Background: Endothelial cell tumors are the most common soft tissue tumor in infants.
Results: Dicer knockdown up-regulated miR-21a-3p that targeted nox-4 mRNA preventing endothelial cell tumor formation in vivo.
Conclusion: Nox-4 silencing inhibits endothelial cell tumor formation. Dicer knockdown up-regulates miR 21a-3p, which targets the Nox-4 3′-UTR.
Significance: Novel drivers of endothelial cell tumor formation are reported.
Keywords: Dicer, Endothelial Cell, MicroRNA, NADPH Oxidase, Tumor, Hemangioendothelioma, miR-21a-3p, Monocyte Chemoattractant Protein-1
Abstract
MicroRNAs (miR) are emerging as biomarkers and potential therapeutic targets in tumor management. Endothelial cell tumors are the most common soft tissue tumors in infants, yet little is known about the significance of miR in regulating their growth. A validated mouse endothelial cell (EOMA) tumor model was used to demonstrate that post-transcriptional gene silencing of dicer, the enzyme that converts pre-miR to mature miR, can prevent tumor formation in vivo. Tumors were formed in eight of eight mice injected with EOMA cells transfected with control shRNA but formed in only four of ten mice injected with EOMA cells transfected with dicer shRNA. Tumors that formed in the dicer shRNA group were significantly smaller than tumors in the control group. This response to dicer knockdown was mediated by up-regulated miR 21a-3p activity targeting the nox-4 3′-UTR. EOMA cells were transfected with miR 21a-3p mimic and luciferase reporter plasmids containing either intact nox-4 3′-UTR or with mutation of the proposed 3′-UTR miR21a-3p binding sites. Mean luciferase activity was decreased by 85% in the intact compared with the site mutated vectors (p < 0.01). Attenuated Nox-4 activity resulted in decreased cellular hydrogen peroxide production and decreased production of oxidant-inducible monocyte chemoattractant protein-1, which we have previously shown to be critically required for endothelial cell tumor formation. These findings provide the first evidence establishing the significance of dicer and microRNA in promoting endothelial cell tumor growth in vivo.
Introduction
Endothelial cell tumors are the most common soft tissue tumor in infants and affect 5–10% of all newborns in the United States and Europe (1–3). The majority of these tumors occur in the head and neck area causing significant visible deformity. Some (10%) of these tumors are known to threaten the development of vital structures, and 1% of all tumors are known to threaten the life of the child (4). A validated mouse model has been developed to identify mechanisms that regulate the growth of hemangioendothelioma, an endothelial cell tumor that occurs in children (5, 6). Murine endothelial (EOMA) cells derived from a spontaneously arising hemangioendothelioma (HE)2 in 129 P/3 mice can be grown in vitro and injected subcutaneously into mice to form HE in vivo. Using this approach, we have previously demonstrated that oxidant-inducible expression of monocyte chemoattractant protein-1 (MCP-1) and macrophage recruitment is required for HE growth (7). Our work has recognized that the primary source of reactive oxygen species in endothelial cells is NADPH oxidase (8). The predominant isoform of the gp91 catalytic subunit of NADPH oxidase present in tumor forming EOMA cells is Nox-4. Abundance of the nox-4 gene, which is constitutively expressed, is elevated 69-fold in EOMA cells compared with non-tumor forming murine aortic endothelial cells (8). When nox-4 transcription is inhibited using post-translational gene silencing techniques or oxidant function is reduced through in vitro or oral administration of antioxidants, HE growth is significantly inhibited (7, 9). Therefore, the Nox-4/MCP-1 axis represents a rational therapeutic target for inhibiting HE growth in vivo.
Although endothelial cell tumors are the most common soft tissue tumors in infants, little is known about the significance of miR in regulating their growth. The objective of this work was to determine how microRNA (miRNA or miR) regulate the Nox-4/MCP-1 pathway that is critical to endothelial cell tumor proliferation. Disruption of miR homeostasis in the tumor cells was achieved by post-transcriptional gene silencing of dicer, a protein that regulates miR biogenesis by converting preliminary miRNA (pre-miR) to mature miRNA capable of binding a target mRNA sequence. Dicer protein and mRNA levels and disease progression have been shown to positively correlate with some tumor types and negatively with other tumors (10–12). Thus, the expression of Dicer and its effects on tumor progression are specific to the cell type and context of miR expression. Of note, dicer expression levels and the contribution of miR to endothelial cell tumor progression have not been previously reported. Post-transcriptional gene silencing of dicer in EOMA cells was performed to test the significance of Dicer in endothelial cell tumor formation in vivo and to determine how changes in dicer levels may affect the Nox-4/MCP-1 axis that is critically required for HE growth.
MATERIALS AND METHODS
The following materials were obtained from the source indicated: hexadimethrine bromide, dimethyl sulfoxide, actinomycin D, and puromycin (Sigma); for cell culture, DMEM, FCS, and penicillin and streptomycin were purchased from Invitrogen. Culture dishes were obtained from Nunc (Rochester, NY).
Cell Culture
Murine endothelial (EOMA) cells were maintained under the same conditions as previously described (13). In brief, EOMA cells were maintained in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin (complete media) and incubated at 37 °C and 5% CO2.
Stable Cell Lines
For generating stable cell lines, EOMA cells were stably transfected with control scrambled shRNA or 58-bp dicer lentiviral shRNA particles (Sigma) (5′-CCGGGCCTCACTTGACCTGAAGTATCTCGAGATACTTCAGCGTCAAGTGAGGCTTTTTG-3′). EOMA cells were plated in 96-well plate (6.0 × 103 well) 24 h prior to infection including hexadimethrine bromide (8 μg/ml). Viruses were added at 10 multiplicity of infection to each well. After 24 h, puromycin (10 mg/ml) was added to cell culture media according to the manufacturer's protocol to select stably transfected clones. Single virus clones were selected to generate monoclonal cell lines. Media with selective antibiotic was replaced at 48-h intervals until cell harvest. Effective knockdown of dicer was confirmed by real time PCR and Western blot.
In Vitro Transfection of miR Mimic or siRNA
EOMA cells (0.1 × 106 cells/well in 12-well plate) were seeded in antibiotic free medium for 18–24 h prior to transfection. DharmaFECTTM 1 transfection reagent was used to transfect cells with miRIDIAN mmu-miR-21a-3p mimic (50 nm), mmu-miR-21a-3p inhibitor (100 nm), or dicer siRNA (100 nm; Thermo Scientific Dharmacon RNA Technologies, Lafayette, CO) per the manufacturer's instructions (14–17). miRIDIAN miR mimic/inhibitor negative controls or siControl nontargeting siRNA pool (Thermo Scientific Dharmacon RNA Technologies) were used for control transfections. Samples were collected after 72 h of miR mimic or siRNA transfection for quantification of miRNA, mRNA, or protein expression.
RNA Extraction and Quantitative Real Time PCR
For detection of mRNA and miRNA, total RNA from tissue and cells was extracted using miRVana miRNA isolation kit according to the manufacturer's protocol (Ambion/Invitrogen) (15, 16). For pre-miRNA isolation, RNA were collected with two column filtration procedure using miRVana miRNA isolation kit according to the manufacturer's protocol (18). Pri and mature miR expression was determined by miR TaqMan assays and TaqMan microRNA reverse transcription kit, followed by quantitative real time PCR using Universal PCR Master Mix (Applied Biosystems/Invitrogen). For mRNA expression studies, cDNA synthesis was achieved by SuperScriptTM III first strand synthesis system (Applied Biosystems/Invitrogen). The abundance of mRNA for genes of interest was quantified by using real time PCR with double-stranded DNA binding dye SYBR green-I (15, 16). The primer sets for JE/MCP-1, nox-1–4, duox1, and duox2 were the same sets as previously reported (8). Additional custom primer sets (Invitrogen) used for these experiments included: M_18s rRNA F, 5′-GTA ACC CGT TGA ACC CCA TT-3′; M_18s rRNA R, 5′-CCA TCC AAT CGG TAG TAG CG-3′; M_DICER F, ACA CGC TCT GGA GAG GTC ACC ATA T-3′; M_DICER R, TCC AAA GTG CCG GAG TCA TTA A-3′; M_Pre-miR-21a-3p F, TGT ACC ACC TTG TCG GAT AGC T; and M_ Pre-miR-21a-3p R, GAT ACC AAA ATG TCA GAC AGC C.
pGL3-NOX-4′-UTR Luciferase Reporter Assay
EOMA cells (0.075 × 106 cells/well in 12-well plate) were seeded in antibiotic free medium for 18–24 h prior to transfection. Cells were transfected as discussed earlier with control and miR-21a-3p mimic for 72 h. After 72 h, cells were transfected with pGL3-nox-4-3′-UTR firefly luciferase expression construct (500 ng/sample; Signosis, Sunnyvale, CA) or construct carrying the mutation of the predicted miR-21a-3p binding site in pGL3-nox-4-3′-UTR (500 ng/sample; Signosis) together with Renilla luciferase pRL-cmv expression construct (10 ng/sample) using LipofectamineTM LTX PLUSTM reagent (Invitrogen). The constructs were designed based on the sequence of miR-21a-3p binding sites, and a total of 959 bp was cloned in the 3′-UTR of constitutively active firefly luciferase construct. Mutation was made in the predicted miR-21a-3p binding site. The cell lysates were assayed with dual luciferase reporter assay kit (Promega, Madison, WI). The data are presented as ratio of firefly to Renilla luciferase activity.
