Background: WWOX encodes a 46-kDa tumor suppressor.
Results: WW1 domain of WWOX mediates its protein-protein interaction with PY motifs that are involved in molecular processes, including transcription, RNA processing, and metabolism.
Conclusion: The WW1 domain of WWOX provides a versatile platform that links WWOX with individual proteins associated with physiologically important networks.
Significance: This study provides a better understanding of WWOX biology in normal and disease states.
Keywords: E3 Ubiquitin Ligase, Mass Spectrometry (MS), Protein-Protein Interactions, Tumor Suppressor Gene, Ubiquitination, WW Domain, WWOX, Itch
Abstract
WW domains are small modules present in regulatory and signaling proteins that mediate specific protein-protein interactions. The WW domain-containing oxidoreductase (WWOX) encodes a 46-kDa tumor suppressor that contains two N-terminal WW domains and a central short-chain dehydrogenase/reductase domain. Based on its ligand recognition motifs, the WW domain family is classified into four groups. The largest one, to which WWOX belongs, recognizes ligands with a PPXY motif. To pursue the functional properties of the WW domains of WWOX, we employed mass spectrometry and phage display experiments to identify putative WWOX-interacting partners. Our analysis revealed that the first WW (WW1) domain of WWOX is the main functional interacting domain. Furthermore, our study uncovered well known and new PPXY-WW1-interacting partners and shed light on novel LPXY-WW1-interacting partners of WWOX. Many of these proteins are components of multiprotein complexes involved in molecular processes, including transcription, RNA processing, tight junction, and metabolism. By utilizing GST pull-down and immunoprecipitation assays, we validated that WWOX is a substrate of the E3 ubiquitin ligase ITCH, which contains two LPXY motifs. We found that ITCH mediates Lys-63-linked polyubiquitination of WWOX, leading to its nuclear localization and increased cell death. Our data suggest that the WW1 domain of WWOX provides a versatile platform that links WWOX with individual proteins associated with physiologically important networks.
Introduction
Specific protein-protein interactions are usually mediated through binding of modular domains to specific motifs within their partners. One example is the small modular WW domains that interact with a variety of distinct peptide ligands, including motifs with core proline-rich sequences (1–3). WW domains are typically 35–40 amino acids in length and fold into a three-stranded, antiparallel β-sheet with two ligand-binding grooves (4, 5). Depending on the nature of their interacting peptide ligands, WW domains are classified into four classes (reviewed in Refs. 6 and 7). Class I WW domains, which include the NEDD4 family proteins and the Yes-associated protein (YAP65), recognize PY3 (PPXY or LPXY) motifs (8–10). Class II WW domains, such as the FE65 WW domain, recognize PPLP motifs, and Class III WW domains (FBP30 WW1) recognize PR motifs. Group IV WW domains, such as those from Pin1, recognize p(S/T)P motifs. These domains are found in many different structural and signaling proteins that are involved in a variety of cellular processes, including RNA transcription and processing, protein trafficking and stability, receptor signaling, and control of the cytoskeleton (reviewed in Refs. 6 and 7).
The WW domain-containing oxidoreductase (WWOX) encodes a tumor suppressor that is commonly inactivated in human cancers (11–14). WWOX inactivation has been linked to genomic and/or epigenetic modifications (13–15). Importantly, WWOX tumor suppressor function is believed to be mediated through its ability to interact with distinct proteins in multiple cellular pathways (16). WWOX contains two N-terminal WW domains and a central short-chain dehydrogenase (SDR) domain. Initial assessment of WWOX WW domains revealed their physical interaction with PPXY-containing proteins (17). The first partner reported was p73 (17). This study was followed by a number of reports validating WWOX physical association with PPXY-containing partners (18) and demonstrating WWOX functional cross-talk with different cellular pathways, including transcription, apoptosis, and cytoskeleton (reviewed in Refs. 7 and 16). Of note, all of the reported interacting WWOX partners investigated so far were interacting with its WW1 domain (12, 13, 16, 17, 19–24). Although the WW2 domain of WWOX has an atypical structure, due to replacement of tyrosine instead of tryptophan, it was suggested that it belongs to Group I (25). Recently, a proteomic screen of the Drosophila orthologue of WWOX revealed putative interacting partners, although none of the proteins identified had PY motifs (26). Nevertheless, a comprehensive interactive proteome of mammalian WWOX WW domains is lacking.
In order to advance our understanding of the functional capacity of WWOX as a tumor suppressor, we investigated the interactions between the WW domains of WWOX and its possible client proteins using mass spectrometry (MS), phage display, and protein interaction analyses in mammalian cells. Our data reveal that the predominant interacting module within WWOX is the WW1 domain. We found that the WW1 domain interacts with PY motifs, including PPXY and LPXY, illuminating the versatility of the WW1 domain of WWOX. Our findings further suggest that the WW1 domain of WWOX acts as a platform that links WWOX with individual proteins associated with physiologically important networks.
EXPERIMENTAL PROCEDURES
Cell Culture and Transient Transfection
HEK293 cells were grown in RPMI1640, supplemented with 10% FBS (Invitrogen), glutamine, and penicillin/streptomycin (Beit-Haemek, Israel). Itch-deficient cells were described elsewhere (27). Transient transfections were achieved using Mirus TransLTi (Mirus Bio LLC, Madison, WI).
Constructs
The expression plasmids encoding the following inserts were described previously: mammalian GST-WWOX (pEBG-WWOX) (23); Myc-WWOX and Myc-p73 (17); ErbB4-HA (20); FLAG-AMOT (angiomotin), FLAG-AMOTL1 (angiomotin-like 1), and FLAG-AMOTL1-PPEA (28); and FLAG-ITCH, Myc-ITCH, Myc-ITCH-C830A, and Ub-HA (29). The FLAG-Ub plasmid is a gift from Dr. Michal Goldberg (Hebrew University). The HA-Ub mutants were purchased from Addgene; the FLAG-DVL1 (disheveled 1) construct was a gift from Prof. Rachel Bar-Shavit (Hebrew University). Cloning of the GST fusion constructs of WWOX domains, ITCH, and NEDD4 into pEBG plasmid was done using a standard procedure. The site-directed mutagenesis procedure was performed using according to the manufacturer's instructions (Stratagene, La Jolla, CA).
GST Pull-down, Immunoprecipitation, and Immunoblot Analysis
Cells were lysed by using Nonidet P-40 lysis buffer containing 50 mmol/liter Tris (pH 7.5), 150 mmol/liter NaCl, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors. In GST pull-down, lysates were mixed with GST beads and rocked for 2 h at 4 ºC. Thereafter, the beads were washed four times with the same buffer containing 0.1% Nonidet P-40. For immunoprecipitation, lysates were precleared with mouse IgG, immunoprecipitations were carried out in the same buffer, and lysates were washed four times with the same buffer containing 0.1% Nonidet P-40. Immunoblotting was conducted under standard conditions. Antibodies used were monoclonal anti-HA (Covance, Princeton, NJ); polyclonal anti-WWOX, monoclonal anti-Myc-HRP, and polyclonal anti-lamin (Santa Cruz Biotechnology, Inc.); monoclonal anti-FLAG and anti-FLAG-HRP (Sigma); monoclonal anti-FK2 (Millipore, Temecula, CA); polyclonal anti-HSP-90 and monoclonal anti-GAPDH (Calbiochem); polyclonal anti-GST (GE Healthcare); and anti-HA-HRP (Roche Applied Science).
Nano-LC-MS/MS Analysis
Precipitations with each GST fusion were repeated at least three times in two independent experiments. The samples were analyzed by 10% SDS-PAGE, stained by Coomassie stain using the blue (R-250) reagent (ThermoFisher Scientific, Rockford, IL), and destained the next day with 10% acetic acid and 20% methanol solution.
Gel lines were cut into equal slices, and individual slices were subject to in-gel reduction, alkylation, and trypsinization based on a protocol described previously (30). Tryptic peptides were extracted with 60% acetonitrile, 1% methanol; evaporated until dryness; resuspended in 1 μl of 60% acetonitrile, 1% methanol; diluted 10-fold with 1% methanol; and analyzed by LC-MS/MS. This peptide separation was performed by using the caplillary C18 column (75-μm inner diameter × 100 mm; Proteoprep, New Objective) on a nano-HPLC system (Eksignet, Dublin, CA) coupled online to an LTQ-Orbitrab mass spectrometer with a nanoelectrospray ion source (ThermoFisher Scientific). To separate the peptides, the column was applied with a linear gradient with a flow rate of 230 nl/min at 25 °C: from 5 to 30% in 10 min, from 30 to 70% in 55 min, from 70 to 80% in 10 min, and held at 80% for an additional 25 min (solvent A is 2% acetonitrile, 0.1% formic acid, and solvent B is 98% acetonitrile, 0.1% formic acid).
Survey MS scans were acquired with a resolution of 30,000, full-scan spectra were collected at 400–2000 m/z, and the most intense ions were submitted for additional collision-induced dissociation fragmentation (normalized collision energy, 35; activation Q, 0.25; time, 30 ms; isolation width, 3.00).
