot of different fractions of the plant FAAH protein purification; B, SDS-PAGE and Western blot of different fractions of the rat FAAH protein purification. Lane 1, supernatant (lysis); lane 2, flow-through; lane 3, wash fraction; lane 4, eluted fraction; MM, molecular marker (molecular mass represented in kDa); lane 5, Western blot (PVDF membrane) probed with mouse monoclonal anti-His antibodies. Proteins in gels were visualized by Coomassie Blue staining.
Utilization of Triton X-100 during protein extraction is mandatory to recover the activity of the rat protein (34) but not for At-FAAH. However, amidohydrolase assays performed with At-FAAH extracted in presence of 1% (v/v) Triton X-100 showed an increase by a factor of about 10 for hydrolysis of NAE to FFA compared with assays performed with protein extracted in 0.2 mm DDM (data not shown). This increase of activity could be explained by improved solubilization of the protein, the lipophilic NAE substrate, and/or FFA product (34). For consistency and optimal activity, extraction of recombinant proteins in Triton X-100 was performed for all subsequent experiments.
Enhanced NAE Amidohydrolase Activity with the Plant or Rat FAAH in the Presence of PDP-EA and Cardanol-EA
Neither of the NAE-like compounds (PDP-EA and cardanol-EA) appeared to be hydrolyzed to their respective acid forms (PDP-acid and cardanol-acid) and ethanolamine by either the plant or animal FAAH enzymes (2 μg of protein and 100–300 μm substrate) (Fig. 3). Somewhat surprisingly, compared with NAE 12:0, the phenoxyacyl-ethanolamides were not suitable substrates for FAAH, as might be anticipated from the promiscuous nature of the FAAH enzymes (3).
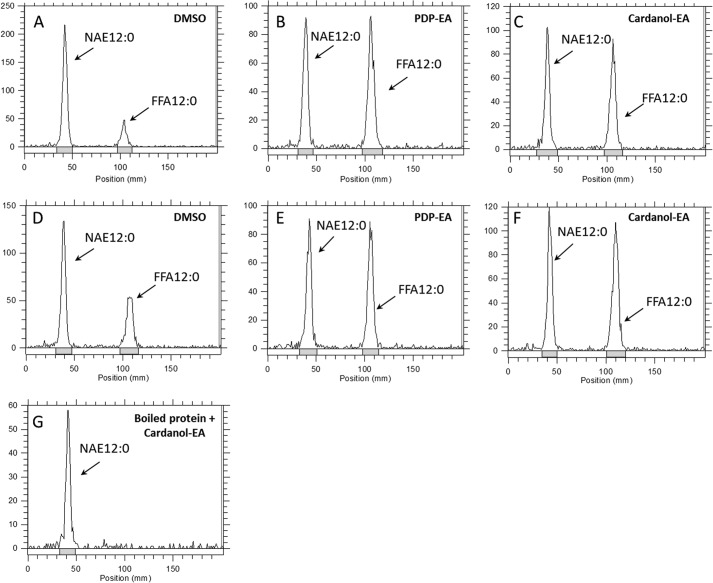
FIGURE 3.
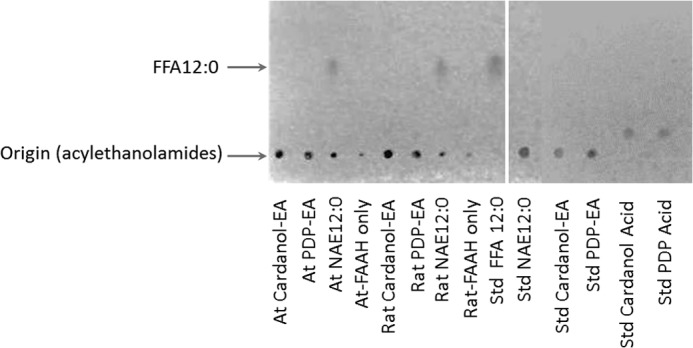
TLC analysis of the lipid composition of the amidohydrolase assay with At-FAAH or rat FAAH and different substrate. Reactions were initiated by the addition of purified At-FAAH or rat FAAH protein (2 μg) with 50 mm BisTris propane-HCl (pH 9.0), at 30 °C for 2 h, 120 rpm, and 200 μm different potential substrates in a final volume of 0.3 ml. Lipids were extracted and then separated by TLC (hexane/diethyl ether/acid acetic, 80:20:2, v/v/v). Positions of the origin (intact acylethanolamides) and the FFA 12:0 are indicated (arrows).
To test whether these phenoxyacyl-ethanolamides might serve as inhibitors of NAE hydrolysis by the FAAH enzymes, we measured the hydrolysis of 100 μm [1-14C]NAE 12:0 with or without 100 μm PDP-EA or cardanol-EA. Instead of reductions in FAAH activity as expected, the amidohydrolase activity of both FAAH enzymes toward NAE was increased in the presence of either phenoxyacyl-ethanolamide compound (Fig. 4). Non-enzymatic hydrolysis of NAE to free fatty acid by PDP-EA or cardanol-EA could be ruled out because no activity was measured in assays using heat-denatured enzyme (Fig. 4G). FAAH activities toward different NAEs, such as N-[1-14C]palmitoylethanolamine (NAE 16:0) or N-[1-14C]arachidonoylethanolamine (NAE 20:4, anandamide), showed a similar enhancement in the presence of the new NAE-like compounds (Fig. 5). An increase of activity by a factor of about 4 ± 1.2 for the recombinant At-FAAH protein was measured in the presence of either PDP-EA or cardanol-EA, by a factor of about 5 ± 2.2 for the rat FAAH protein for the unsaturated NAE (NAE 16:0 and NAE 12:0), and by up to a factor 7 ± 1.1 for the rat FAAH protein and NAE 20:4 (Fig. 5).