Western Blot Analysis
Immunoblotting was performed using EOMA cell lysates. After protein extraction, the protein concentration was determined by BCA protein assay. The samples (25–40 μg of protein/lane) were separated on a 4–12% SDS-polyacrylamide gel electrophoresis and probed with rabbit polyclonal anti-Nox-4 antibody (1:1000 dilution; Epitomics, Burlingame, CA), rabbit polyclonal anti-dicer 1 antibody (1:400 dilution; Abcam, Cambridge, MA), lamin A (1:500 dilution; Sigma), and anti-mouse β-actin (1:10,000 dilution; Sigma). Bands were visualized by horseradish peroxidase-conjugated anti-rabbit IgG raised in donkey and horseradish peroxidase-conjugated anti-mouse IgG raised in sheep (Amersham Biosciences) at 1:2,000 dilution and the enhanced chemiluminescence assay (Amersham Biosciences), according to the manufacturer's instructions.
Capillary Electrophoresis Immunoassay
Capillary electrophoresis immunoassay or SimpleWestern analyses were performed using the SimonTM machine (ProteinSimple, Santa Clara, CA) according to the manufacturer's protocol. In brief, 7.5-μg samples were mixed with a master mix (ProteinSimple) to a final concentration of 1× sample buffer, 1× fluorescent molecular weight markers, and 40 mm DTT and then heated at 95 °C for 5 min. The samples, blocking reagent, primary antibodies, HRP-conjugated secondary antibodies, chemiluminescent substrate, and separation and stacking matrices were also dispensed to designated wells in a 384-well plate. After plate loading, the separation electrophoresis and immunodetection steps took place in the capillary system and were fully automated. Simple Western analysis is carried out at room temperature, and instrument default settings were used. Nox-4 (1:50 dilution), Lamin A (1:50 dilution), and β-actin (1:100 dilution) primary antibody were diluted with antibody diluent (ProteinSimple). The digital image was analyzed with Compass software (ProteinSimple), and the quantified data of the detected protein were reported as molecular weight (19, 20).
Determination of Intracellular Peroxides
Intracellular peroxides were detected using dichlorodihydrofluorescein diacetate as described previously (7). Control and miR-21a-3p mimic transfected cells were washed three times with PBS. Cells were detached from monolayer using trypsin and centrifuged (600 × g for 5 min). Next, the cells were washed with PBS and centrifuged, then resuspended in PBS, and incubated with dichlorodihydrofluorescein diacetate (40 μm) for 30 min at 37 °C. 5-(and-6)-Carboxy-2′,7′ dichlorofluorescein diacetate oxidation was detected using an Accuri C6 Flow Cytometer (Accuri, Ann Arbor, MI) at 530-nm excitation with a gated sample size of 10,000 cells. For hydrogen peroxide detection, Amplex Red assay (Invitrogen) was used according to the manufacturer's instructions (Invitrogen) as described previously (7). Spectrophotometric readings were obtained at 530-nm excitation and 590-nm fluorescence detection using a Bio-TEK ELX 808IU micro plate reader (Bio-TEK Instruments).
In Vitro Angiogenesis Assay
Four-well plates were coated with 100 μl of Matrigel® (Cultrex® Basement membrane extract reduced growth factor; R&D Systems, Minneapolis, MN) and let to solidify for 30 min at 37 °C. Next, stably transfected control and dicer shRNA cells were seeded (3 × 104 cells/well) on top of the solidified Matrigel® and maintained in a cell culture incubator. After 24 h of cell seeding on Matrigel, cells were stained with 3 μm calcein-AM (Invitrogen) 20–30 min at 37 °C and 5% CO2. Endothelial tube formation was observed and digitally photographed under an inverted light microscope at 5× magnification (Axiovert 200M; Zeiss, Oberkochen, Germany). Tube formation was quantified by AxioVisionRel 4.8 software (Zeiss) to measure total area within the tubes (17).
Cell Migration Assay
For migration assay, control shRNA and dicer shRNA lentiviral particle stable infected EOMA cells (3 × 105 cells/0.6 ml of complete medium) were seeded in the lower chamber of the 24-well plates, incubated for 8 h at 37 °C and 5% CO2 to allow adherence. After 8 h, the medium was changed to low serum medium (0.6 ml). Equilibrated Transwell inserts (8-μm pore size; Costar, Corning, NY) were placed, and RAW cells were seeded (5 × 105 cells/0.1 ml of low serum medium) on upper chamber, and cells were kept in cell culture incubator for 5 h (8). After 5 h, cells were fixed using 10% formalin at room temperature for 10 min and stained with DAPI (1:10,000; Vector Laboratories, Burlingame, CA) at room temperature for 10 min. After staining cells on the upper surface of the Transwell membrane were removed by rubbing with a sterile cotton swab, and cells on the lower surface were visualized by microscopy. Cell migration was measured by AxioVisionRel 4.8 software (Zeiss) for taking the pictures and counted cell number for cell migration rates.
MCP-1 ELISA
EOMA cells (0.1 × 106 cells) were seeded in 12-well plates. MCP-1 level in the medium was determined using commercially available ELISA kit (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Absorbance was detected at 450 nm. BCA protein assay was performed on an aliquot of all tested samples, and the results were standardized per milligram of protein.
Immunocytochemistry
Cells (0.1 × 106 cells/well) were seeded on a coverslip placed in 12-well plates for 24 h and then transfected with control and miR21a-3p mimic for 72 h. After 72 h of transfection cells were washed with PBS three times and then fixed in 10% buffered formalin for 20 min. Next, the cells were washed three times with PBS followed by permeabilization using 0.1% Triton X-100/PBS for 15 min. Next, the cells were washed three times with PBS, incubated with 10% goat serum (Vector Laboratories) for 1 h at room temperature, and incubated with Nox-4 antibody (1:100; Epitomics, Burlingame, CA) overnight at 4 °C. The cells were washed with PBS three times and incubated with an Alexa Flour 488 goat anti-rabbit antibody (1:200 dilution; Invitrogen) for 1 h at room temperature. Cells were counterstained with rhodamine phalloidin (1:250 dilution; Invitrogen) for 20 min and mounted in gel mount with DAPI (aqueous mount; Vector Laboratories). The images were captured by confocal microscope, and quantification of fluorescent intensity of image was measured using FV10-ASW 3.0 software (Olympus, Tokyo, Japan).
Immunoprecipitation and Total RNA Extraction
EOMA cells were seeded in a 12-well plate at a density of 0.1 × 106 cells/well 24 h before treatment. Cells were transfected as described above with control and dicer siRNA for 72 h. After 72 h, cells (3 wells/sample) were washed with ice-cold phosphate-buffered saline (pH 7.4) and lysed with lysis buffer (150 mm KCl, 25 mm Tris-HCl, 5 mm EDTA, 0.5% IgePal, 1 mm PMSF, 1× protease inhibitor) as described previously (21). Protein concentration was determined using the BCA protein assay kit (Pierce). TrueBlot anti-rabbit Ig IP beads (eBiosciences, Inc., San Diego, CA) were prewashed with lysis buffer for 40 min. Cell lysates (500 μg) were incubated with prewashed beads for 1 h at 4 °C on a rotisserie shaker (Barnstead/Thermolyne, Dubuque, IA). 5–10 μg of anti-human Ago2 antibody (Abcam) were added with the lysate and incubated overnight in a rotisserie shaker at 4 °C. The beads were then washed three times with ice-cold lysis buffer (centrifugation at 2500 × g at 4 °C for 5 min) and separated into two portions: one portion for RNA isolation to identify miRNA target genes and another portion for Western blotting to check for successful IP of Ago2. Immunoprecipitated complexes were washed three times with lysis buffer (centrifugation at 2500 × g at 4 °C for 5 min). For Western blot, samples were subjected to SDS/PAGE after reduction with 1 m DTT (21, 22). RNA were isolated using TRIzol and glycogen (Invitrogen) (21).