Database Searching and Criteria for Protein Identification
Peak lists were generated by using the Bioworks version 3.3 package (ThermoFisher Scientific), and the peak lists from each LC-MS/MS run were merged into one file for the Mascot search (Matrix Science) against the NCBInr 20100120 database. Only precursor charge states +2, +3, and +4 were considered. For database searches, Cys carbamidomethylation was set as a fixed modification, and Met oxidation and one missed cleavage were set as variable modifications. Mass tolerance for peptides identified by the MS and MS/MS analysis was set to 0.6 and 0.2 Da, respectively. A significant Mascot probability (p ≤ 0.05) was used for positive peptide identification. The Mascot identity threshold score was defined as 35 for p < 0.05. The Mascot results were filtered manually to validate differences between observed and calculated masses in the protein match (up to 0.01 Da) and in the MS/MS match (up to 0.2 Da), alone with evaluation of adherence of the mass errors. An initial list of identified proteins with a minimum of two unique peptides where at least one unique peptide has a score higher than 35 was created for the GST-WW1-WFPA and the GST-WW1 pull-down experiments. Sequences from the initial protein list were searched against the cRAP (Common Repository of Adventitious Proteins) list of common laboratory contaminants (available on the global proteome machine [gpm] website). To remove sequence redundancy and nonspecific binders, sequences were clustered by using the CD-HIT server, using a sequence identity cut-off of 90%. Similar sequences were removed, and proteins identified in the GST-WW1-WFPA were excluded from the final list of the GST-WW1 binders (data not shown).
Identification of the Ubiquitinated Proteins
Sample preparation and the LC-MS/MS analysis was performed as described above. To identify ubiquitinated lysines of WWOX, we used the Proteome Discoverer program, version 1.4 (Thermo Fisher Scientific) and searched against the Uniprot/Swiss-Prot (human) and the cRAP databases. Search parameters included one missed cleavage site; Cys carbamidomethylation was set as a fixed modification, and Met oxidation and Lys ubiquitination (diglycine, 114 Da) were set as a variable modification. Mass tolerance for peptides identified by the MS/MS analysis was set to at 0.5 Da. Only peptides with a significant Xcorr score were considered.
Protein Functional Annotation and Enrichment Analysis
For functional analysis of the WWOX-interacting proteins identified by the mass spectrometry analysis, we applied the DAVID annotation tool (31, 32) by using default parameters and the GO-FAT annotation vocabulary. Functional annotation clustering was done by using DAVID version 6.7, and the GO enrichment analyses was based on the human genome database.
Phage Display
To map the specificity of the human WW1 domain of WWOX, we used phage display technology (33), where very large libraries of random peptides can be expressed on the surface of phage articles. The phage colonies were then sequenced with Illumina deep sequencing. We used only the following: 1) sequences with mean PHRED score over 35 (for whole read) and 2) sequences that do not include cysteine (due to disulfide bond) or a premature stop codon. In this way, a total of 2733 unique peptides (from 80,605 sequences which passed two filters) were identified to bind to the WWOX WW1 domain. To generate a sequence logo and detect multiple binding specificity, MUSI was used (30). We scanned both proteome and the WWOX-interacting proteins with the position weight matrix from the phage display experiment. We obtained the best position weight matrix score for each protein from both lists (human proteome and MS proteins) and plotted the cumulative distribution function (CDF). WWOX-interacting proteins identified by mass spectrometry have much higher position weight matrix scores compared with random proteins (p value <2.2E−18 using rank sum test).
Motif Analyses
We have scanned LPXY and PPXY motifs in both proteome (Ensembl version 64) and WWOX-interacting proteins from mass spectrometry (240 proteins (MS-set-1) or 144 proteins (MS-set-2)). Overlap of LPXY- and PPXY-containing proteins was assessed statistically using bootstrapping. To compare enriched motifs in MS analysis with the human proteome, we ran a χ2 test.
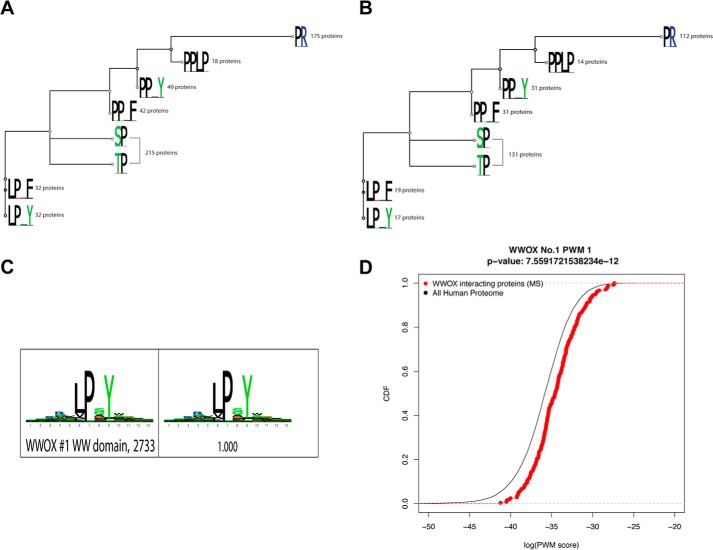
To generate the tree, we used MAFFT, version 7 (K. Katoh and D. M. Standley) to align the sequences and then Geneious, version 5.1 (Biomatters) to build a tree (neighbor-joining method). The number beside each logo (see Fig. 2) indicates the number of proteins with the motif from our mass spectrometry data (of 240 proteins). The branch length is calculated from pairwise comparisons of sequences (an 8 × 8 matrix is calculated); a shorter branch length indicates more sequence similarity.
FIGURE 2.
A and B, tree motif analysis for putative interacting partners of WWOX obtained from the MS study. The tree was built based on the alignment of eight different motifs of interest. We have used MAFFT to align the sequences and then Geneious to build the tree (neighbor-joining method). The number beside each logo indicates the number of proteins with the motif from our mass spectrometry data (of 240 proteins; MS-SET-1 (A); or of the 144 proteins; MS-SET-2 (B)). C, sequence logo for WW1 domain from phage display. High affinity peptides were identified by phage display, and subsequent sequencing of the phage colonies was performed. These sequences were then filtered, aligned, and clustered using the MUSI software to obtain a sequence logo. D, obtained WW1 binding partners are enriched in hits to the phage-derived specificity profile. Distributions of similarity scores to the phage-derived profile are shown in the observed WW1 binders as compared with the whole proteome. The observed binders are highly enriched in sequences that match well to the phage-derived logo when compared with the background. The p value as from a rank sum test is indicated.
Subcellular Fractionation
Nuclear and cytoplasmic extracts were prepared as follows. First, cells were scraped in PBS, and after centrifugation, the cell pellet was reconstituted in a hypotonic lysis buffer (10 mmol/liter HEPES (pH 7.9), 10 mmol/liter KCl, 0.1 mmol/liter EDTA) supplemented with 1 mmol/liter DTT and a broad-spectrum mixture of protease inhibitors (Sigma-Aldrich). The cells were allowed to swell on ice for 15 min, and then Nonidet P-40 was added, and cells were lysed by vortex. After centrifugation, the cytoplasmic fraction was collected. Afterward, nuclear extracts were obtained by incubating nuclei in a hypertonic nuclear extraction buffer (20 mmol/liter HEPES (pH 7.9), 0.42 mol/liter KCl, 1 mmol/liter EDTA) supplemented with 1 mmol/liter DTT for 15 min at 4 °C. The nuclear fraction was collected after centrifugation.
In Vivo Ubiquitination Assay
HEK293 cells were cotransfected with the different indicated expression vectors as indicated in the figures. After 24 h, cells were treated or untreated with MG-132 (20 μmol/liter; Sigma-Aldrich) for 4 h. Lysates were subjected to GST pull-down or immunoprecipitated as appropriate, washed four times, and immunoblotted with the indicated antibodies.
Immunofluorescence
Cells were seeded on round slide coverslips in 12-well plates. Twenty-four hours later, cells were transfected with expression plasmids encoding FLAG-ITCH, Myc-WWOX, or both together. Twenty-four hours posttransfection, cells were fixed in 3.7% PBS-buffered formaldehyde and permeabilized with 0.05% Triton X-100 at room temperature. Cells were then incubated in 10% goat serum (Invitrogen) with primary antibody (anti-FLAG and/or anti-WWOX) each for 1 h and for 1 h with secondary antibody. Alexa Fluor-647 anti-mouse or Alexa Fluor- 488 anti-rabbit antibody (Molecular Probes, Inc.) was used to detect ITCH (red) and WWOX (green), respectively (see Fig. 7). Cells were examined by confocal microscopy (Olympus, Tokyo, Japan) under ×60 magnification. DAPI was used as a counterstain for nuclei.
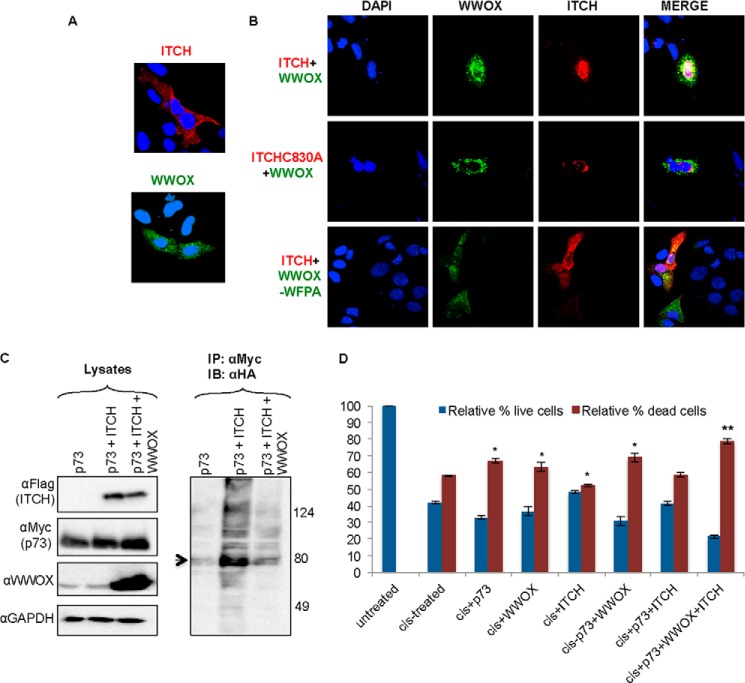
FIGURE 7.