FIGURE 4.
Representative radiochromatograms of the total [1-14C]lipid components of following FAAH reactions in the presence (or absence) of 100 μm PDP-EA or cardanol-EA. Reactions were initiated by the addition of purified rat or At-FAAH protein (0.3 μg) with 50 mm BisTris propane-HCl (pH 9.0) and 100 μm [1-14C]NAE 12:0 in a final volume of 0.15 ml. Reactions proceeded at 30 °C with shaking (120 rpm) for 25 min. A–C, reaction with At-FAAH protein, solvent control (DMSO), with 100 μm PDP-EA, and with 100 μm cardanol-EA; D–F, reaction with rat FAAH protein, solvent control (DMSO), with 100 μm PDP-EA, and with 100 μm cardanol-EA; G, assays with heat-denatured At-FAAH protein (5 min at 100 °C) plus cardanol-EA (100 μm). Lipids were extracted and separated by TLC (60:40:5; v/v/v). Chromatograms were obtained by radiometric scanning of the TLC plate.
FIGURE 5.
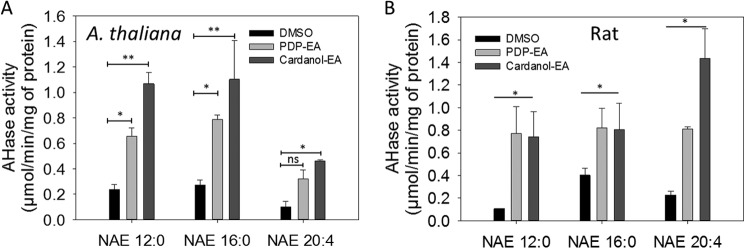
Amidohydrolase activity (μmol/min/mg of protein) of purified At-FAAH or rat FAAH protein with different NAEs in the presence of 100 μm PDP-EA or cardanol-EA. A, assays with purified At-FAAH (0.3 μg); B, assays with purified rat FAAH (0.3 μg). Reactions were carried out in 50 mm BisTris propane-HCl (pH 9.0) at 30 °C, 30 min, with shaking (120 rpm) in a final volume of 0.15 ml. Data points represent means ± S.D. (error bars) of triplicate assays. Plots were generated with SigmaPlot software version 12.0. *, p < 0.05; **, p < 0.01, as determined by Student's t test. ns, not significant.
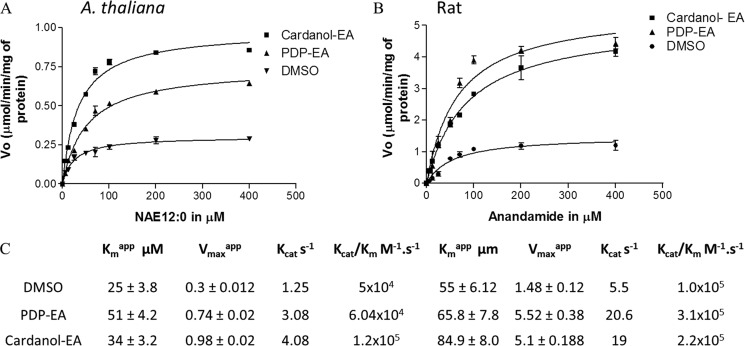
Kinetic Parameters of A. thaliana and Rat FAAH Proteins
To determine the effect of PDP-EA and cardanol-EA on the activity of the plant and rat FAAH proteins, we measured the kinetic parameters of each enzyme with the respective substrates they probably encounter endogenously (NAE 12:0 or NAE 20:4, respectively). Each enzyme exhibited typical Michaelis-Menten-type kinetics when initial velocity measurements were made at increasing concentrations of NAE 12:0 or NAE 20:4 substrates for At-FAAH or rat FAAH, respectively. Both apparent Vmax (Vmaxapp) and apparent Km (Kmapp) were calculated for each enzyme and summarized (Fig. 6). No statistical difference (t test, confidence level 95%) for the Kmapp values of the At-FAAH was observed with or without a 100 μm concentration of either PDP-EA or cardanol-EA (Fig. 6). However, catalytic efficiency (Kcat/Km) of the At-FAAH enzyme increased in the presence of both phenoxyacyl-ethanolamides particularly for the cardanol-EA compounds (1.2 × 105 m−1/s−1) compared with solvent control of 5 × 104 m−1/s−1). The Kmapp obtained in our assays for the plant FAAH was similar to Km values determined elsewhere (17.6 μm) (7).
FIGURE 6.
Kinetic characterization of At-FAAH1p and rat FAAH in the presence of 100 μm PDP-EA or cardanol-EA. Initial velocities were measured at increasing concentrations of [1-14C]NAE 12:0 for At-FAAH (A) or [1-14C]-NAE 20:4 for rat FAAH (B). Reactions were initiated by the addition of purified protein (0.3 μg). C, apparent kinetic parameters of the enzymes estimated by transformations of these original data (i.e. double-reciprocal plots). Reactions were carried out in 50 mm BisTris propane- HCl (pH 9.0) at 30 °C in a final volume of 0.15 ml. Data points represent means ± S.D. (error bars) of triplicate assays. Velocity is given in μmol/min/mg of protein. Plots were generated with Prism software version 3.0 (GraphPad Software), and data were fitted to a nonlinear regression (curve fit) using a one-site binding (hyperbola) equation with a R2 between 0.93 and 0.98.