In Vivo Endothelial Cell Tumor Studies
All animal protocols were approved by the Institutional Animal Care and Use Committee of the Ohio State University (Columbus, OH). The mice were maintained under standard conditions at 22 ± 2 °C with 12:12 dark:light cycles with access to food and water ad libitum. 129P/3 mice (6–8 weeks, female; Jackson Laboratories, Bar Harbor, ME) were subcutaneous injected with EOMA cells, as previously described (13). Tumor volume was determined using calipers to measure length, width, and height of each tumor. Tumor mass was determined by draining the blood from the tumor and weighing the residual solid tumor mass dissected free from any surrounding soft tissue as previously described (13). Mice were killed on days 4 and 7 after the injection, and tumor tissue was either embedded in OCT compound and frozen for histological analyses or snap frozen in liquid nitrogen for RNA isolation.
Statistical Methods
At least three independent replicates were conducted for all experiments. Two-sided two-sample t test was used to compare the difference between two groups, and analysis of variance for comparison among more than three groups with Tukey's adjustment for the multiple pairwise comparisons among groups. Nonparametric procedures were used when normality assumptions of the data were violated even after proper data transformation. A p value of ≤0.05 was considered statistically significant.
RESULTS
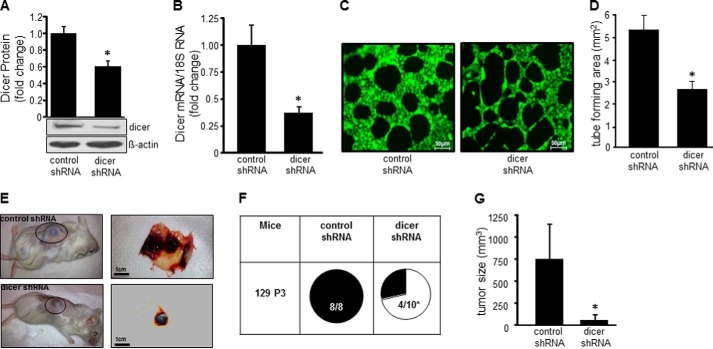
To determine whether microRNA biogenesis plays a significant role in endothelial cell tumor development, stable transduction of EOMA cells with dicer shRNA was performed. Western blot confirmed successful knockdown as manifested by significantly decreased Dicer protein and gene expression (Fig. 1, A and B). A MatrigelTM assay was performed to test the potential anti-angiogenic effects of Dicer knockdown on EOMA cells. There was a significant decrease in EOMA cell tube forming capacity in dicer shRNA transfected cells compared with control shRNA transfected cells (Fig. 1, C and D). Endothelial cell tumor formation following subcutaneous injection of EOMA cells is a validated in vivo model of angiogenesis, because it requires tumor forming endothelial cells to establish a connection with host vasculature to develop into a tumor that is perfused and filled with blood. When EOMA cells were injected into syngeneic 129 P/3 mice, HE tumor formation occurred in all mice receiving control transfected EOMA cells, but only in 4 of 10 mice receiving dicer shRNA transfected EOMA cells (Fig. 1E). These mice were euthanized, and HE were collected after 7 days because that is the time when the mice will start to succumb to high output cardiac failure from platelet and red blood cell sequestration within the tumor (Kassabach-Merritt phenomenon). The HE that did develop from the dicer shRNA transfected cells were significantly smaller in size compared with the tumors observed in the control transfection group (Fig. 1F). Thus, dicer and its essential function in miR biogenesis play a significant role in promoting HE formation in vivo.
FIGURE 1.
MicroRNA biogenesis is essential for endothelial cell tumor formation. EOMA cell stable transfectants were generated using lentiviral shRNA delivery. A and B, targeted post-transcriptional gene silencing of dicer was confirmed by Western blot (A) and real time PCR (B). C and D, Matrigel angiogenesis assay was used to evaluate the functional effects of dicer knockdown in vitro. Cells were stained with calcein-AM (C), and the area within the formed tubes was quantitated by analyzing three high powered fields per well using AxioVision Rel 4.8 software (D). Matrigel tube formation was compromised in dicer knockdown EOMA cells. E, reduced tumor size in response to dicer knockdown. F, incidence of tumor formation in mice injected with EOMA cells transfected with controls shRNA or dicer shRNA. G, tumor volume as quantified using calipers (length × width × height). The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
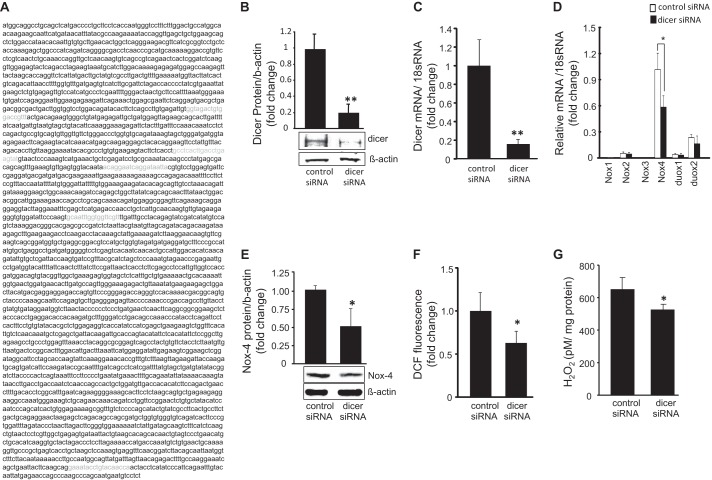
In vitro experiments aimed at testing whether the effects of Dicer on HE growth were mediated through the Nox-4/MCP-1 axis were conducted using a siRNA-dependent dicer knockdown approach. The murine dicer mRNA sequence (NCBI reference sequence NM_148948.2) from nucleic acids 280–3959 is shown and binding sites of the dicer siRNA (amino acids 1021–1029, 1457–1475, 1841–1859, and 3870–3888) and dicer shRNA (amino acids 1345–1356) were identified and confirmed not to target the same mRNA sequence to eliminate the possibility that off target effects were responsible for the observed results (Fig. 2A). Successful post-transcriptional gene silencing of dicer following siRNA transfection was tested by Western blot and real time PCR (Fig. 2, B and C). NADPH oxidase is known to be the primary source of ROS in endothelial cells, and gp91 is the catalytic subunit in NADPH oxidase responsible for ROS production. To determine whether Dicer knockdown affected NADPH oxidase expression, real time PCR was performed on all known isoforms of gp91 observed in mice. Nox-4 was the predominant isoform detected in EOMA cells, and it was significantly inhibited by Dicer knockdown, whereas all other isoforms were represented by trivial levels of expression and were not affected by dicer knockdown (Fig. 2D). Nox-4 Western blot confirmed the real time PCR results demonstrating a significant decrease in Nox-4 protein levels in response to Dicer knockdown (Fig. 2E). To determine whether there was a loss of Nox-4 function after Dicer knockdown, ROS production was measured. Dicer knockdown significantly decreased ROS production (Fig. 2F). The bioactive form of ROS produced by NADPH oxidase in EOMA cells is H2O2. H2O2 levels were significantly lower in response to dicer knockdown (Fig. 2G).
FIGURE 2.