WWOX-ITCH functional association. A and B, ITCH coexpression with WWOX mediates WWOX nuclear localization. HeLa cells were transiently transfected with FLAG-ITCH and GFP-WWOX, FLAG-ITCH-C830A and GFP-WWOX, or FLAG-ITCH and GFP-WWOX-WFPA as indicated. At 24 h, cells were fixed and immunostained with anti-FLAG followed by Alexa Fluor-647 secondary conjugated antibodies. Cells were visualized by confocal microscopy at ×60. DAPI, nuclei; red, ITCH; green, GFP-WWOX; yellow, co-localization of WWOX and ITCH. C, WWOX suppresses ITCH-mediated ubiquitination of p73. HEK293 cells were transiently cotransfected with HA-Ub, MYC-p73, FLAG-ITCH, and WWOX as indicated. At 24 h, cells were lysed, and immunoprecipitation (IP) was performed using anti-Myc antibody. Lysates and precipitates were immunoblotted (IB) using the indicated antibodies. Arrow, ubiquitinated p73 (right blot). D, coexpression of WWOX and ITCH enhances p73-mediated cell death. HEK293 cells were transiently transfected with HA-p73, GST-WWOX, or FLAG-ITCH or in the combination indicated. At 24 h, cells were treated with cisplatin (25 μm) for an additional 24 h. The number of live and dead cells was determined using trypan blue. The relative percentage of cells is shown. Shown is the average of three independent experiments. *, p < 0.05; **, p < 0.01. Error bars, S.E.
Cell Death
Cells were seeded at a density of 2.5 × 105 in 6-well plates. Twenty-four hours after transfection with the indicated expression plasmids, cells were treated with 25 μm cisplatin for an additional 24 h. Floating and attached cells were collected and combined together, washed with PBS, and counted after staining with trypan blue using a TC10 cell counter (Bio-Rad). The relative numbers of live and dead cells are presented.
Statistical Analysis
Data were expressed as mean ± S.E., as indicated in the figure legends. Two-tailed Student's t test was used for comparisons between two groups as applicable, where indicated, and a level of significance of p < 0.05 was considered statistically significant. In general, all experiments described throughout were repeated at least three times.
RESULTS
Generation and Characterization of Truncated Mammalian GST-WWOX Fusions
To identify cellular proteins that interact with WWOX, we constructed mammalian GST fusion proteins of the full-length protein or the different WWOX domains (Fig. 1A). This set included GST-WWOX, GST-WW1, GST-WW2, GST-WW1,2 and GST-SDR. To examine the expression of these fusions, they were individually transfected into HEK293 cells, and 24 h later, cells were lysed. Immunoblot analysis of these fusions revealed the proper size of each of the domains (Fig. 1. A and B). We noted that expression of GST-WW1 was stronger than that of the other fusions, probably due to stability- and/or half-life-related reasons. To validate the functionality and specificity of these domains, we tested their ability to interact with a well known partner of WWOX, ErbB4. ErbB4, through its PPXY motifs, has been shown to specifically interact with WW1 of WWOX (20, 34). To this end, HEK293 cells were transiently co-transfected with ErbB4-HA and each of the GST fusions. Twenty hours later, cells were lysed, and GST pull-down was performed followed by immunoblot analysis. As shown in Fig. 1C, fusions containing the WW1 domain (WWOX, WW1, and WW1,2), but not with GST-WW2 or GST-SDR, interacted with ErbB4, consistent with our previous observations (18). These results indicate that these fusions are functionally relevant.
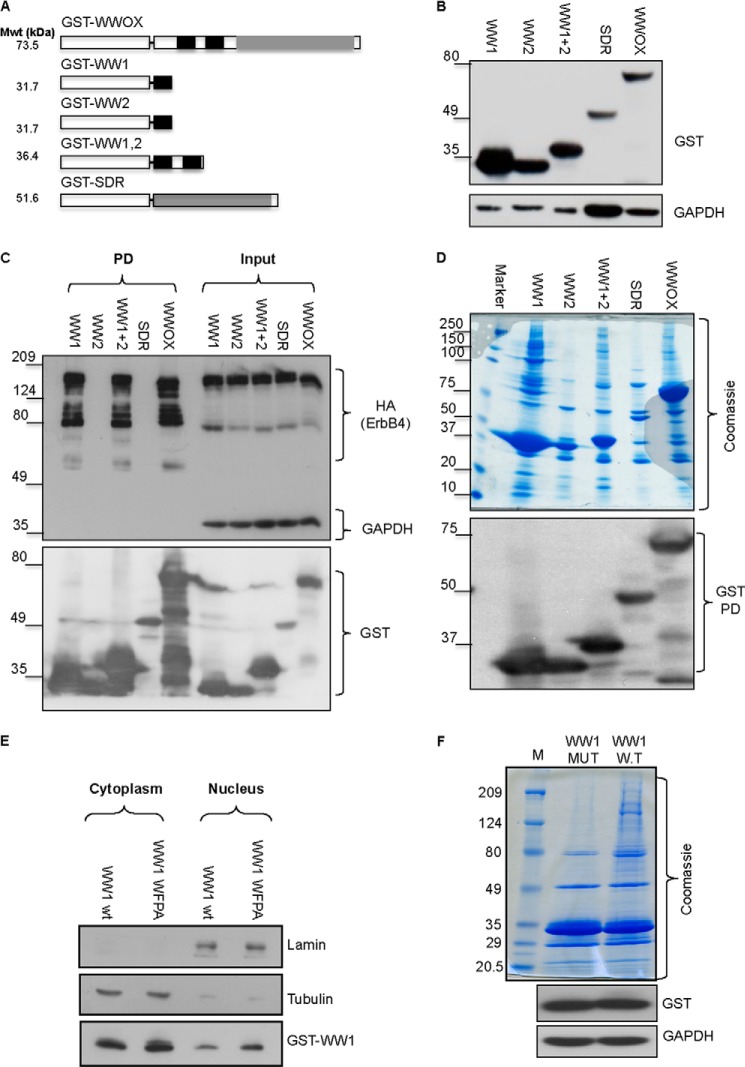
FIGURE 1.
Generation and characterization of WWOX interacting domains. A, schematic illustration of WWOX domains and predicted molecular weight (Mwt). GST is shown as a white box. WW domains of WWOX are shown in black boxes; SDR domains of WWOX are shown as gray boxes. B, generation of full-length and truncated mammalian GST fusions of WWOX. HEK293 cells were transiently transfected with the different GST fusions. After 24 h, cells were lysed, and immunoblot analysis using anti-GST was performed. GAPDH was used for normalization. C, validation of interaction ability of GST-WW1. HEK293 cells were transiently co-transfected with ErbB4-HA and the different GST fusions. At 24 h, cells were lysed, and GST pull-down (PD) was performed. Immunoblot analysis using anti-HA (ErbB4), GST, and GAPDH revealed specific interaction between WW1-containing GST fusions. D, characterization of interaction potential of the different GST-WWOX fusions. HEK293 cells were transiently transfected with the different GST fusions. After 24 h, cells were lysed, and GST pull-down was performed. Top, Coomassie Blue staining of the precipitated proteins of the different fusions. Note that the full-length WWOX lane is most similar to WW1,2 and WW1. Bottom, immunoblot of the different GST fusions using anti-GST antibody. E, characterization of WW1 and WW1-WFPA mutant. HEK293 cells were transiently transfected with GST-WW1 or GST-WW1-WFPA mutant. After 24 h, cells were subfractionated into cytoplasmic and nuclear fractions. Immunoblot analysis, using anti-GST antibody, revealed similar levels and subcellular localization of both proteins. Lamin and tubulin were used to indicate successful nuclear and cytoplasmic subfractionation, respectively. F, characterization of interaction potential of the WW1 domain of WWOX. HEK293 cells were transiently transfected with GST-WW1 or GST-WW1-WFPA mutant. After 24 h, cells were lysed, and GST pull-down was performed. Top, Coomassie Blue analysis of the precipitated proteins of the different fusions. Note that GST-WW1-WFPA (MUT) lost the potential of precipitating protein complexes as compared with intact WW1 (W.T). The lower blots show expression levels of the GST fusions.
Next, we determined the ability of these fusions to associate with protein complexes in vivo. HEK293 cells were transfected with each of the GST fusions, and GST pull-down was performed. Precipitated complexes were run on SDS-PAGE followed by Coomassie Blue staining (Fig. 1D). A representative gel shows that only GST fusions containing the WW1 domain are able to precipitate proteins, whereas WW2 and SDR fusions were unable to do so (Fig. 1D). Intriguingly, GST-WW1,2 showed less interaction ability as compared with GST-WW1. Immunoblot analysis validated expression of all domains (Fig. 1D, bottom blot). These data further suggest that the WW1 of WWOX is the main functional interacting domain, at least under the conditions examined in our settings.