For the rat FAAH, there was a modest increase in the Km estimated in the presence of both phenoxyacyl-ethanolamide compounds (66 ± 5.52 and 85 ± 8 μm) compared with that measured in solvent controls (55 ± 6.12 μm). Similar to At-FAAH, rat FAAH exhibited an increase in Kcat in the presence of PDP-EA and cardanol-EA, indicating an increase in turnover rate of the recombinant protein with respect to NAE. An increase by a factor of 2–3 in the ratio Kcat/Km was calculated for the rat FAAH with the phenoxyacyl-ethanolamides, suggesting an increase in the catalytic efficiency of the rat enzyme with these compounds. Although there is variation in the reported kinetic parameters for rat FAAH, those measured here were similar to those reported elsewhere (37).
Protection by Phenoxyacyl-Ethanolamides from Ethanolamine Product Inhibition for both At-FAAH and Rat FAAH
We noted a statistical dose-dependent reduction in NAE hydrolase activity at increasing ethanolamine concentrations (Fig. 7). Although enzyme regulation by product inhibition is not uncommon, feedback inhibition of FAAH by ethanolamine has not been described, neither for rat nor for At-FAAH. We demonstrated that this regulatory feature is evident for both At-FAAH and rat FAAH. Perhaps even more interestingly, both PDP-EA and cardanol-EA relieved this ethanolamine inhibition almost completely at concentrations up to 10 mm ethanolamine (Fig. 7). No dramatic change in pH of the reaction was measured following the addition of ethanolamine (up to 10 mm, pH 9.0; at 100 mm, pH 9.7; 25 °C), indicating the inhibitory effects of ethanolamine were not due to alterations in reaction pH.
FIGURE 7.
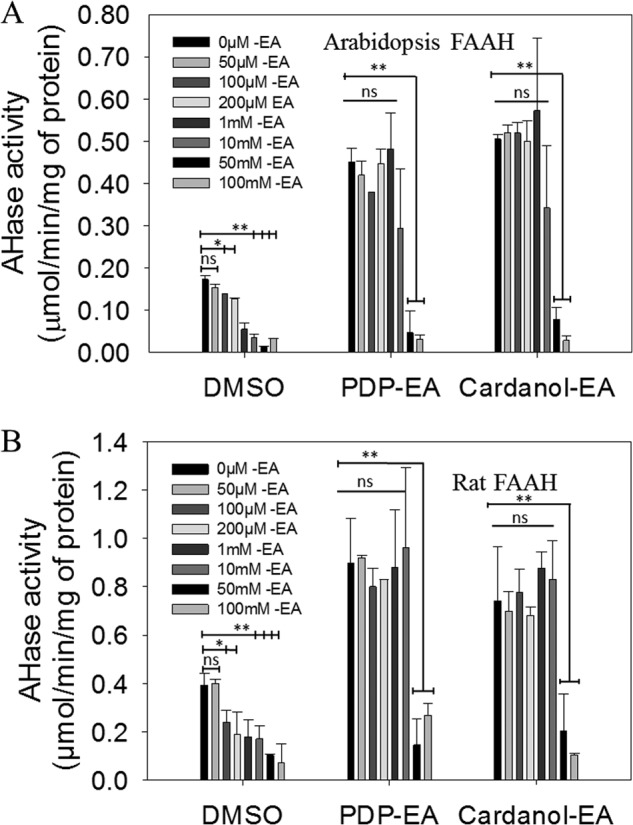
Inhibition of At-FAAH1 or rat FAAH proteins at increasing concentrations of ethanolamine and protection from inhibition by 100 μm of PDP-EA or cardanol-EA. Reactions were initiated by the addition of 0.3 μg of At-FAAH (A) or with rat FAAH (B) purified proteins. Reactions were carried out in 50 mm BisTris propane-HCl (pH 9.0) at 30 °C with 100 μm [1-14C]NAE 12:0 in a final volume of 0.15 ml. Data points represent means ± S.D. (error bars) of triplicate assays. Plots were generated with SigmaPlot software version 12.0.
Effects of PDP-EA and Cardanol-EA on Plant Growth
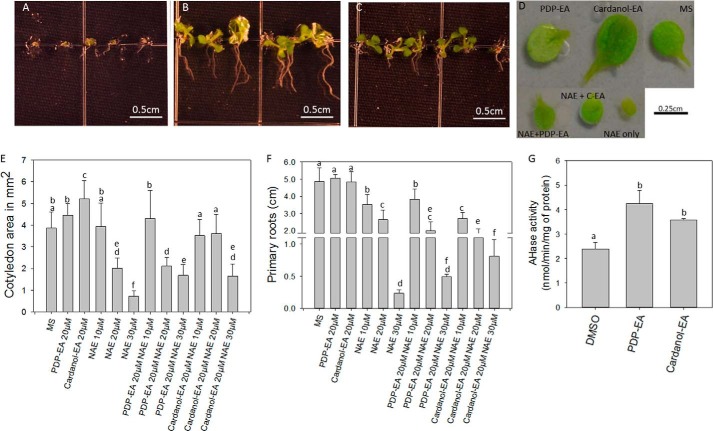
Negative effects on seedling growth by exogenous NAE 12:0 are well documented (13–15). PDP-EA and cardanol-EA were able to reverse partially these negative growth effects (Fig. 8). Representative images of Arabidopsis seedlings germinated and grown in medium containing NAE 12:0 alone or containing NAE 12:0 with PDP-EA or cardanol-EA are shown in Fig. 8 (A–C). Quantitative measurements of seedling growth (cotyledon size and primary root elongation) are summarized in Fig. 8, E and F. Despite a structure somewhat similar to that of NAE 12:0, PDP-EA alone and especially cardanol-EA alone showed a positive impact on seedling growth (cotyledon size, Fig. 8, D and E) opposite to the effects of NAE 12:0. And both compounds partially reversed the negative effects of NAE 12:0 with respect to cotyledon size. Cardanol-EA reversed negative growth effects of NAE 12:0 in primary roots. One potential explanation for these effects on seedling growth by PDP-EA and cardanol-EA is through their biochemical enhancement of At-FAAH activity in vivo. FAAH overexpression in transgenic Arabidopsis (with increased FAAH activity) conferred enhanced seedling growth as well as tolerance of the negative growth effects of NAE (13), with effects on cotyledon size and primary root length very similar to those observed here by adding these phenoxyacyl-ethanolamides to non-transgenic seedlings (Fig. 8). As expected, NAE amidohydrolase activity from crude seedling extracts showed an enhancement when assayed in the presence of PDP-EA or cardanol-EA and [1-14C]NAE 12:0 (Fig. 8G). Therefore, it is possible that exogenous application of these phenoxyacyl-ethanolamides can stimulate FAAH activity in planta and positively influences plant growth.