Dicer knockdown attenuated Nox-4 activity in tumor forming EOMA cells. A, murine dicer sequence identification of dicer siRNA binding sites in green and shRNA binding site in yellow confirm distinct binding sites within dicer mRNA. B and C, transfection with dicer siRNA decreased dicer protein (B) and mRNA (C) expression. D, mRNA expression of gp91 isoforms present in EOMA cells. Only Nox-4 was present at significant levels and was affected significantly by dicer knockdown. E, dicer knockdown lowered Nox-4 protein expression. F and G, the effect of dicer knockdown on Nox-4 function was measured using flow cytometry (F) to detect oxidation of 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (DCF) and by the Amplex Red (Invitrogen) assay to measure H2O2 production (G). The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
In EOMA cells, Dicer knockdown limited both MCP-1 protein, as well as gene expression (Fig. 3, A and B). The functional activity of MCP-1 was tested using a Transwell assay in which murine RAW macrophages were placed in the upper chamber and EOMA cells transfected with either control or dicer siRNA were placed in the lower chamber. Dicer knockdown in EOMA cells resulted in a significant decrease in RAW cell migration toward EOMA cells (Fig. 3, C and D).
FIGURE 3.
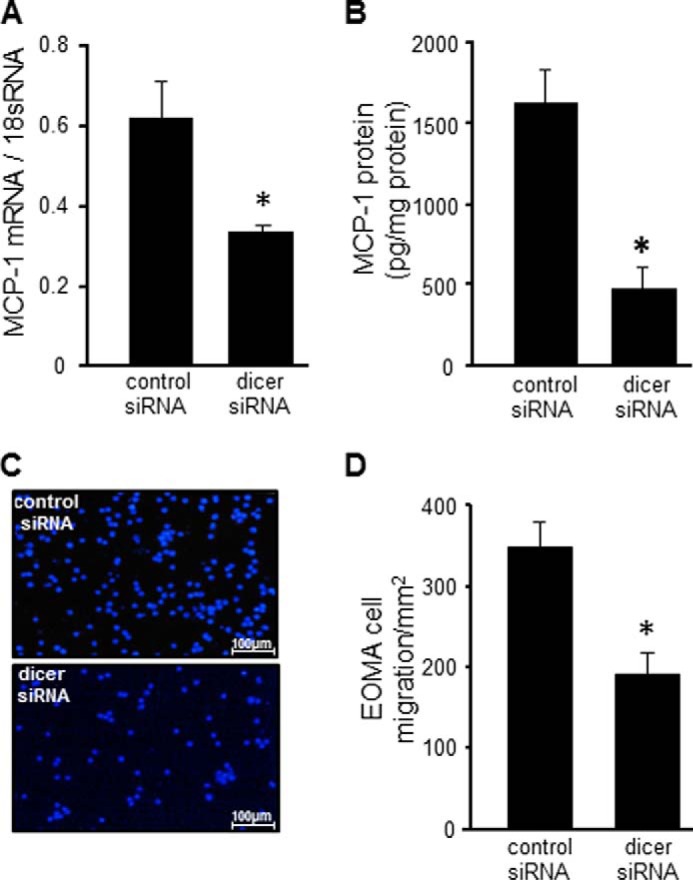
Dicer knockdown compromised MCP-1 protein expression and function. A and B, effect of dicer knockdown on MCP-1 mRNA expression (A) and MCP-1 protein expression (B). C, loss of MCP-1 function was confirmed using a Transwell migration assay. Lower chambers contained EOMA cells transduced with either control shRNA or dicer shRNA, and upper wells contained RAW cells, a murine macrophage cell line. D, migrated cells were fixed with paraformaldehyde and stained with DAPI, and the numbers of migrating cells were scored. Cells were counted using total cells from each image divided with total area (2.4 mm2) to obtain final migrated EOMA cells/mm2. The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
The observation that Dicer knockdown in EOMA cells resulted in attenuated Nox-4-derived cellular H2O2 production, MCP-1 expression, and blunted angiogenic property of EOMA cells in vitro can be explained by two possible mechanisms. First, compromised miR maturation resulted in desilencing of target protein(s), which in turn repressed Nox-4 expression. In this case, the effect of dicer on Nox-4 would therefore be indirect. However, Nox-4 is constitutively expressed, and endogenous proteins that repress Nox-4 in endothelial cells have not yet been reported. Second, perturbation in mature miR homeostasis as caused by dicer knockdown resulted in transient increase in abundance of a specific miR that targeted nox-4, providing a direct mechanism to inhibit HE formation, and this was the line of investigation that was pursued.
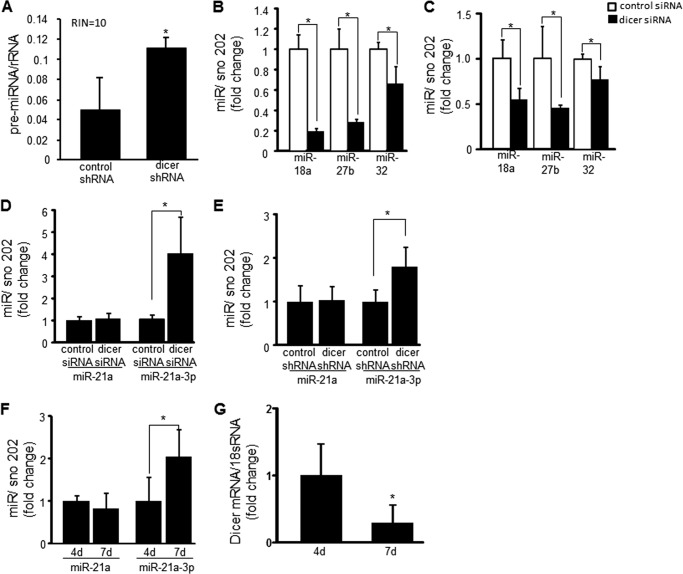
Dicer functions by cleaving preliminary miRNA (pre-miRNA) sequences into mature miR that are 20–24 nucleotides in length. To confirm that Dicer cleavage of pre-miRNA was decreased, pre-miRNA of 50–100 nucleotides in length were quantified. RNA integrity was also qualitatively assessed to rule out the option that the 50–100-nucleotide RNA fragments were merely products of RNA degradation. A significant increase in the accumulation of RNA of 50–100 nucleotides in length was observed after dicer shRNA transfection compared with control, and the RNA integrity was excellent (Fig. 4A). Although 18 and 26 S RNA are also in the size range for pre-miRNA, they have separately identifiable peaks that allow them to be excluded when calculating the pre-miRNA concentration. We sought to quantitate the amount of RNA of 20–40 nucleotides in length, because mature miR are 20–24 nucleotides in length. However, the amount of RNA of that size fell below the 50 pg/μl threshold level of detection for the Bioanalyzer 2100. Therefore, real time PCR was used to demonstrate a quantitative decrease in miR-18a, 27b, and 32, because they are known to promote angiogenesis in endothelial cells (23–25). There was a significant decrease in the amount of the above-mentioned mature miR after Dicer knockdown in vitro (Fig. 4B) and in vivo from HE tumors generated using EOMA transfected with Dicer shRNA (Fig. 4C), confirming the loss of Dicer functional activity in EOMA cells both in vitro and in vivo.
FIGURE 4.
Dicer knockdown compromised processing of pre-miR to mature miR, resulting in decreased abundance of angiogenic miRs 18a, 27b, and 32 but increased abundance of miR-21a-3p. A, the amount of RNA 50–100 nucleotides in size, consistent with the size of pre-miR, was measured in response to dicer knockdown. Prior to analysis, the RNA integrity (RIN) of the samples was analyzed on a scale of 1–10 (where 10 is the highest integrity) to ensure that fragments were not derived from sample degradation. Dicer knockdown resulted in the accumulation of RNA fragments the size of pre-miR. B and C, real time PCR analyses showed that dicer knockdown in EOMA cells resulted in decreased processing of pre-miR to mature miR as shown with miR18a, miR27b, and miR32 both in vitro (B) and in vivo (C) from HE tumor extracts generated using EOMA transfected with dicer shRNA. D and E, dicer knockdown did not alter cellular levels of miR21a but did increase miR21a-3p in dicer siRNA transfected (D) and dicer shRNA transfected cells (E). F and G, HE specimens showed increased miR-21a-3p expression between days 4 and 7 after injection with untreated EOMA cells with a corresponding decrease in dicer mRNA levels confirming in vitro findings. The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
miR-21 was selected as a potential candidate for increased expression in EOMA cells after Dicer knockdown as increased abundance of miR-21 has been observed in many cancers (26–29). Real time PCR was used to measure miR-21a levels in EOMA cells, because this represents the murine homolog for human miR-21. The classic miR-21a target strand was identified, but it did not vary in response to Dicer knockdown. When we screened for the passenger strand (miR-21a-3p) using real time PCR, it was noted that miR-21a-3p expression increased after dicer knockdown using both siRNA and shRNA techniques (Fig. 4, D and E). The increase in miR21a-3p expression in response to Dicer knockdown, coupled with decreased levels of other miRs, suggests that miR-21a-3p is preferentially processed when Dicer levels are low. To confirm the validity of this in vitro observation, real time PCR was used to measure the levels of miR-21a and miR-21a-3p in homogenized HE tumor specimens collected 4 and 7 days after injection of untreated EOMA cells. There was no change in miR-21a levels in HE over time, but there was a significant increase in miR-21a-3p levels from days 4 to 7 (Fig. 4F), and there was a corresponding decrease in the level of dicer mRNA expression observed in HE between days 4 and 7 (Fig. 4G).