The above results prompted us to examine the nature of the WW1-interacting proteins. To this end, HEK293 cells expressing intact WW1 or WW1-WFPA, a specific mutation in the conserved tryptophan and proline within WW domains that should abolish WW domain-interacting ability, were further examined by proteomics. First, we confirmed that GST-WW1-WFPA is expressed in a similar fashion as GST-WW1. Immunoblot and subcellular fractionation analyses revealed comparable levels and compartment localization for both fusions, respectively (Fig. 1E). Next, we examined by Coomassie Blue staining the incompetence of WW1-WFPA in interaction ability. As shown in Fig. 1F, whereas GST-WW1 was able to pull down protein complexes, WW1-WFPA mutant was not. These results further indicate that WW1 interactions are specific and could reflect the putative interacting proteins of WWOX.
Proteomic Screening Identifies Novel, Physiologically Relevant WW Domain-associated Proteins
Proteins that were precipitated by GST-WW1 and GST-WW1-WFPA (Fig. 1F) were then excised from the gel, digested by trypsin, extracted from the gel, and analyzed by LC-MS/MS analysis, and interacting proteins were identified (see “Experimental Procedures”). Precipitations with each GST fusion were repeated at least three times in two independent experiments (see “Experimental Procedures”). To exclude nonspecific interactions, all proteins identified in GST-WW1-WFPA were excluded from the list of GST-WW1-associated proteins. Using these criteria, a total of 240 unique proteins were identified as being precipitated by WW1 but not WW1-WFPA fusion (data not shown). The matched proteins were divided into two sets (supplemental Table S1). MS-Set-1 proteins comprised at least two unique peptides, but at least one passed the identity threshold score (144 proteins (labeled in black) + 96 proteins (labeled in blue)). MS-Set-2 proteins comprised at least two unique peptides that passed the identity threshold score (144 peptides, black). As shown below, putative interaction with WWOX was conformed from both data sets (below). To characterize protein function classes of the WW1-interacting proteins, we performed protein annotation enrichment analysis by using the DAVID comprehensive data set (DAVID version 6.7), which clusters different annotation groups mainly from GO, KEGG, and other databases (31, 32) (see “Experimental Procedures”). Functional annotation and enrichment scores and p values were calculated for the WWOX interacting partners (supplemental Table S2). This analysis showed that the majority of the WWOX-interacting partners are involved in processes such as transcription, RNA processing and splicing, chromatin remodeling, metabolism, and signaling pathways (supplemental Table S2).
WW1 Domain Ligands Are Enriched for Specific WW Domain-binding Motifs
We next examined whether the proteins identified in our MS study are enriched for the different WW domain classes (supplemental Table S1). Because WW domains of WWOX were previously shown to preferentially bind PPXY motifs (7, 14), we next examined whether PPXY motifs are enriched in the WW1-binding proteins in our MS experiment. Here we present data on MS-Set-1, but the same analysis was done on the more stringent data set (MS-Set-2), and we found no major differences (supplemental material and Fig. 2, A and B). Analysis of over 92,000 reference proteins in the human proteome (Ensemble version 64) showed that only ∼5.0% of these proteins contained the PPXY motif (Table 1). In contrast, ∼20% of the proteins isolated in association with the WW1 domain of WWOX in our MS-Set-1 possess one such motif (p = 5.54E−28), a significant enrichment of 4.2-fold (supplemental Tables S1 and S3). Notably, we found a strong enrichment for the LPXY motif (2.47-fold; P = 1.28E−07). We also found enrichment, although less significant compared with (L/P)PXY, for other polyproline motifs belonging to other classes of WW domains, including Class II PPLP (2.61-fold, P = 4.31E−05) and, to a lesser extent, Class III PR (1.28-fold; P = 5.54E−07) and Class IV (S/T)P (1.14-fold; P = 2.37E−05) (supplemental Tables S1 and S4 –S6) (Fig. 2A).
We also observed that proteins containing PPXF (3.12-fold, p = 4.28E−15) and LPXF (1.83-fold, p = 5.0E−04) motifs are enriched in WWOX putative interacting partners (supplemental Tables S1, S7, and S8). There were 12 proteins that contain both LPXY and PPXY motifs among WWOX WW1 domain-interacting proteins (supplemental Table S3, p value ∼0.0001). All proteins that are PPLP-containing also contain PPXY or LPXY motifs (supplemental Table S9). Almost all PPXY or LPXY motifs also contained PR or (S/T)P motifs (supplemental Tables S10 and S11). Taken together, these results suggest that the binding properties of the WW1 domain of WWOX are not limited to PPXY alone but are more comprehensive to include other proline-rich motifs (i.e. LPXY).
Phage Display Profiling Reveals WWOX-WW1 Interaction with LPXY Motifs
To gain further confidence in the identified interactions, we sought to elucidate the binding specificity of WW1 using phage display. Phage display provides an accurate and unbiased in vitro method to study the specificity of modular peptide recognition domains (35, 36). This technology uses bacteriophage to express libraries of up to 10 billion random peptides as genetic fusions to phage coat proteins (33). To this end, phage particles were repeatedly incubated with WW1 domain of WWOX followed by washing away non-interacting phage and subsequent sequencing of the phage-encapsulated DNA. Our analysis revealed that the PY motif, LPXY variant, peptides strongly interacted with the WW1 domain of WWOX (Fig. 2C). Furthermore, we found that WWOX-interacting proteins match the phage-derived profile (when represented as a position weight matrix) much better than random proteins (Fig. 2D, p value 7.56E–12 using rank sum test). Taken together, our MS and phage display data suggest that an extended consensus sequence of (L/P)PXY is recognized by the WWOX WW1 domain.
Validation of WWOX WW1-interacting PPXY-containing Partners
Several of the PPXY-containing proteins identified in our MS study have previously been shown to co-immunoprecipitate with WWOX. For example, our MS analysis revealed that p73 (GI number 2370177) is among the hits identified. p73 was one of the first identified partners of WWOX (17). In fact, it has been shown that WWOX, via its WW1, associates with the PPXY motif of p73 and enhances its proapoptotic function in SaOS2 cells (17). Perhaps due to low protein abundance of p73, we succeeded in identifying only one unique peptide matching this protein (supplemental Table S1). Another known partner of WWOX that we identified in our MS assay was WBP-2 (GI number 4205086, Mascot score 145, matching 10 unique peptides). Recently, it has been shown that the WW1 domain of WWOX physically interacts with WBP1 and WBP2 (37). Our finding of those known partners provided proof-of-concept evidence that our MS experiment successfully identified putative partners of the WW1 domain of WWOX. Nevertheless, the majority of the proteins we identified using MS have not been shown previously to interact with WWOX through its WW1 domain. Therefore, we assessed whether selected proteins identified in our MS analysis associate with full-length WWOX.
DVL2 has recently been reported to bind with WWOX through its SDR domain (38). However, our MS analysis revealed that proteins DVL1 and DVL2 (GI numbers 4758216 and 1706528, Mascot scores 159 and 47, matching 15 and 4 unique peptides, respectively) are candidate interacting partners of WWOX through its WW1 domain. Our validation analysis indeed confirmed WW1, but not mutant WW1-WFPA, interaction with DVL1 (Fig. 3A). Furthermore, we examined whether full-length WWOX associates with DVL1. As shown in Fig. 3B, WWOX is able to precipitate DVL1 when both are co-expressed in HEK293 cells. By contrast, point mutations in the WW1 domain of WWOX and, to a lesser extent, in the PPXY motif in DVL1 attenuated this interaction, thus suggesting that this association is mediated through WW1-PPXY interaction (Fig. 3B). Nevertheless, we cannot exclude the possibility that other motifs could be responsible for this interaction or that the interaction is indirect. To examine whether DVL1 binds the SDR domain of WWOX, we repeated the experiment in the presence of GST-SDR. As shown in Fig. 3C, GST-SDR interacted very weakly with DVL1 as compared with GST-WWOX and GST-WWOX-WFPA, suggesting that WW1 is the main interacting module with DVL1.