FIGURE 8.
Characterization of NAE sensitivity and NAE amidohydrolase activity for Arabidopsis seedlings in the presence or absence of PDP-EA or cardanol-EA. A, A. thaliana wild type (Col-0) seedlings with 30 μm NAE 12:0; B, seedlings plus 30 μm NAE 12:0 and 20 μm cardanol-EA; C, seedlings plus 30 μm NAE 12:0 and 20 μm PDP-EA; D, images of the cotyledon sizes of seedlings in presence of 20 μm phenoxyacyl-ethanolamide compounds with or without 30 μm NAE 12:0. Seedlings were grown with these different treatments for 11 days under long day conditions. E and F, sizes of the cotyledons or roots as a function of different NAE, PDP-EA, or cardanol-EA concentrations. G, FAAH activity reactions were initiated by the addition of 6 μg of crude plant protein extract from 11-day-old seedlings in 50 mm BisTris propane-HCl (pH 9.0) at 30 °C for 2 h with 200 μm [1-14C]NAE 12:0 and 200 μm NAE-like compounds in a final volume of 0.3 ml. Data points represent means ± S.D. (error bars) of triplicate assays. Plots were generated with SigmaPlot software version 12.0. N + C-EA, NAE 12:0 + cardanol-EA. Means with different letters are significantly different (p < 0.005) as determined by one-way analysis of variance with Tukey's post-test.
Effects of Cardanol-EA on Viability of Cultured Primary Neurons
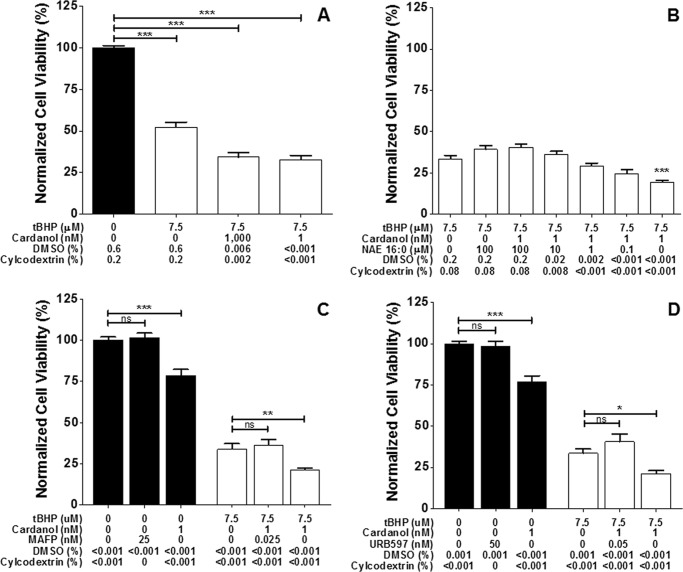
Treatment of cultured embryonic primary neurons with cardanol-EA exacerbated tBHP cell death (Fig. 9). Following overnight exposure to oxidative stress induced by treatment with 7.5 μm tBHP, neuronal cultures pretreated with 1 μm cardanol-EA contained 18% fewer viable cells than cultures pretreated with vehicle alone (Fig. 9A). Incubation of neurons with the exogenously administered FAAH substrate, NAE 16:0 (N-palmitoylethanolamine) (38) for 1 h prior to the addition of 1 nm cardanol-EA resulted in a dose-dependent reversal of cardanol-induced exacerbation of tBHP-mediated cell death (Fig. 9B). Primary neurons treated with cardanol-EA and incubated in antioxidant-free medium overnight demonstrated a significant reduction in viability compared with neurons incubated overnight in antioxidant free medium with vehicle or with the specific, irreversible FAAH inhibitors (26) MAFP and URB597 (Fig. 9, C and D, respectively). Additional experiments at different concentrations of vehicle confirmed these results (not shown). Incubation of neurons with these inhibitors for 1 h prior to the addition of cardanol-EA completely reversed cardanol-induced exacerbation of tBHP-mediated cell death, suggesting that this ex vivo effect of cardanol-EA is a result of FAAH activation.
FIGURE 9.
Cardanol pretreatment sensitizes primary cortical neurons to oxidative insult. Rat embryonic day 18 cortical neurons were pretreated for 1–2 h with cardanol prior to an overnight (16–18-h) tBHP exposure. The cardanol stock solution was prediluted 1:5 in 2-hydroxypropyl-β-cyclodextrin prior to dilution in medium to enhance delivery. Neuronal viability was measured by calcein fluorescence and normalized to the vehicle control. A, cardanol pretreatment exacerbates tBHP-mediated cell death. B, the bioactive lipid NAE 16:0 (palmitoylethanolamide, N-(2-hydroxyethyl)hexadecanamide), a substrate of FAAH, reverses this effect. Cardanol treatment significantly reduces neuronal viability in antioxidant-free medium and exacerbates tBHP-mediated cell death. C and D, preincubation with the specific, irreversible FAAH inhibitors MAFP and URB597 block this effect of cardanol. Data points represent means ± S.E. (error bars) of triplicate assays. Plots were generated with GraphPad Prism version 5.0. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001, as determined by one-way analysis of variance with Dunnett's post-test.