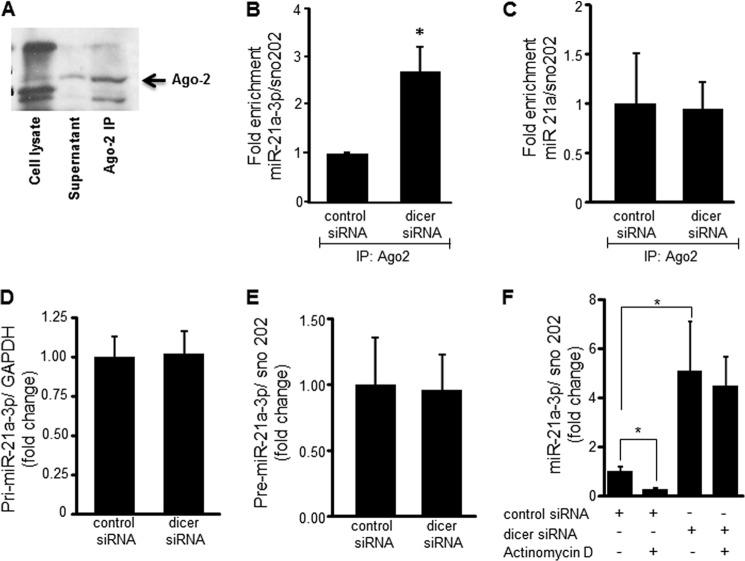
For mature miR to bind to the target mRNA sequence, it must be loaded into the RISC complex and specifically onto the catalytic protein, Argonaute-2 (Ago-2). The functional readiness of miR-21a-3p in EOMA cells was analyzed by performing Ago-2 immunoprecipitation and measuring the amount of Ago-2 bound miR-21a-3p that was poised to bind target mRNA. The immunoblot in Fig. 5A shows that immunoprecipitation was able to specifically isolate Ago-2 protein from EOMA cell lysate. Dicer knockdown resulted in significantly increased binding of miR-21a-3p to Ago-2 (Fig. 5B), but there was no change in miR-21a binding to Ago-2 (Fig. 5C). This confirmed the specificity of miR-21a-3p as mediating the observed effects of Dicer knockdown and indicated enhanced loading of miR21a-3p into the RISC complex under conditions of Dicer knockdown. Next, we sought to determine whether increased binding of miR-21a-3p to Ago-2 protein was caused by increased biogenesis or decreased degradation of miR-21a-3p. miR biogenesis entails production of a long primary nucleotide sequence (pri-miRNA) in the nucleus that undergoes cleavage to yield a shorter sequence called preliminary miRNA (pre-miRNA) that is exported into the cytoplasm. No difference was observed in pri or pre levels of miR-21a-3p between controls and Dicer knockdown cells (Fig. 5, D and E). It is thus established that Dicer knockdown does not cause an increase in miR-21a-3p biogenesis.
FIGURE 5.
miR-21a-3p loading into the RISC complex and half-life were increased in response to dicer knockdown. Ago-2 immunoprecipitation (IP) was performed to identify miR loaded into the RISC complex and ready to bind mRNA targets. A, immunoblot for Ago-2 immunoprecipitation showed significant levels of Ago-2 protein in the immunoprecipitate, but not in cell lysate or cell supernatant fractions. B, miR-21a-3p abundance after Ago-2 immunoprecipitation was significantly increased after dicer knockdown. C, real time PCR analysis of miR-21a after Ago-2 immunoprecipitation remained unchanged in response to dicer knockdown. D and E, measurement of pri-miR-21a-3p (D) and pre-miR-21 (E) by real time PCR did not show any significant increase in abundance following dicer knockdown. F, miR-21a-3p decay was examined 6 h after addition of actinomycin D (2.5 μg/ml) in control and dicer siRNA transfected EOMA cells. A significant decrease in miR-21a-3p in control siRNA transfected cells but not in dicer siRNA transfected cells was noted. The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
To test miR-21a-3p stability, transfected EOMA cells were treated for 6 h with actinomycin D to inhibit de novo oligonucleotide synthesis. There was a low basal level of miR-21a-3p present in transfection controls, which significantly decreased in response to treatment with actinomycin D, suggesting rapid use and degradation of miR-21a-3p over the 6-h exposure examined. EOMA cells transfected with dicer siRNA had significantly increased basal levels of miR-21a-3p compared with corresponding controls, consistent with results observed in Fig. 4 (D and E). However, in dicer siRNA transfected cells, there was no significant change in miR-21a-3p levels in response to actinomycin D treatment (Fig. 5F). Collectively, these results indicate that under the condition of Dicer knockdown, miR-21a-3p is protected from degradation while being loaded in the RISC complex, thereby extending its half-life.
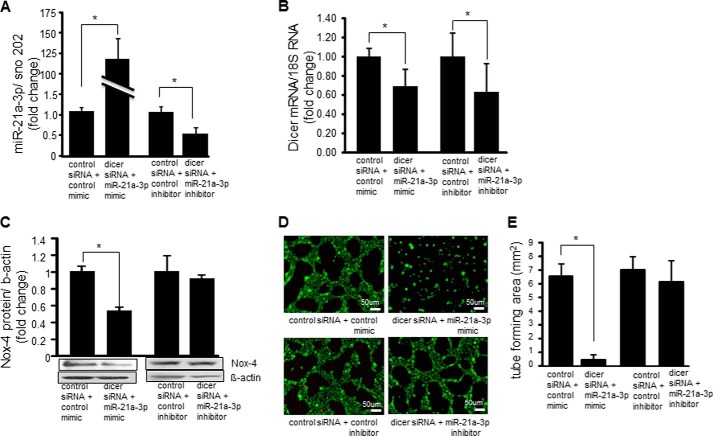
With the goal of testing a causal relationship between miR-21a-3p and endothelial cell tumor formation, rescue experiments were performed using miR-21a-3p mimics and inhibitors in dicer siRNA transfected EOMA cells to demonstrate the impact of this miR on the angiogenic phenotype of tumor forming endothelial cells (Fig. 6A). Dicer mRNA levels were measured in these same cells treated with either miR-21a-3p mimic or inhibitor to confirm that they did not affect the siRNA knockdown of dicer (Fig. 6B). Western blot showed a significant decrease in Nox-4 protein level after treatment with the miR-21a-3p mimic, although the miR-21a-3p inhibitor did not increase Nox-4 protein levels as expected (Fig. 6C). This observation may be attributable to a combination of extremely high levels of constitutively expressed Nox-4 protein and the relative lack of potency of the miR inhibitor compared with the miR mimic. MatrigelTM experiments were used to visualize the extent of miR-21a-3p inhibition on the angiogenic phenotype of EOMA cells. The addition of miR mimics to dicer siRNA transfected EOMA cells markedly inhibited tube formation, whereas the addition of the miR-21a-3p inhibitor to dicer siRNA transfected cells was able to restore the tube forming capacity of the EOMA cells back to dicer siRNA baseline levels (Fig. 6, D and E; for effects of dicer siRNA alone on EOMA cell tube formation see Fig. 1, C and D).