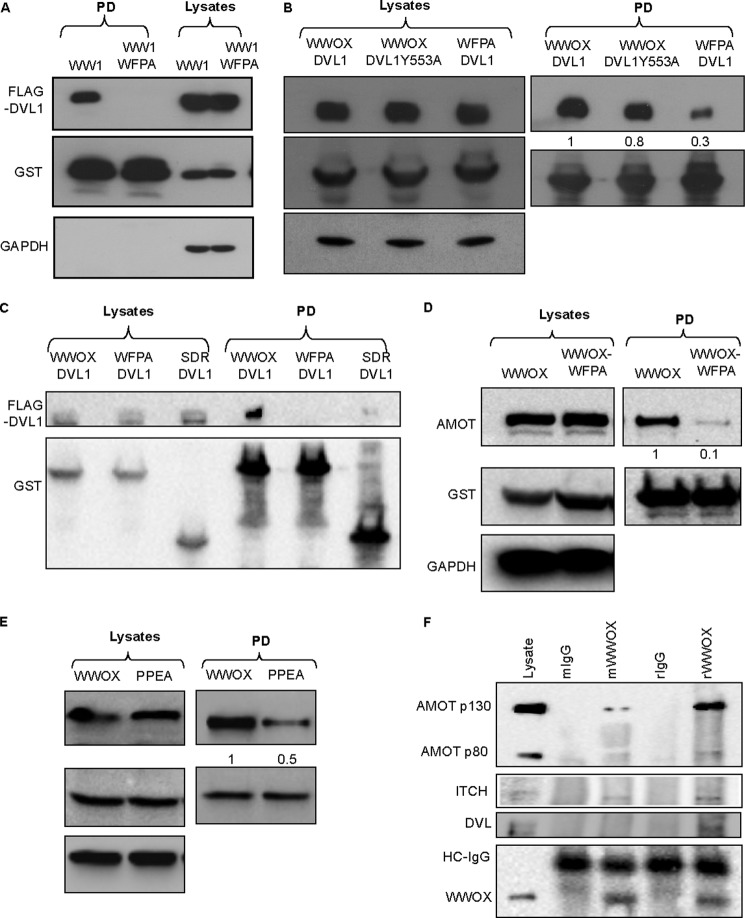
FIGURE 3.
Validation of putative PPXY-containing partners of WWOX identified by MS. A, HEK293 cells were transiently cotransfected with FLAG-DVL1 and GST-WW1 or GST-WW1-WFPA. After 24 h, cells were lysed, and GST pull-down was performed. Precipitates and lysates (input) were immunoblotted with the indicated antibodies. B, HEK293 cells were transiently cotransfected with FLAG-DVL1 or FLAG-DVL1Y553A and GST-WWOX or GST-WWOX-WFPA. After 24 h, cells were lysed, and GST pull-down was performed. Precipitates and lysates (input) were immunoblotted with the indicated antibodies. Numbers below the DVL1 blots represent quantification of bands relative to the GST blot. C, HEK293 cells were transiently cotransfected with FLAG-DVL1 and GST-WWOX or GST-WWOX-WFPA or GST-SDR. Cells were treated as in B. D, HEK293 cells were transiently transfected with FLAG-AMOTL1 and GST-WWOX or GST-WWOX-WFPA. After 24 h, cells were lysed, and GST pull-down was performed. Precipitates and lysates (input) were immunoblotted with the indicated antibodies. Densitometry shows band quantification. E, HEK293 cells were transiently transfected with GST-WWOX and FLAG-AMOTL1 or FLAG-AMOTL1-PPEA. After 24 h, cells were lysed, and GST pull-down was performed. Precipitates and lysates (input) were immunoblotted with the indicated antibodies. Densitometry shows band quantification. F, endogenous interaction. HEK293 cells were lysed and immunoprecipitated using rabbit (r) or mouse (m) anti-WWOX antibody. Precipitates were blotted using anti-WWOX, anti-angiomotin, anti-ITCH, or anti-DVL antibodies. Anti-IgG (r or m) was used as a control.
Another interesting and novel candidate that was revealed from our MS analysis is angiomotin (AMOT) (GI number 63102901, Mascot score 52, matching 2 unique peptides) and angiomotin-like 1 (AMOTL1) (GI number 22027646, Mascot score 99, matching 11 unique peptides) (39, 40). Both AMOT and AMOTL1 contain two PPXY motifs and one LPXY motif and have been shown to associate with the WW domains of YAP, the most downstream effector of the Hippo pathway (28, 41–43). To test whether AMOT associates with WWOX, HEK293 cells were transfected with full-length GST-WWOX or GST-WWOX-WFPA. After 24 h, cells were lysed, and GST-pull-down (PD) was performed. Immunoblot analysis revealed specific interaction between WWOX and endogenous AMOT (Fig. 3D). This interaction was significantly diminished upon expression of WWOX-WFPA. Point mutation in both PPXY motifs of AMOT-L1 significantly diminished physical interaction with WWOX (Fig. 3E).
To validate that these proteins interact under physiological conditions, we examined endogenous protein interaction. Lysates of HEK293 cells were immunoprecipitated using polyclonal or monoclonal anti-WWOX, and complexes were immunoblotted using anti-angiomotin, anti-DVL1, or anti-WWOX antibody. As seen in Fig. 3F, endogenous specific interaction can be detected between WWOX and AMOT (long isoform (p130), which contains PY motifs), DVL1, and ITCH (see below). Of note, the rabbit antibody against WWOX was more efficient than the mouse one in immunoprecipitating the endogenous complexes, probably due to the polyclonality of the antibody. Taken together, these findings indicate that our MS analysis revealed that WWOX, via its WW1 domain, is able to associate with multiple PPXY-containing proteins involved in various cellular networks.
WWOX, via Its WW1 Domain, Physically Associates with ITCH, an LPXY-containing Partner
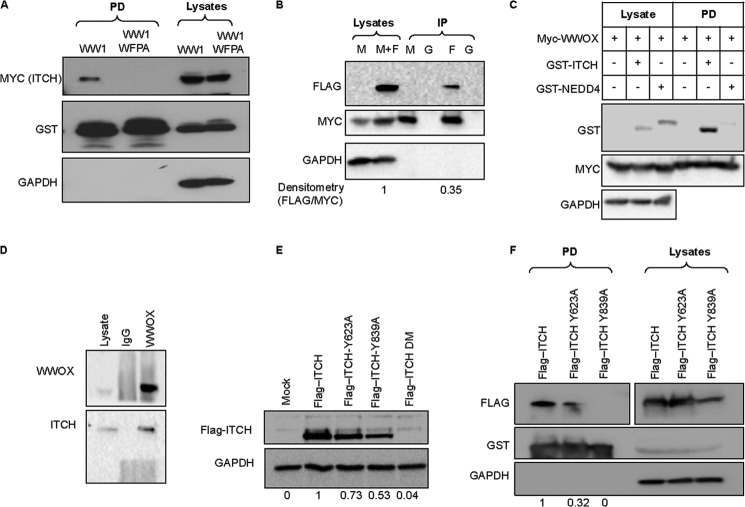
We next set to validate WWOX interaction with the new PY variant, LPXY, which we discovered in this study to associate with the WW1 domain. Among those is ITCH (also known as AIP4) (GI number 27477109, Mascot score 66, matching 7 unique peptides). ITCH, which belongs to the NEDD4 family proteins, is a ubiquitin E3 ligase that contains two LPXY motifs as well as four WW domains (27, 29, 44–48). To validate the WW1 domain of WWOX interaction with ITCH, HEK293 cells were transiently transfected with GST-WW1 or GST-WW1-WFPA and MYC-ITCH. GST pull-down indeed confirmed physical interaction between the WW1 domain, but not WW1-WFPA, and ITCH (Fig. 4A). We next validated that full-length WWOX can immunoprecipitate ITCH in HEK293 cells and found that WWOX can indeed co-immunoprecipitate ∼35% of the expressed ITCH (Fig. 4B). For further validation, we examined whether NEDD4, another related E3 ligase, can interact with WWOX. As shown in Fig. 4C and unlike GST-ITCH, GST-NEDD4 was unable to bind WWOX. Finally, we examined endogenous protein interactions and observed specific complexes between endogenous WWOX and ITCH (Figs. 3F and 4D).
FIGURE 4.
Physical association between ITCH and WWOX. A, HEK293 cells were transiently cotransfected with MYC-ITCH and GST-WW1 or GST-WW1-WFPA. After 24 h, cells were lysed, and GST pull-down was performed. Precipitates and lysates (input) were immunoblotted with the indicated antibodies. B, HEK293 cells were transiently transfected with MYC-WWOX (M) or MYC-WWOX and FLAG-ITCH (M+F). After 24 h, cells were lysed, and immunoprecipitation (IP) using anti-MYC (M), anti-FLAG (F), or anti-IgG (G) was performed. Precipitates and lysates (input) were immunoblotted with antibodies against FLAG (ITCH) or MYC (WWOX). C, HEK293 cells were co-transfected with MYC-WWOX and GST-ITCH or GST-NEDD4. Cells were treated as in A. Densitometry shows band quantification. D, endogenous interaction. Lysates of HEK293 cells were immunoprecipitated with anti-WWOX or anti-IgG, and complexes were immunoblotted with anti-WWOX and anti-ITCH. E, HEK293 cells were transiently cotransfected with the indicated expression vector (top). At 24 h, cell lysates were probed with anti-FLAG (ITCH). GAPDH was used for normalization. DM, double mutant (Y623A,Y839A). Densitometry shows band quantification. F, HEK293 cells were transiently cotransfected with GST-WWOX and the indicated ITCH expression vector (top). At 24 h, cells were lysed, and GST pull-down was performed. Precipitates and lysates (input) were immunoblotted with the indicated antibodies. Densitometry shows band quantification.
To examine whether ITCH binds WWOX via its LPXY motifs, we generated constructs harboring point mutations in the terminal tyrosine of the two LPXY motifs (Y623A and Y839A). FLAG-ITCH-Y623A, FLAG-ITCH-Y839A, and double mutant (ITCH-Y623A,Y839A) vectors were each expressed in HEK293 cells. As seen in Fig. 4E, expression levels of FLAG-ITCH mutants were markedly affected, especially ITCH-Y839A and DM, suggesting that these mutations affect half-life and/or stability of ITCH. In fact, the Tyr-839 mutant lies near the critical Cys residue in the ITCH HECT domain that is crucial for ITCH catalytic function, which might affect ITCH conformation and thus its autoubiquitination (49). Importantly, the ability of GST-WWOX to precipitate FLAG-ITCH-Y623A and FLAG-ITCH-Y839A was significantly diminished when compared with FLAG-ITCH (Fig. 4F). These results indicate that ITCH, via its LPXY motifs, associates with WWOX.