DISCUSSION
Despite the structural similarity between the phenoxyacyl-ethanolamide compounds synthesized for these studies (Fig. 1) and the naturally occurring NAE lipids that are present in essentially all multicellular eukaryotes, neither of the FAAH proteins (rat or Arabidopsis) were able, in our conditions, to utilize these compounds efficiently as substrates for hydrolysis (Fig. 3). This was somewhat surprising, given the broad range of acyl amides and acyl esters that can be hydrolyzed by FAAH (3). On the other hand, previous studies demonstrated that substitutions at the α-position of the acyl chain of primary amide or anandamide analogue compounds rendered the compound resistant to hydrolysis by FAAH (39, 40). The incorporation of a bulky phenoxy-group near the amide moiety may serve a similar structural hindrance to the enzyme's active site and restrict hydrolysis. However, even if these newly synthesized lipids were not used as substrates by FAAH, we speculated that these compounds would act as inhibitors toward NAE hydrolysis. Instead, a rather unique characteristic was identified for these compounds; we found that these phenoxyacyl-ethanolamides functioned to stimulate hydrolysis of NAEs by FAAH (Figs. 4–6). These compounds stimulated the activity of FAAH from both plant and mammalian sources, suggesting a more general feature of FAAH, not specific to the type of organism. There were subtle differences between plant and mammalian FAAH, such as the impact of the phenoxyacyl-ethanolamides on the affinity of the enzyme toward NAEs (raised the Km for NAE 20:4 in rat FAAH substantially in the presence of the cardanol-EA and the Km for NAE 12:0 of At-FAAH in the presence of PDP-EA). On the other hand, in both plant and mammalian FAAH, the turnover number of the enzyme was increased by the addition of the phenoxyacyl-ethanolamides (Fig. 6). Moreover, a new negative feedback property of FAAH activity by ethanolamine was discovered for both FAAH proteins, and this was prevented to a substantial degree by the addition of the phenoxyacyl-ethanolamides (Fig. 7).
The detergent Triton X-100 has been used extensively for the solubilization and to enhance the recoverable activity of recombinant rat FAAH (2, 34, 41). This non-ionic detergent probably mimics somewhat the endogenous membrane environment of FAAH and maintains the functional amidase and esterase activities of the enzyme toward lipophilic substrates. In the case of At-FAAH, Triton X-100 appeared to be better than the alkylglycoside detergent, DDM, historically used for solubilizing active At-FAAH enzyme (7). However, Km values measured here were generally similar for At-FAAH solubilized in DDM (26 ± 5.09 μm (Fig. 6) versus 13–50 μm (7)), and this procedural change allowed for more consistent comparisons between plant and animal FAAH enzymes for our comparative studies. Even so, both PDP-EA and cardanol-EA enhanced FAAH activities from both plant and animal sources (Figs. 4–6) and prevented product inhibition by ethanolamine at least up to 10 mm (Fig. 7). The increase in FAAH turnover number measured in Fig. 6 in the presence of phenoxyacyl-ethanolamides may be a direct result of prevention of product inhibition by ethanolamine, even in the case of rat FAAH, where the affinity for NAE substrate appeared to be reduced by the phenoxyacyl-ethanolamide analogues. It still remains to be clarified directly whether the effect of the phenoxyacyl-ethanolamides is via a specific binding site on FAAH or is more general in terms of influencing FAAH or substrate solubility/accessibility. PDP-EA or cardanol-EA could displace the NAE from micelles, making it more accessible to the enzyme. Alternatively, PDP-EA or cardanol-EA could influence the ratio ethanolamine/protein and alter the product inhibition of FAAH. Although unlikely, it is possible that another minor protein contaminates both the rat and At-FAAH protein preparations, influencing the results. Interestingly, there are some structural similarities between Triton X-100 and both phenoxyacyl ethanolamides. However, the rat-FAAH protein purified with a lysis buffer containing 15 mm PDP-EA or cardanol-EA instead of 15 mm Triton X-100 (0.1%) showed no recoverable activity (unlike protein purified with the original detergent; data not shown). Therefore, despite structural similarities between the phenoxyacyl ethanolamides and Triton X-100, they are insufficient to act as detergents and solubilize active FAAH on their own. On the other hand, the effects of these compounds on NAE-mediated inhibition of seedling growth or in the modulation of neuronal cell death would suggest that these compounds indeed act through a specific effect on the FAAH enzyme per se.
NAE 12:0 inhibits seedling growth when applied exogenously (see Fig. 8 and also Refs. 11, 13, and 15). Co-application of PDP-EA and especially cardanol-EA reversed these inhibitory growth effects of NAE 12:0 (Fig. 8). Similarly, the overexpression of FAAH in transgenic Arabidopsis seedlings also results in an NAE-tolerant phenotype (13). It is possible that the new phenoxyacyl-ethanolamides are able to enhance endogenous FAAH activity in wild-type seedlings, to confer some tolerance to the growth inhibition by NAE 12:0. Certainly there are other possible mechanisms by which these phenoxyacyl-ethanolamides might be acting, and this area will require further experimentation, but to date, only increased activity of FAAH has been shown to confer tolerance toward NAE 12:0, and this is consistent with the in vitro action of PDP-EA and cardanol-EA on purified recombinant FAAH enzymes (Figs. 4–7) and in crude seedling homogenates (Fig. 8G).