FIGURE 6.
miR-21a-3p inhibitors rescued angiogenic phenotype of dicer knocked-down EOMA cells. A, effectiveness of miR-21a-3p mimic and inhibitor in EOMA cells transfected with control siRNA or dicer siRNA was confirmed by real time PCR. B, miR-21a-3p mimic and inhibitor did not have off target effects that alter dicer mRNA levels. C, addition of miR-21a-3p mimic to EOMA cells transfected with dicer siRNA resulted in decreased Nox-4 protein levels that were restored to basal (control siRNA) levels following addition of miR-21a-3p inhibitor. D, MatrigelTM tube formation showed profound loss of angiogenic response in dicer siRNA transfected EOMA cells with addition of miR-21a-3p mimic and restoration of angiogenic response with addition of miR-21a-3p inhibitor. E, quantification of length of tube formation. The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
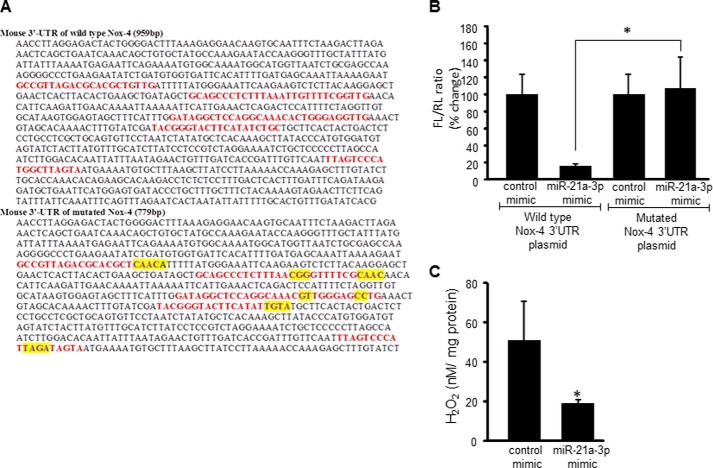
Having identified an increase in miR-21a-3p abundance relative to other miR following Dicer knockdown, the next step was to determine whether this miR strand targeted Nox-4. To test this hypothesis, potential binding sites for miR-21a-3p in the 3′-UTR of nox-4 were identified, and firefly luciferase reporter plasmids were constructed containing either the wild type/intact 959-bp 3′-UTR sequence or a 779-bp sequence containing mutations at each of the five potential binding sites at positions 239–260, 327–355, 444–474, 501–521, and 710–729 (Fig. 7A). Nox-4 reporter plasmids were co-transfected with Renilla firefly luciferase plasmids into EOMA cells previously treated with either control mimics or miR-21a-3p mimics. There was a significant decrease in luciferase levels with intact nox-4 3′-UTR in the presence of elevated miR-21a-3p and no difference in the luciferase levels between control and miR-21a-3p mimics observed in cells transfected with the mutated nox-4 3′-UTR sequence (Fig. 7B). To confirm the loss of Nox-4 function with elevated miR-21a-3p, cellular levels of H2O2 were measured using an Amplex Red assay and were significantly lower in cells treated with miR-21a-3p mimic (Fig. 7C).
FIGURE 7.
miR-21a-3p targets Nox-4 in EOMA cells. A, proposed binding sites for miR-21a-3p were identified in the Nox-4 3′-UTR that was inserted into a firefly luciferase plasmid vector (wild type). The binding sites were altered as indicated by yellow highlights to generate a second (mutated) firefly luciferase plasmid vector. B, EOMA cells were co-transfected with either the wild type or mutated Nox-4 3′-UTR firefly luciferase plasmids and a Renilla luciferase plasmid. Nox-4 transcriptional activation as measured by luciferase assay demonstrated significant loss of luciferase activity in wild type vectors in the presence of elevated miR21a-3p and no difference in the luciferase levels in cells transfected with the mutated Nox-4 3′-UTR vector. C, Amplex Red assay demonstrated loss of Nox-4 function with significantly decreased H2O2 levels observed in miR-21a-3p mimic delivered cells. The results are means ± S.D. of at least three independent experiments. *, p < 0.05.
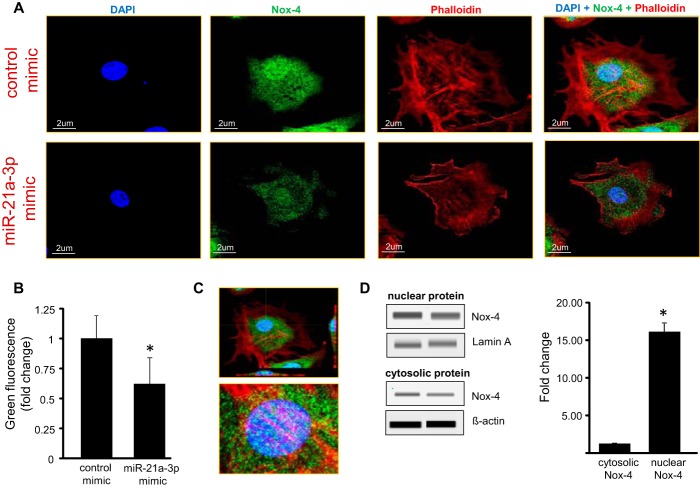
Because miR-21a-3p binding to nox-4 has not been previously described, confocal microscopy was used to provide additional evidence testing the effects of miR-21a-3p on Nox-4 protein expression. EOMA cells were incubated with fluorescently labeled antibodies to identify the nucleus (DAPI), Nox-4, and the cytoskeleton (phalloidin) within EOMA cells, and comparisons were made between cells treated with control mimics versus miR-21a-3p mimics (Fig. 8A). In both groups, Nox-4 expression was localized to the perinuclear area with a significant decrease in Nox-4 fluorescence intensity in the miR21a-3p-treated cells (Fig. 8B). Higher resolution images revealed that Nox-4 was localized directly on the nucleus and perhaps within the nucleus or located on invaginations of the nuclear membrane based on cross-sectional images of EOMA cells (Fig. 8C). Additional data to support this observation were obtained using Western blot to compare levels of Nox-4 protein detected in nuclear versus cytosolic fractions of EOMA cells with approximately a 15-fold increase in nuclear Nox-4 protein compared with cytosolic levels (Fig. 8D). These results visualize and quantify the strong association between Nox-4 and the nucleus of tumor forming endothelial cells.
FIGURE 8.
Lowered Nox-4 in response to treatment with miR-21a-3p mimic. A, EOMA cells with or without miR-21a-3p mimic were labeled with fluorescent probes DAPI (blue) for nucleus, phalloidin (red) for actin and (green) for Nox-4. Confocal microscopy showed that Nox-4 was localized in perinuclear area in EOMA cells, and there was significant decrease in Nox-4 fluorescence intensity in miR-21a-3p delivered cells. B, fluorescence intensity of images was quantified using Olympus FV10-ASW software. C, Z-stack image showing Nox-4 localized mostly in the perinuclear space. D, immunoblot analyses of nuclear and cytosolic fractions of EOMA cell lysates compare relative distribution of Nox-4 protein. The results are expressed as means ± S.D. of at least three independent experiments. *, p < 0.05.
DISCUSSION
MicroRNA are gaining increasing recognition for their role in regulating solid tumor formation (30–33). However, there is scanty literature addressing the significance of miR in the types of endothelial cell tumors commonly seen in infants and children. A commonly accepted paradigm in cancer biology views microRNA as providing a stabilizing role (“applying the brakes”) that maintain cellular differentiation and that loss of Dicer activity and miR production (“removing the brakes”) allows cells to go back to a less differentiated and unchecked proliferative state. Low Dicer levels are commonly reported in solid tumors including breast (34), ovarian (35), prostate (12), and skin (36) cancers when compared with normal tissue. Targeted silencing of dicer is known to perturb miR biogenesis in a manner where the levels of most mature miRs go down desilencing their targets. Interestingly, even with such a perturbation in miR biogenesis, the cellular levels of some miR remain relatively unchanged (37, 38). Outcomes of Dicer knockdown in the endothelial cell tumor model are consistent with that paradigm. When Dicer was knocked down using post-translational gene silencing techniques, tumor-forming capability was lost, indicating that there is a threshold level of Dicer activity necessary to maintain miR production and promote tumor formation. These results establish the central role of Dicer activity and related mature miR homeostasis in the formation of hemangioendothelioma.