WWOX Ubiquitination Mediated by ITCH
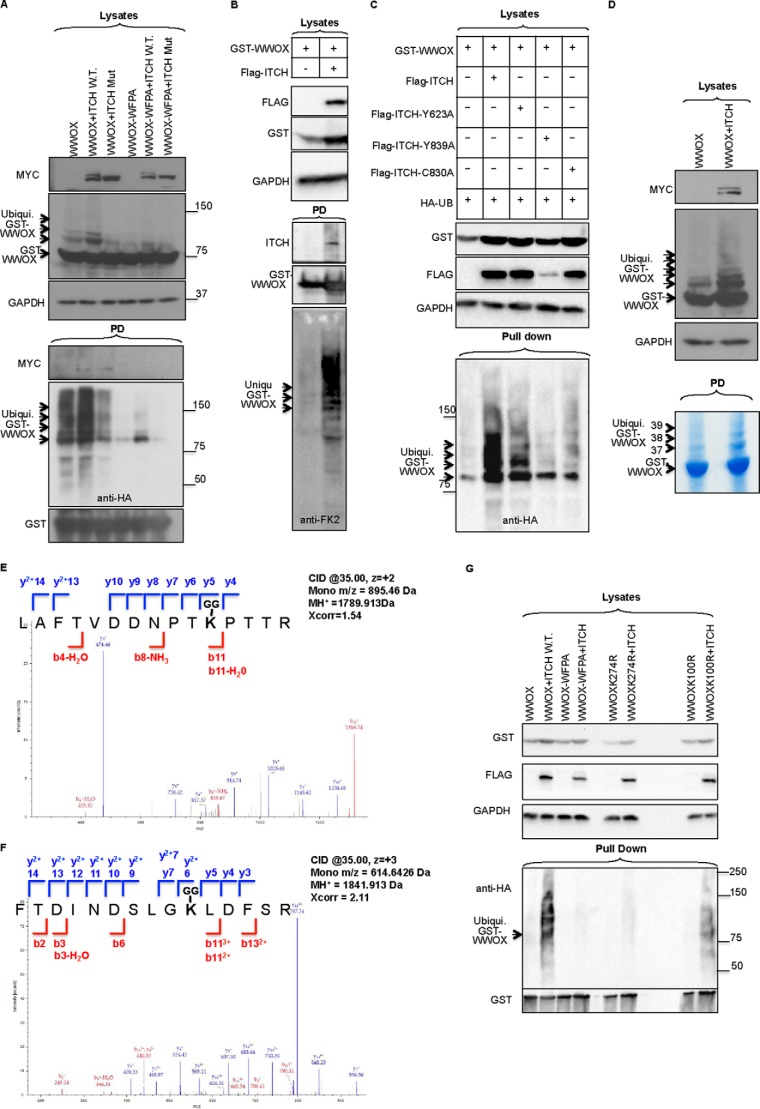
To analyze the functional significance of the interaction between WWOX and ITCH, the role of ITCH in ubiquitination of WWOX was analyzed. Extracts of HEK293 cells transfected with plasmids expressing HA-Ub, GST-WWOX, and FLAG-ITCH or the catalytically inactive FLAG-ITCH-C830A mutant were subjected to GST pull-down followed by immunoblotting. Expression of ITCH increased polyubiquitination of WWOX, unlike the inactive C830A mutant of ITCH, which was still capable of binding WWOX (Fig. 5A). To determine whether ITCH can polyubiquitinate WWOX without overexpression of exogenous ubiquitin, we co-expressed WWOX and ITCH and used a specific antibody (anti-FK2) to detect endogenous ubiquitin. As shown in Fig. 5B, expression of ITCH led to enhancement of polyubiquitination of GST-WWOX, further confirming WWOX as a substrate of ITCH catalytic function.
FIGURE 5.
ITCH mediates polyubiquitination of WWOX. A, HEK293 cells were transiently cotransfected with HA-Ub, GST-WWOX or GST-WWOX-WFPA, and MYC-ITCH (WT) or catalytic mutant (Mut) ITCH (C830A). At 24 h, cells were lysed, and GST pull-down was performed. Lysates and precipitates were immunoblotted using the indicated antibodies. The top blots show lysates (input) using the indicated antibodies. GST-WWOX (∼73.5 kDa) and ubiquitinated GST-WWOX protein are observed and indicated by arrows. In the bottom pull-down (PD) blot, arrows indicate ubiquitinated GST-WWOX protein. B, HEK293T cells were transfected with GST-WWOX and FLAG-ITCH as indicated. At 24 h post-transfection, cells were lysed, and GST pull-down was performed overnight. Lysates and pulled down complexes were detected using the indicated antibodies. Anti-FK-2 antibody was used to detect endogenous ubiquitin. C, HEK293 cells were transiently cotransfected with GST-WWOX, HA-Ub, FLAG-ITCH, or FLAG-ITCH-Y623A, FLAG-ITCH-Y839A, or FLAG-ITCH-C830A. At 24 h, cell lysates were subjected to GST pull-down and subsequently probed with anti-HA-HRP. Lysates were immunoblotted with anti-FLAG and anti-GST, and GAPDH was used for normalization. D, HEK293 cells were transiently cotransfected with HA-Ub, MYC-ITCH, and GST-WWOX. At 24 h, cells were lysed, and GST pull-down was performed. The top blots show lysates (input) using the indicated antibodies. GST-WWOX and ubiquitinated GST-WWOX protein are observed. Bottom, Coomassie Blue staining of self-GST pull-down. Bands were digested and submitted to MS analysis. For all panels, an arrow on the left indicates GST-WWOX, whereas arrows on the right indicate ubiquitinated GST-WWOX protein. Numbers indicate sample ID submitted to MS-MS analysis. E and F, annotated MS/MS spectra of the identified WWOX ubiquitinated peptides is shown for LAFTVDDNPTK100PTTR (C) and FTDINDSLGK274LDFSR (D). The Xcorr scores of these peptides are 1.54 and 2.11, and the MH+ masses are 1789.913 and 1841.913, respectively. Matched b and y ions are shown in red and blue, respectively (E). G, HEK293 cells were transiently cotransfected with HA-Ub, MYC-ITCH (WT), GST-WWOX, GST-WWOX-WFPA, GST-WWOX-K274R, or GST-WWOX-K100R. At 24 h, cells were lysed, and GST pull-down was performed. Lysates and precipitates were immunoblotted using the indicated antibodies.
Next, we examined whether mutation of the LPXY motifs of ITCH affects WWOX ubiquitination. As seen in Fig. 5C, the ITCH-Y623A mutant significantly reduced the potential of ITCH to ubiquitinate WWOX, the same as for ITCH-Y839A, although the expression level of this mutant was very low as compared with wild type, and thus it is hard to make conclusions about the effect of this mutant. Nevertheless, these results may indicate that specific interaction between ITCH, through its LPXY, and WWOX, mediated through its WW1 domain, is responsible for WWOX ubiquitination.
To determine the potential lysine that is ubiquitinated in the WWOX sequence, we expressed GST-WWOX in HEK293 cells, performed GST pull-down, subjected it to SDS-PAGE, and stained with Coomassie Brilliant Blue (Fig. 5D). The WWOX band and those potentially ubiquitinated bands above it were cut and analyzed by MS. MS analysis revealed that those modified bands (Fig. 5D, 37, 38, and 39) are indeed WWOX-ubiquitin conjugates (data not shown). Next, we examined the potential lysine(s) within the WWOX sequence that could be potentially ubiquitinated by ITCH. To do this, we repeated the above experiment and utilized LC-MS/MS technology to identify the post-translation modification on WWOX following expression of ITCH. MS analysis revealed two post-translation modifications associated with ubiquitination (diglycine modification of lysine, 114 Da) and identified two potential lysine residues that are ubiquitinated by ITCH, Lys-100 and Lys-274 (peptides LAFTVDDNPTKPTTR and FTDINDSLGKLDFSR, respectively) (Fig. 5, E and F).
To further confirm that Lys-100 and Lys-274 are ubiquitinated by ITCH, we performed site-directed mutagenesis to replace each lysine with arginine and then examined the effect of WWOX ubiquitination mediated by ITCH. We found that whereas WWOX was ubiquitinated by ITCH, the WWOX-K274R mutant was not as compared with the WWOX-WFPA construct (Fig. 5G). Of note, ITCH could still ubiquitinate the WWOX-K100R mutant, although to a lesser extent. Altogether, these data suggest that ITCH predominantly mediates polyubiquitination of WWOX at Lys-274.