One intriguing aspect of these compounds is their growth-promoting properties in seedlings (Fig. 8). This is especially evident for cardanol-EA and its influence on cotyledon size (Fig. 8, D and E). This is the opposite of the effect of NAEs, which appear to retard growth and reduce cotyledon size. Besides their antagonistic effects on NAE treatment in seedlings, it seems that these compounds have their own inherent growth-regulating properties. Interestingly, seedlings overexpressing FAAH showed significant increases in cotyledon size (and other organs as well (13)), again suggesting that these phenoxyacyl-ethanolamides may be acting through modulation of endogenous FAAH activity.
The cardanol-EA-mediated reduction of cellular viability and exacerbation of oxidative stress-induced cell death in primary neurons (Fig. 9) are potentially mediated by the depletion of neuroprotective NAEs (21, 22, 42, 43) through an increase of FAAH activity (Fig. 5). The reversal of cardanol's sensitizing effect on neuronal cell death by NAE 16:0 begins well below its reported IC50 of 5.1 μm for anandamide hydrolysis by rat brain FAAH (44, 45), which may indicate that cardanol-EA does not affect binding of NAE 16:0 to rat FAAH in the same way that it does with anandamide (3.1-fold increase in Km; Fig. 6). The specificity of the pharmacological inhibitors of FAAH (26) used to reverse cardanol-induced exacerbation of tBHP-mediated cell death (Fig. 9, C and D) suggests FAAH activation as the underlying mechanism of action.
Cardanol-EA is a mixture of different phenoxyacyl-ethanolamides, all with 15 carbon alkyl chains (Fig. 1). Fifty percent of this mixture is composed of an alkyl chain with one double bond, 16% with two double bonds, 29% with three double bonds, and 5% of PDP-EA with no double bonds (saturated form). The PDP-EA compound is a single species with a 15-carbon saturated alkyl chain. There may be some slight differences in terms of action on FAAH between the two phenoxyacyl-ethanolamide preparations, but generally, the stimulatory activity was observed for both compounds. The advantage of the cardanol-EA mixture is that it is synthesized from natural materials found in many plant sources. The starting materials for these phenoxyacyl-ethanolamides are derived from cardanol, which is a major constituent of cashew nut shell waste streams, and ethanolamides synthesized from these phenolic lipids may find many applications beyond their chemical properties as lipophilic hydrocarbon polymers. Given the plant growth-promoting properties, especially of cardanol-EA, there may be agricultural applications for these compounds, or based on their action on FAAH activity, these compounds may find therapeutic applications where manipulation of localized endogenous NAE levels might be desired.
Overall, the activities of these two new NAE-like compounds (PDP-EA and cardanol-EA) open a new interesting and unexplored approach for the in situ regulation of NAE metabolism in plant and animal systems. For example, these molecules might be used as pharmacological agents to modulate appetite by decreasing the endogenous levels of acylethanolamide agonists in animal systems (39), as chemo-sensitizing agents targeted at lipid-signaling pathways affected by disease processes, or as a modulator of endocannabinoid signaling in applications ranging from cytoprotection to cellular development and excitable cell function. In plants, NAE metabolism has been shown to be associated with biotic and abiotic stresses as well as seedling and reproductive growth and development. Hence, these compounds might find applications in agriculture. Further experiments will need to be done to define the utility of these FAAH enhancers in vivo to modulate the numerous effects of NAEs in plants and animals.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NEI, Grant EY022774; NIH, NIA, Grants AG010485, AG022550, and AG027956; the NIH NCRR; and NIH, NIGMS, Grant RR027093 (to P. K.). This work was also supported by the United States Department of Energy, Office of Science, Division of Basic Energy Sciences, Grant DE-FG02-05ER15647 (to K. D. C. and E. B. B.).
- FAAH
- fatty acid amide hydrolase
- NAE
- N-acylethanolamine
- PDP
- 3-(n-pentadecyl)-phenol
- PDP-EA
- 3-n-pentadecylphenolethanolamide
- cardanol-EA
- cardanol-ethanolamide
- BisTris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- MAFP
- 5Z,8Z,11Z,14Z-eicosatetraenyl-methyl ester phosphonofluoridic acid
- URB597
- 3′-(aminocarbonyl)[1,1′-biphenyl]-3-yl)-cyclohexylcarbamate
- DDM
- N-dodecyl-β-d-maltoside
- tBHP
- tert-butyl hydroperoxide
- FFA
- free fatty acid.