Cellular oxidants are commonly implicated in neoplastic transformation (39, 40). However, the source of these oxidants and how they initiate or support cellular transformation remains poorly understood for many malignancies. We have previously demonstrated that Nox-4 is a critical source of reactive oxygen species production that drives HE tumor formation (8). Nox-4 is constitutively expressed in endothelial cells, and expression of endogenous proteins that inhibit Nox-4 or miR that target Nox-4 in endothelial cells has not been previously reported. miR-25 has been reported to target Nox-4 in renal cells (41). Our earlier work have identified that in healthy microvascular endothelial cells, Nox-dependent angiogenic property is subject to control by miRs (17). This work provides the first evidence demonstrating that in tumor forming endothelial cells, Nox-dependent angiogenic property is regulated by a novel miR pathway. The identification of miR-21a-3p for its ability to target nox-4 is intriguing because the significance may extend to other health and disease settings. miR-21 is widely recognized as an “oncomiR” because many of its targets are tumor suppressors, and elevated miR-21 levels have been observed in many forms of cancer (42, 43). Additionally, our previous work has identified miR-21 as a key regulator of inflammation (44, 45). One plausible explanation for the dual function of miR-21 is that the target strand targets tumor suppressors, and the passenger strand targets inflammatory proteins. Another possible explanation for the significance of the 3p strand is that low Dicer levels result in alternative miR processing with an expression bias to 3p strands as has been recently reported in mice with a mutation in a single allele for the dicer 1 gene (46). The observation that miR-21a-3p targets nox-4 indicates that it functions as a key regulator at the intersection of inflammation, redox biology, and cancer biology.
MicroRNA biogenesis may be broadly explained by three distinct steps: (i) transcription of pri-mIR, (ii) processing of pri-miR to pre-miR, and (iii) processing of pre-miR to mature miR, followed by decay. Gantier et al. (37) noted that miR-21 had a relatively long half-life after dicer knockdown. This work presents the first results identifying the mechanism for elevated miR-21 under conditions of lowered dicer levels. We demonstrate that miR-21a-3p is specifically loaded into the RISC complex, and this protects it from rapid decay. There is evidence in the literature to support the conclusion that loading in the RISC complex slows miR decay (47). This work is the first to demonstrate that such a principle of slower miR turnover applies to miR-21 under conditions of dicer knockdown. Although low Dicer levels and elevated miR-21 have been previously reported for solid tumors, this work presents the first evidence of a dynamic relationship between increasing abundance of miR-21a-3p and decreasing levels of dicer using in vivo tumor specimens. Further studies to delineate the mechanisms regulating this observed effect may provide greater insight into how Dicer and miR-21 promote neoplastic transformation in those tumors in which Dicer levels are low and miR-21 levels are high.
We have previously demonstrated that Nox-4-derived H2O2 is present in the nucleus of EOMA cells and that mice with HE have elevated levels of oxidized DNA, detected as 8-hydroxy-2-deoxyguanosine, in their urine. This observation is significant because the nucleus is weak in peroxidase capabilities (8), and the close proximity to Nox-4, a major source of cellular oxidants, implies that DNA within the nucleus is at high risk for oxidative damage. Removal of oxidized DNA through base excision repair puts cell at increased risk for single-strand breaks with low fidelity DNA repair and accumulation of mutations that promote tumorigenesis (40, 48). Elevated levels of oxidized 8-hydroxy-2-deoxyguanosine have been reported in the clinical setting for several different kinds of tumors including breast (49), lung (50), and bladder (51). The selective localization of Nox-4 to the perinuclear space as reported in this work indicates that this co-localization is likely a significant contributor to endothelial cell transformation.
In summary, this work presents first evidence demonstrating the essential role of Dicer and miR biogenesis to endothelial cell tumor formation. It also provides the first evidence identifying miR-21a-3p as an endothelial cell-derived endogenous inhibitor of Nox-4 activity. This is significant because Nox-4 is known to mediate pathologic inflammatory conditions in endothelial cells such as pulmonary artery fibrosis (52) and atherosclerosis (53). This work also provides the first evidence on the relationship between Dicer and miR-21a-3p in endothelial cell tumors and specifically identifies the mechanism for prolonged miR-21a-3p half-life under conditions of Dicer knockdown. These findings pave the way to broader applications to many other types of tumors where low cellular levels of Dicer and elevated miR-21 levels have been observed (42, 43, 54).
This work was supported, in whole or in part, by National Institutes of Health Grants GM095657 (to G. M. G.), DK076566 (to S. R.), and GM069589 and GM077185 (to C. K. S.).
- HE
- hemangioendothelioma
- miR or miRNA
- microRNA
- MCP-1
- monocyte chemoattractant protein-1
- RISC
- RNA-inducing silencing complex.
REFERENCES
- 1. Haggstrom A. N., Drolet B. A., Baselga E., Chamlin S. L., Garzon M. C., Horii K. A., Lucky A. W., Mancini A. J., Metry D. W., Newell B., Nopper A. J., Frieden I. J. (2006) Prospective study of infantile hemangiomas. Clinical characteristics predicting complications and treatment. Pediatrics 118, 882–887 [DOI] [PubMed] [Google Scholar]
- 2. Jacobs A. H., Walton R. G. (1976) The incidence of birthmarks in the neonate. Pediatrics 58, 218–222 [PubMed] [Google Scholar]
- 3. Mulliken J. B. (1988) Diagnosis and natural history of hemangiomas. In Vascular Birthmarks: Hemangiomas and Malformations (Mulliken J. B., Young A. E., eds) pp. 41–62, WB Saunders, Philadelphia, PA [Google Scholar]
- 4. Paller A. S. (2000) Responses to anti-angiogenic therapies. J. Investig. Dermatol. Symp. Proc. 5, 83–86 [DOI] [PubMed] [Google Scholar]
- 5. Hoak J. C., Warner E. D., Cheng H. F., Fry G. L., Hankenson R. R. (1971) Hemangioma with thrombocytopenia and microangiopathic anemia (Kasabach-Merritt syndrome). An animal model. J. Lab. Clin. Med. 77, 941–950 [PubMed] [Google Scholar]
- 6. Warner E. D., Hoak J. C., Fry G. L. (1971) Hemangioma, thrombocytopenia, and anemia. The Kasabach-Merritt syndrome in an animal model. Arch. Pathol. 91, 523–528 [PubMed] [Google Scholar]
- 7. Gordillo G. M., Onat D., Stockinger M., Roy S., Atalay M., Beck F. M., Sen C. K. (2004) A key angiogenic role of monocyte chemoattractant protein-1 in hemangioendothelioma proliferation. Am. J. Physiol. Cell Physiol. 287, C866–C873 [DOI] [PubMed] [Google Scholar]
- 8. Gordillo G., Fang H., Park H., Roy S. (2010) Nox-4-dependent nuclear H2O2 drives DNA oxidation resulting in 8-OHdG as urinary biomarker and hemangioendothelioma formation. Antioxid. Redox. Signal. 12, 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordillo G., Fang H., Khanna S., Harper J., Phillips G., Sen C. (2009) Oral administration of blueberry inhibits angiogenic tumor growth and enhances survival of mice with endothelail cell neoplasm. Antioxid. Redox Signal. 11, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jakymiw A., Patel R. S., Deming N., Bhattacharyya I., Shah P., Lamont R. J., Stewart C. M., Cohen D. M., Chan E. K. (2010) Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer 49, 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dudek H., Wong D. H., Arvan R., Shah A., Wortham K., Ying B., Diwanji R., Zhou W., Holmes B., Yang H., Cyr W. A., Zhou Y., Shah A., Farkiwala R., Lee M., Li Y., Rettig G. R., Collingwood M. A., Basu S. K., Behlke M. A., Brown B. D. (2014) Knockdown of β-catenin with dicer-substrate siRNAs reduces liver tumor burden in Vivo. Mol. Ther. 22, 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma X., Fan Y., Gao Y., Zhang Y., Huang Q., Ai Q., Ni D., Chen W., Zhang P., Song E., Wang B., Shi T., Zheng T., Zhang X. (2014) Dicer is down-regulated in clear cell renal cell carcinoma and in vitro Dicer knockdown enhances malignant phenotype transformation. Urol. Oncol. 32, 46. [DOI] [PubMed] [Google Scholar]
- 13. Gordillo G. M., Atalay M., Roy S., Sen C. K. (2002) Hemangioma model for in vivo angiogenesis. Inducible oxidative stress and MCP-1 expression in EOMA cells. Methods Enzymol. 352, 422–432 [DOI] [PubMed] [Google Scholar]