ITCH Mediates Lys-63-linked Ubiquitination of WWOX
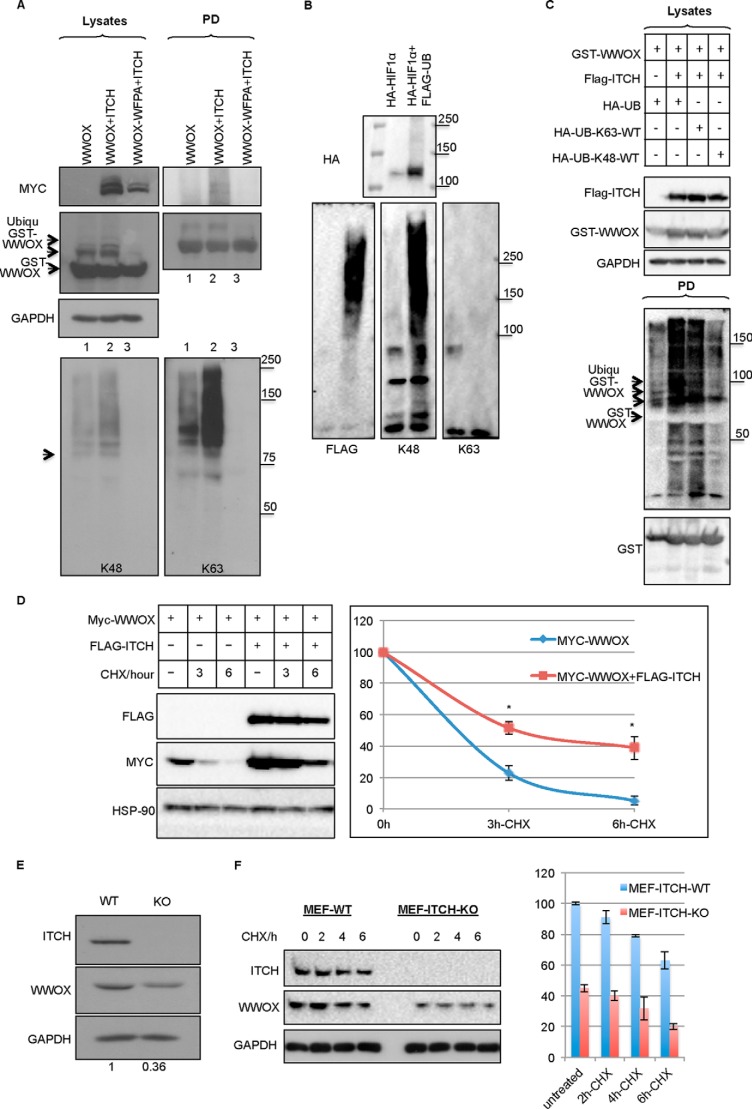
To determine whether ITCH-mediated ubiquitination of WWOX is Lys-48 or Lys-63-linked polyubiquitin, we used specific antibodies that could differentiate between the two modifications (50). We found that ITCH-mediated WWOX ubiquitination is predominantly Lys-63-linked (Fig. 6A). To confirm the validity of these antibodies, we overexpressed HIF1α, a well known protein that undergoes proteasomal degradation that is Lys-48-linked (51). As shown in Fig. 6B, HIF1α ubiquitination was detected using antibodies that recognize Lys-48-linked polyubiquitination but not the antibody that recognizes Lys-63-linked chains. To further confirm that ITCH mediates Lys-63-linked ubiquitination of WWOX, we used HA-Ub constructs that are either Lys-63 only or Lys-48 only (all other lysines are mutated to arginines). Using these constructs, we again demonstrated that ITCH mediates Lys-63-linked polyubiquitination of WWOX (Fig. 6C).
FIGURE 6.
ITCH mediates Lys-63-linked polyubiquitination of WWOX. A, HEK293 cells were transiently cotransfected with HA-Ub, MYC-ITCH, and GST-WWOX or GST-WWOX-WFPA. At 24 h, cells were lysed, and GST pull-down was performed. Lysates and precipitates were immunoblotted using the indicated antibodies. Bottom, immunoblotting with anti-HA (left), anti-Ub-Lys-48 (middle), and anti-Ub-Lys-63 (right). B, HEK293T cells were transfected with HA-HIF1α alone or together with FLAG-Ub. At 24 h post-transfection, cells were treated with 20 μm MG-132 for 4 h. Cells were then lysed, and immunoprecipitation was performed overnight. Lysates and precipitates were detected using anti-FLAG (left), anti-Ub-Lys-48 (middle), and anti-Ub-Lys-63 (right). C, HEK293T cells were transfected with GST-WWOX, FLAG-ITCH, and HA-Ub constructs that are either Lys-63 only or Lys-48 only (all other lysines are mutated to arginines). At 24 h post-transfection, cells were lysed, and GST pull-down was performed overnight. Lysates and pulled down complexes were detected using the indicated antibodies. D and E, ITCH expression is associated with WWOX stabilization. D, HEK293 cells were transiently cotransfected with MYC-WWOX and FLAG-ITCH as indicated at the top. After 24 h, cells were incubated with 20 mg/ml CHX for the indicated times, lysed, and probed with anti-Myc and FLAG antibodies. HSP90 was used for normalization. Quantification of three experiments is shown. p value is 10−5. E, wild-type or Itch-deficient mouse embryonic fibroblasts (MEF) were lysed and immunoblotted with the indicated antibodies. F, wild-type or Itch-deficient mouse embryonic fibroblasts were incubated with 20 mg/ml CHX for the indicated times, lysed, and probed with anti-ITCH and WWOX antibodies. GAPDH was used for normalization. In E and F, densitometry analysis (ratio of WWOX/GAPDH (y axis)) indicates shorter half-life of WWOX in the absence of ITCH. Quantification of three experiments is shown. Data are expressed as mean ± S.E. (error bars), p < 0.001.
Because Lys-48 linkages are mostly associated with commitment for proteasomal degradation, whereas Lys-63-linked polyubiquitination plays established roles in DNA damage repair, protein kinase activation, and trafficking (52), we set out to determine the significance of ITCH-mediated WWOX ubiquitination. To determine whether ITCH-mediated ubiquitination of WWOX leads to WWOX degradation, we analyzed the half-life of WWOX in the presence or absence of ITCH using the protein synthesis inhibitor cycloheximide (CHX). Expression of ITCH in the presence of CHX extended the half-life of WWOX as compared with CHX alone (Fig. 6D). To further validate the significance of WWOX as a target of ITCH, we examined WWOX levels in Itch-knock-out (Itch−/−) mouse embryonic fibroblasts (27). We found that WWOX levels were decreased in Itch−/− mouse embryonic fibroblasts (Fig. 6E). Moreover, WWOX half-life is shorter in the absence of ITCH (Fig. 6F). These results suggest that ITCH-mediated ubiquitination of WWOX is associated with WWOX stabilization.
Functional Relevance of ITCH-WWOX Association
We next determined the functional relevance of WWOX-ITCH association. First, we examined the effect of WWOX-ITCH interaction on their subcellular localization. To address this, we studied the localization of both proteins with the aid of confocal microscopy. ITCH alone or with GFP-WWOX was transiently expressed in HeLa cells. Localization of the FLAG-tagged ITCH and GFP-WWOX was then determined by immunofluorescent staining, as described under “Experimental Procedures.” As shown in Fig. 7A, ITCH alone or WWOX alone localizes in the cytoplasm. Coexpression of WWOX and ITCH resulted in enhanced expression of WWOX and colocalization of WWOX and ITCH in the nucleus (Fig. 7B, top). By contrast, coexpression of ITCHC830A and WWOX and, to a lesser extent, ITCH and WWOX-WFPA did result in nuclear WWOX localization (Fig. 7B, middle and bottom, respectively). These results indicate that expression of ITCH enhances WWOX ubiquitination and translocation into the nucleus, although we cannot exclude the opposite scenario where WWOX also affects ITCH expression and localization.
Second, we investigated the effect of WWOX expression on ITCH-mediated ubiquitination of other target proteins. Previous characterization of ITCH and WWOX partners revealed their association with p73. Whereas ITCH ubiquitinates and degrades p73 (27), WWOX has been shown to enhance p73 proapoptotic function in SaOS2 cells (17). These data prompted us to determine whether coexpression of WWOX and ITCH together with p73 affects its ubiquitination. To this end, HEK293 cells were transiently transfected with MYC-p73α or with HA-Ub together with FLAG-ITCH and/or WWOX. Prior to cell lysis, cells were treated with MG132 for 4 h. Immunoprecipitation using anti-Myc antibody followed by immunoblotting with anti-HA-HRP-conjugated antibody revealed that ITCH enhances p73 polyubiquitination (Fig. 7C). By contrast, coexpression of WWOX significantly reduces ITCH-mediated polyubiquitination of p73. This finding might suggest that WWOX competes with ITCH for p73 interaction and ubiquitination, but it might also indicate that WWOX competes with p73 for interaction with ITCH.
Finally, we set out to determine the functional outcome of WWOX-ITCH coexpression on p73-mediated cell death. Expression of p73 enhances cisplatin-mediated cell death, whereas ITCH reduces this effect (27). Therefore, we aimed to examine whether ITCH-mediated WWOX ubiquitination enhances p73-mediated cisplatin sensitivity. HEK293 cells were transfected with p73, WWOX, or ITCH alone or in combination as indicated. Twenty-four hours later, cells were treated with cisplatin for an additional 24 h, and the relative number of live and dead cells was determined. As shown in Fig. 7D, we found that whereas p73 enhances cisplatin mediated-cell death, expression of ITCH reduces the p73 effect. Importantly, expression of WWOX rescued this later effect of ITCH (Fig. 7D). Altogether, these findings might indicate that WWOX-ITCH interaction renders cells more sensitive to p73 proapoptotic function.