REFERENCES
- 1. Chapman K. (2004) Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog. Lipid Res. 43, 302–327 [DOI] [PubMed] [Google Scholar]
- 2. Cravatt B. F., Giang D. K., Mayfield S. P., Boger D. L., Lerner R. A., Gilula N. B. (1996) Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 384, 83–87 [DOI] [PubMed] [Google Scholar]
- 3. McKinney M. K., Cravatt B. F. (2005) Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 74, 411–432 [DOI] [PubMed] [Google Scholar]
- 4. Kilaru A., Blancaflor E. B., Venables B. J., Tripathy S., Mysore K. S., Chapman K. D. (2007) The N-acylethanolamine-mediated regulatory pathway in plants. Chem. Biodivers. 4, 1933–1955 [DOI] [PubMed] [Google Scholar]
- 5. Katayama K., Ueda N., Katoh I., Yamamoto S. (1999) Equilibrium in the hydrolysis and synthesis of cannabimimetic anandamide demonstrated by a purified enzyme. Biochim. Biophys. Acta 1440, 205–214 [DOI] [PubMed] [Google Scholar]
- 6. Coulon D., Faure L., Salmon M., Wattelet V., Bessoule J. (2012) N-Acylethanolamines and related compounds: aspects of metabolism and functions. Plant Sci. 184, 129–140 [DOI] [PubMed] [Google Scholar]
- 7. Shrestha R., Dixon R. A., Chapman K. D. (2003) Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis thaliana. J. Biol. Chem. 278, 34990–34997 [DOI] [PubMed] [Google Scholar]
- 8. Wei B. Q., Mikkelsen T. S., McKinney M. K., Lander E. S., Cravatt B. F. (2006) A second fatty acid amide hydrolase with variable distribution among placental mammals. J. Biol. Chem. 281, 36569–36578 [DOI] [PubMed] [Google Scholar]
- 9. Bracey M. H., Hanson M. A., Masuda K. R., Stevens R. C., Cravatt B. F. (2002) Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science 298, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 10. Shrestha R., Kim S. C., Dyer J. M., Dixon R. A., Chapman K. D. (2006) Plant fatty acid (ethanol) amide hydrolases. Biochim. Biophys. Acta 1761, 324–334 [DOI] [PubMed] [Google Scholar]
- 11. Kim S. C., Kang L., Nagaraj S., Blancaflor E., Mysore K. S., Chapman K. D. (2009) Mutations in Arabidopsis fatty acid amide hydrolase reveal that catalytic activity influences growth but not sensitivity to abscisic acid or pathogens. J. Biol. Chem. 284, 34065–34074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blancaflor E. B., Hou G., Chapman K. D. (2003) Elevated levels of N-lauroylethanolamine, an endogenous constituent of desiccated seeds, disrupt normal root development in Arabidopsis thaliana seedlings. Planta 217, 206–217 [DOI] [PubMed] [Google Scholar]
- 13. Wang Y. S., Shrestha R., Kilaru A., Wiant W., Venables B. J., Chapman K. D., Blancaflor E. B. (2006) Manipulation of Arabidopsis fatty acid amide hydrolase expression modifies plant growth and sensitivity to N-acylethanolamines. Proc. Natl. Acad. Sci. U.S.A. 103, 12197–12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teaster N. D., Motes C. M., Tang Y., Wiant W. C., Cotter M. Q., Wang Y. S., Kilaru A., Venables B. J., Hasenstein K. H., Gonzalez G., Blancaflor E. B., Chapman K. D. (2007) N-Acylethanolamine metabolism interacts with abscisic acid signaling in Arabidopsis thaliana seedlings. Plant Cell 19, 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cotter M. Q., Teaster N. D., Blancaflor E. B., Chapman K. D. (2011) N-Acylethanolamine (NAE) inhibits growth in Arabidopsis thaliana seedlings via ABI3-dependent and -independent pathways. Plant Signal. Behav. 6, 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Teaster N. D., Keereetaweep J., Kilaru A., Wang Y. S., Tang Y., Tran C. N., Ayre B. G., Chapman K. D., Blancaflor E. B. (2012) Overexpression of fatty acid amide hydrolase induces early flowering in Arabidopsis thaliana. Front. Plant Sci. 3, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tripathy S., Venables B. J., Chapman K. D. (1999) N-Acylethanolamines in signal transduction of elicitor perception: attenuation of alkalinization response and activation of defense gene expression. Plant Physiol. 121, 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang L., Wang Y. S., Uppalapati S. R., Wang K., Tang Y., Vadapalli V., Venables B. J., Chapman K. D., Blancaflor E. B., Mysore K. S. (2008) Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 56, 336–349 [DOI] [PubMed] [Google Scholar]
- 19. Luchicchi A., Pistis M. (2012) Anandamide and 2-arachidonoylglycerol: pharmacological properties, functional features, and emerging specificities of the two major endocannabinoids. Mol. Neurobiol. 46, 374–392 [DOI] [PubMed] [Google Scholar]
- 20. Liu J., Wang L., Harvey-White J., Osei-Hyiaman D., Razdan R., Gong Q., Chan A. C., Zhou Z., Huang B. X., Kim H. Y., Kunos G. (2006) A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. U.S.A. 103, 13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duncan R. S., Chapman K. D., Koulen P. (2009) The neuroprotective properties of palmitoylethanolamine against oxidative stress in a neuronal cell line. Mol. Neurodegener. 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duncan R. S., Xin H., Goad D. L., Chapman K. D., Koulen P. (2011) Protection of neurons in the retinal ganglion cell layer against excitotoxicity by the N-acylethanolamine, N-linoleoylethanolamine. Clin. Ophthalmol. 5, 543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Devane W. A., Hanus L., Breuer A., Pertwee R. G., Stevenson L. A., Griffin G., Gibson D., Mandelbaum A., Etinger A., Mechoulam R. (1992) Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 258, 1946–1949 [DOI] [PubMed] [Google Scholar]