- 14. Chan Y. C., Roy S., Huang Y., Khanna S., Sen C. K. (2012) The microRNA miR-199a-5p down-regulation switches on wound angiogenesis by derepressing the v-ets erythroblastosis virus E26 oncogene homolog 1-matrix metalloproteinase-1 pathway. J. Biol. Chem. 287, 41032–41043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan Y. C., Roy S., Khanna S., Sen C. K. (2012) Downregulation of endothelial microRNA-200b supports cutaneous wound angiogenesis by desilencing GATA binding protein 2 and vascular endothelial growth factor receptor 2. Arterioscler. Thromb. Vasc. Biol. 32, 1372–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park H. A., Kubicki N., Gnyawali S., Chan Y. C., Roy S., Khanna S., Sen C. K. (2011) Natural vitamin E α-tocotrienol protects against ischemic stroke by induction of multidrug resistance-associated protein 1. Stroke 42, 2308–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shilo S., Roy S., Khanna S., Sen C. K. (2008) Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler. Thromb. Vasc. Biol. 28, 471–477 [DOI] [PubMed] [Google Scholar]
- 18. Schmittgen T. D. (2008) Regulation of microRNA processing in development, differentiation and cancer. J. Cell Mol. Med. 12, 1811–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J. Q., Heldman M. R., Herrmann M. A., Kedei N., Woo W., Blumberg P. M., Goldsmith P. K. (2013) Absolute quantitation of endogenous proteins with precision and accuracy using a capillary Western system. Anal. Biochem. 442, 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rustandi R. R., Loughney J. W., Hamm M., Hamm C., Lancaster C., Mach A., Ha S. (2012) Qualitative and quantitative evaluation of Simon, a new CE-based automated Western blot system as applied to vaccine development. Electrophoresis 33, 2790–2797 [DOI] [PubMed] [Google Scholar]
- 21. Tan L. P., Seinen E., Duns G., de Jong D., Sibon O. C., Poppema S., Kroesen B. J., Kok K., van den Berg A. (2009) A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res. 37, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu H., He Y., Zhang X., Peng Z., Yang Y., Zhu R., Bai J., Tian Y., Li X., Chen W., Fang D., Wang R. (2011) The rat IgGFcγBP and Muc2 C-terminal domains and TFF3 in two intestinal mucus layers bind together by covalent interaction. PLoS One 6, e20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fish J. E., Srivastava D. (2009) MicroRNAs. Opening a new vein in angiogenesis research. Sci. Signal. 2, pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urbich C., Kuehbacher A., Dimmeler S. (2008) Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 79, 581–588 [DOI] [PubMed] [Google Scholar]
- 25. Wang S., Olson E. N. (2009) AngiomiRs. Key regulators of angiogenesis. Curr. Opin. Genet. Dev. 19, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hinkal G. W., Grelier G., Puisieux A., Moyret-Lalle C. (2011) Complexity in the regulation of Dicer expression. Dicer variant proteins are differentially expressed in epithelial and mesenchymal breast cancer cells and decreased during EMT. Br J. Cancer 104, 387–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kitagawa N., Ojima H., Shirakihara T., Shimizu H., Kokubu A., Urushidate T., Totoki Y., Kosuge T., Miyagawa S., Shibata T. (2013) Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 104, 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krichevsky A. M., Gabriely G. (2009) miR-21. A small multi-faceted RNA. J. Cell Mol. Med. 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumarswamy R., Volkmann I., Thum T. (2011) Regulation and function of miRNA-21 in health and disease. RNA Biol. 8, 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pesta M., Klecka J., Kulda V., Topolcan O., Hora M., Eret V., Ludvikova M., Babjuk M., Novak K., Stolz J., Holubec L. (2010) Importance of miR-20a expression in prostate cancer tissue. Anticancer Res. 30, 3579–3583 [PubMed] [Google Scholar]
- 31. Slack F. J., Weidhaas J. B. (2008) MicroRNA in cancer prognosis. N. Engl. J. Med. 359, 2720–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su X., Chakravarti D., Cho M. S., Liu L., Gi Y. J., Lin Y. L., Leung M. L., El-Naggar A., Creighton C. J., Suraokar M. B., Wistuba I., Flores E. R. (2010) TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taulli R., Bersani F., Foglizzo V., Linari A., Vigna E., Ladanyi M., Tuschl T., Ponzetto C. (2009) The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Invest. 119, 2366–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grelier G., Voirin N., Ay A. S., Cox D. G., Chabaud S., Treilleux I., Léon-Goddard S., Rimokh R., Mikaelian I., Venoux C., Puisieux A., Lasset C., Moyret-Lalle C. (2009) Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J. Cancer 101, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merritt W. M., Lin Y. G., Han L. Y., Kamat A. A., Spannuth W. A., Schmandt R., Urbauer D., Pennacchio L. A., Cheng J. F., Nick A. M., Deavers M. T., Mourad-Zeidan A., Wang H., Mueller P., Lenburg M. E., Gray J. W., Mok S., Birrer M. J., Lopez-Berestein G., Coleman R. L., Bar-Eli M., Sood A. K. (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 359, 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sand M., Gambichler T., Skrygan M., Sand D., Scola N., Altmeyer P., Bechara F. G. (2010) Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest. 28, 649–653 [DOI] [PubMed] [Google Scholar]
- 37. Gantier M. P., McCoy C. E., Rusinova I., Saulep D., Wang D., Xu D., Irving A. T., Behlke M. A., Hertzog P. J., Mackay F., Williams B. R. (2011) Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39, 5692–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koscianska E., Starega-Roslan J., Krzyzosiak W. J. (2011) The role of Dicer protein partners in the processing of microRNA precursors. PLoS One 6, e28548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 40. Lord C. J., Ashworth A. (2012) The DNA damage response and cancer therapy. Nature 481, 287–294 [DOI] [PubMed] [Google Scholar]
- 41. Fu Y., Zhang Y., Wang Z., Wang L., Wei X., Zhang B., Wen Z., Fang H., Pang Q., Yi F. (2010) Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am. J. Nephrol. 32, 581–589 [DOI] [PubMed] [Google Scholar]
- 42. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 43. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Das A., Ganesh K., Khanna S., Sen C. K., Roy S. (2014) Engulfment of apoptotic cells by macrophages. A role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 192, 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roy S., Khanna S., Hussain S. R., Biswas S., Azad A., Rink C., Gnyawali S., Shilo S., Nuovo G. J., Sen C. K. (2009) MicroRNA expression in response to murine myocardial infarction. miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc. Res. 82, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anglesio M. S., Wang Y., Yang W., Senz J., Wan A., Heravi-Moussavi A., Salamanca C., Maines-Bandiera S., Huntsman D. G., Morin G. B. (2013) Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J. Pathol. 229, 400–409 [DOI] [PubMed] [Google Scholar]
- 47. Berger S. M., Pesold B., Reber S., Schönig K., Berger A. J., Weidenfeld I., Miao J., Berger M. R., Gruss O. J., Bartsch D. (2010) Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs. Nucleic Acids Res. 38, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ziech D., Franco R., Pappa A., Panayiotidis M. I. (2011) Reactive oxygen species (ROS)–induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res. 711, 167–173 [DOI] [PubMed] [Google Scholar]
- 49. Kuo H. W., Chou S. Y., Hu T. W., Wu F. Y., Chen D. J. (2007) Urinary 8-hydroxy-2′-deoxyguanosine (8-OHdG) and genetic polymorphisms in breast cancer patients. Mutat Res. 631, 62–68 [DOI] [PubMed] [Google Scholar]
- 50. Yano T., Shoji F., Baba H., Koga T., Shiraishi T., Orita H., Kohno H. (2009) Significance of the urinary 8-OHdG level as an oxidative stress marker in lung cancer patients. Lung Cancer 63, 111–114 [DOI] [PubMed] [Google Scholar]
- 51. Soini Y., Haapasaari K. M., Vaarala M. H., Turpeenniemi-Hujanen T., Kärjä V., Karihtala P. (2011) 8-hydroxydeguanosine and nitrotyrosine are prognostic factors in urinary bladder carcinoma. Int. J. Clin. Exp. Pathol. 4, 267–275 [PMC free article] [PubMed] [Google Scholar]
- 52. Pache J. C., Carnesecchi S., Deffert C., Donati Y., Herrmann F. R., Barazzone-Argiroffo C., Krause K. H. (2011) NOX-4 is expressed in thickened pulmonary arteries in idiopathic pulmonary fibrosis. Nat. Med. 17, 31–33 [DOI] [PubMed] [Google Scholar]
- 53. Chamberlain J., Francis S., Brookes Z., Shaw G., Graham D., Alp N. J., Dower S., Crossman D. C. (2009) Interleukin-1 regulates multiple atherogenic mechanisms in response to fat feeding. PLoS One 4, e5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]