- 24. Di Marzo V., De Petrocellis L., Fezza F., Ligresti A., Bisogno T. (2002) Anandamide receptors. Prostaglandins Leukot. Essent. Fatty Acids 66, 377–391 [DOI] [PubMed] [Google Scholar]
- 25. Cota D., Genghini S., Pasquali R., Pagotto U. (2003) Antagonizing the cannabinoid receptor type 1: a dual way to fight obesity. J. Endocrinol. Invest. 26, 1041–1044 [DOI] [PubMed] [Google Scholar]
- 26. Cota D., Marsicano G., Lutz B., Vicennati V., Stalla G. K., Pasquali R., Pagotto U. (2003) Endogenous cannabinoid system as a modulator of food intake. Int. J. Obes. Relat. Metab. Disord. 27, 289–301 [DOI] [PubMed] [Google Scholar]
- 27. Busquets-Garcia A., Puighermanal E., Pastor A., de la Torre R., Maldonado R., Ozaita A. (2011) Differential role of anandamide and 2-arachidonoylglycerol in memory and anxiety-like responses. Biol. Psychiatry 70, 479–486 [DOI] [PubMed] [Google Scholar]
- 28. Cravatt B. F., Lichtman A. H. (2003) Fatty acid amide hydrolase. An emerging therapeutic target in the endocannabinoid system. Curr. Opin. Chem. Biol. 7, 469–475 [DOI] [PubMed] [Google Scholar]
- 29. Cravatt B. F., Demarest K., Patricelli M. P., Bracey M. H., Giang D. K., Martin B. R., Lichtman A. H. (2001) Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc. Natl. Acad. Sci. U.S.A. 98, 9371–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trox J., Vadivel V., Vetter W., Stuetz W., Scherbaum V., Gola U., Nohr D., Biesalski H. K. (2010) Bioactive compounds in cashew nut (Anacardium occidentale L.) kernels: effect of different shelling methods. J. Agric. Food Chem. 58, 5341–5346 [DOI] [PubMed] [Google Scholar]
- 31. Kozubek A., Tyman J. H. (1999) Resorcinolic lipids, the natural non-isoprenoid phenolic amphiphiles and their biological activity. Chem. Rev. 99, 1–26 [DOI] [PubMed] [Google Scholar]
- 32. Hillard C. J., Wilkison D. M., Edgemond W. S., Campbell W. B. (1995) Characterization of the kinetics and distribution of N-arachidonylethanolamine (anandamide) hydrolysis by rat brain. Biochim. Biophys. Acta 1257, 249–256 [DOI] [PubMed] [Google Scholar]
- 33. Shrestha R., Noordermeer M. A., van der Stelt M., Veldink G. A., Chapman K. D. (2002) N-Acylethanolamines are metabolized by lipoxygenase and amidohydrolase in competing pathways during cottonseed inhibition. Plant Physiol. 130, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patricelli M. P., Lashuel H. A., Giang D. K., Kelly J. W., Cravatt B. F. (1998) Comparative characterization of a wild type and transmembrane domain-deleted fatty acid amide hydrolase: identification of the transmembrane domain as a site for oligomerization. Biochemistry 37, 15177–15187 [DOI] [PubMed] [Google Scholar]
- 35. Balachandran V. S., Jadhav S. R., Pradhan P., De Carlo S., John G. (2010) Adhesive vesicles through adaptive response of a biobased surfactant. Angew. Chem. Int. Ed. Engl. 49, 9509–9512 [DOI] [PubMed] [Google Scholar]
- 36. Balachandran V. S., Jadhav S. R., Vemula P. K., John G. (2013) Recent advances in cardanol chemistry in a nutshell: from a nut to nanomaterials. Chem. Soc. Rev. 42, 427–438 [DOI] [PubMed] [Google Scholar]
- 37. Mileni M., Johnson D. S., Wang Z., Everdeen D. S., Liimatta M., Pabst B., Bhattacharya K., Nugent R. A., Kamtekar S., Cravatt B. F., Ahn K., Stevens R. C. (2008) Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc. Natl. Acad. Sci. U.S.A. 105, 12820–12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Natarajan V., Schmid P. C., Reddy P. V., Schmid H. H. (1984) Catabolism of N-acylethanolamine phospholipids by dog brain preparations. J. Neurochem. 42, 1613–1619 [DOI] [PubMed] [Google Scholar]
- 39. Boger D. L., Fecik R. A., Patterson J. E., Miyauchi H., Patricelli M. P., Cravatt B. F. (2000) Fatty acid amide hydrolase substrate specificity. Bioorg. Med. Chem. Lett. 10, 2613–2616 [DOI] [PubMed] [Google Scholar]
- 40. Lang W., Qin C., Lin S., Khanolkar A. D., Goutopoulos A., Fan P., Abouzid K., Meng Z., Biegel D., Makriyannis A. (1999) Substrate specificity and stereoselectivity of rat brain microsomal anandamide amidohydrolase. J. Med. Chem. 42, 1682. [DOI] [PubMed] [Google Scholar]
- 41. Patricelli M. P., Patterson J. E., Boger D. L., Cravatt B. F. (1998) An endogenous sleep-inducing compound is a novel competitive inhibitor of fatty acid amide hydrolase. Bioorg. Med. Chem. Lett. 8, 613–618 [DOI] [PubMed] [Google Scholar]
- 42. Garg P., Duncan R. S., Kaja S., Koulen P. (2010) Intracellular mechanisms of N-acylethanolamine-mediated neuroprotection in a rat model of stroke. Neuroscience 166, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garg P., Duncan R. S., Kaja S., Zabaneh A., Chapman K. D., Koulen P. (2011) Lauroylethanolamide and linoleoylethanolamide improve functional outcome in a rodent model for stroke. Neurosci. Lett. 492, 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jonsson K. O., Vandevoorde S., Lambert D. M., Tiger G., Fowler C. J. (2001) Effects of homologues and analogues of palmitoylethanolamide upon the inactivation of the endocannabinoid anandamide. Br. J. Pharmacol. 133, 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vandevoorde S., Saha B., Mahadevan A., Razdan R. K., Pertwee R. G., Martin B. R., Fowler C. J. (2005) Influence of the degree of unsaturation of the acyl side chain upon the interaction of analogues of 1-arachidonoylglycerol with monoacylglycerol lipase and fatty acid amide hydrolase. Biochem. Biophys. Res. Commun. 337, 104–109 [DOI] [PubMed] [Google Scholar]