DISCUSSION
To explore the range of cellular processes and ligand-binding preferences of WWOX, we have employed an MS-based screen to identify proteins from HEK293 cells that associate in vivo with individual mammalian GST-WWOX domains. Using this approach, we identified the WW1 domain of WWOX to be the predominant functional interacting domain. Two hundred forty proteins [MS-Set-1] or 144 proteins [MS-Set-2], when using more stringent conditions, were identified. Previous characterization of WWOX partners revealed interacting proteins containing proline-rich sequences typical of peptide ligands for class-I WW domains, mainly PPXY (7, 16). Our current data confirm that many proteins from the MS list are enriched for PPXY motifs and further exposed specific interaction with LPXY PY variant-containing proteins, such as ITCH. The fact that the LPXY motif was identified in our phage display approach further confirms WWOX-specific interaction with PY motifs. Nevertheless, the majority of the pulled down proteins were non-PY-containing motifs. Additionally, PPXF and LPXF motifs (9, 53) were found enriched in our MS study. These findings might suggest that the WW1 domain of WWOX binds non-canonical proline-rich motifs or that these interactions are not direct, although these findings shall be further validated in future studies. Nevertheless, our findings are consistent with a recent report highlighting the plasticity of the first WW domain of YAP in being able to recognize either the canonical PY motif or a phosphorylated serine-proline (pSP) motif, which comprise Class IV of WW domains (54, 55). It is also worth noting that several reports have demonstrated WWOX association with non-PPXY members through its SDR domain, such as hyaluronidase (56), Jnk1 (57), and Tau (58, 59).
Our findings also reveal that the main interacting domain within WWOX is WW1. Interestingly, our data demonstrate that WW2 and SDR domains of WWOX exhibit much less interacting ability, if any. We are aware of the different expression levels of these domains as compared with the GST-WW1 domain. Attempts to enhance the expression level of GST-WW2 and GST-SDR fusions failed (data not shown), suggesting stability- and/or half-life-related behavior. Intriguingly, complexes pulled down via GST-WW1,2 were much less than those by GST-WW1 alone, suggesting that WW2 might have an inhibitory effect on the interaction ability of WWOX. In agreement with these observations, McDonald et al. (37) recently demonstrated that whereas the WW1 domain of WWOX binds to PPXY motifs within WBP1 and WBP2 in a physiologically relevant manner, the WW2 domain exhibits no affinity toward any of these PPXY motifs. This impaired ability was suggested to be due to the presence of Glu-66/Tyr-85 residues within the WW2 domain binding pocket as compared with Arg-25/Trp-44 within the WW1 domain. Introduction of an E66R/Y85W double substitution within the WW2 domain of WWOX results in gain of function and restores binding to WBP1 and WBP2 (37). Furthermore, the Y85W substitution within the WW2 domain restores its binding to PPXY motifs of ErbB4 in a manner very similar to the binding of the WW1 domain of WWOX. Altogether, these data indicate that WW1 domain is the predominant interacting module of WWOX and that the WW2 atypical nature explains its failure to mediate protein-protein interaction (60). Future analysis shall further decipher the significance of WW2 domain of WWOX for its interacting ability.
Although some of the interacting proteins in our analysis have been described as binding partners for specific WW domain proteins, most of these putative binding partners have not been previously identified. We therefore further examined the interaction of the WW1 domain of WWOX with LPXY-containing ITCH as a novel functional protein-protein interaction. We found that WWOX directly binds to the ubiquitin E3 ligase ITCH in a fashion that requires the functional WW1 domain of WWOX and the PY motif in ITCH. As a functional outcome, ITCH ubiquitinates WWOX independent of degradation. In fact, we observed that coexpression of WWOX and ITCH stabilizes WWOX levels and enhances p73-mediated cell death. We also show that ITCH stimulates Lys-63-linked ubiquitination of WWOX, and this is associated with subtle nuclear localization of WWOX. Because Lys-63-linked polyubiquitination plays established roles in DNA damage response, protein kinase activation, and trafficking (52), we assume that WWOX translocation into the nucleus might be associated with novel functions that remain to be identified. Nevertheless, WWOX translocation into the nucleus was previously suggested, such as in the case of RUNX2 (21) and p53 (61), and could facilitate specific interactions related to cell cycle regulation and transcription. Our MS analysis for the specific lysine that is ubiquitinated by ITCH revealed Lys-100 and Lys-274 as candidate targets. Indeed, we confirmed that Lys-274 and, to a much lesser extent, Lys-100 are direct targets of ITCH catalytic function. Intriguingly, Mahajan et al. (62) observed that full-length WWOX but not a truncated form of WWOX that lacks the C terminus, WWOXΔ5–8, is polyubiquitinated and degraded. Intriguingly, exons 5–8 contain the SDR domain, which harbors Lys-274 that is Lys-63-linked polyubiquitinated by ITCH. Whether Lys-274 is the same lysine in the WWOX C terminus that also targets WWOX for degradation is not known and would be of great interest to determine.
ITCH, which contains four WW domains, has many substrates, and its function has been shown to require intact WW domains. ITCH promotes the ubiquitination and degradation of several transcription factors, such as c-Jun and Jun-B (63), Notch (64), p73 (27), p63 (29), ErbB4 (46, 48), Gli (47), and proteins involved in cell death regulation, such as c-FLIP (44, 45). Specific ITCH interactions with its partners typically involve its WW domains and PY motifs within its substrates.
Recently, we showed that WWOX binds the terminal PY motif of the p73 transcription factor and demonstrated that this functional interaction promotes apoptosis (17). Interestingly, ITCH, via its WW domains, also associates with the same PY motif of p73, and this interaction causes the enhanced ubiquitination and degradation of p73 (27). It has been shown previously that WW domain proteins can compete with each other to determine the functional outcome of their interacting partners (19, 20, 65, 66). In a recent report, we demonstrated that WWOX competes with ITCH for binding ΔNp63α and that WWOX attenuates ITCH-mediated degradation of ΔNp63α (19). In fact, we also observed a physical interaction between WWOX and ITCH, perhaps releasing ΔNp63α from ITCH and mediating its stabilization. We thus assumed that WWOX and ITCH might also compete for interaction with p73 and protect it from degradation. Indeed, when coexpressed with ITCH and p73, WWOX was able to reduce ITCH-mediated p73 ubiquitination. A proposed model is that both WW domain-containing proteins, ITCH and WWOX, in the cytoplasm are competing for interaction with PPXY-containing target proteins. WWOX interaction with ITCH releases p73 where ITCH can ubiquitinate WWOX and affects its localization and function, leading to enhanced cell death. This model also suggest that WWOX can tune the function of ITCH in different cell contexts (i.e. when WWOX is absent as in the case of many cancer cells, ITCH has more access to its partners, for example to p73, rendering cancer cells more chemoresistant) (67).
The spectrum of proteins identified in our MS analysis suggests that WWOX binds an extensive network of protein-protein interaction in vivo. Our findings revealed that WWOX signaling could affect critical pathways in cancer, including the Hippo tumor suppressor pathway and Wnt-β-catenin. The MS analysis further revealed a number of multiprotein complexes with central functions involved in transcription, splicing, adhesion, ubiquitination, and chromatin remodeling. It is likely that WW1 domain of WWOX serves to bridge or regulate such cellular processes, thus potentially functioning to establish networks of interactions. It remains to be seen how different physiological contexts and stimuli affect WWOX-interacting ability. In conclusion, our experiments show that the WW1 domain of WWOX associates with multiprotein cellular complexes, many of which have not been described previously.
Supplementary Material
Acknowledgments
We are grateful to all members of the Aqeilan laboratory for technical help and fruitful discussion, to Dr. Iddo Friedberg (Miami University, Miami, OH) for assistance in the protein annotation analysis, and to Alexandra Eliassaf (Hebrew University-Hadassah Medical School) for assistance with the MS data analyses.
This work was supported in part by funds from Israel Science Foundation Grant 12-542 and German Israel Foundation Grant 1101-60.11/2010 (to R. I. A.); Pennsylvania Breast Cancer Coalition Grants 60707 and 920093 and the Geisinger Clinic (to M. S.); and the Lejwa Fund for Biochemistry (to D. R.).

This article contains supplemental Tables S1–S11.
- PY
- proline-tyrosine-rich
- WWOX
- WW domain-containing oxidoreductase
- WW1 and WW2
- first and second WW domain, respectively
- Ub
- ubiquitin
- CHX
- cycloheximide
- SDR
- short-chain dehydrogenase domain.
REFERENCES
- 1. Bork P., Sudol M. (1994) The WW domain: a signalling site in dystrophin? Trends Biochem. Sci. 19, 531–533 [DOI] [PubMed] [Google Scholar]
- 2. Sudol M., Bork P., Einbond A., Kastury K., Druck T., Negrini M., Huebner K., Lehman D. (1995) Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 270, 14733–14741 [DOI] [PubMed] [Google Scholar]
- 3. Sudol M., Chen H. I., Bougeret C., Einbond A., Bork P. (1995) Characterization of a novel protein-binding module: the WW domain. FEBS Lett. 369, 67–71 [DOI] [PubMed] [Google Scholar]
- 4. Macias M. J., Hyvönen M., Baraldi E., Schultz J., Sudol M., Saraste M., Oschkinat H. (1996) Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382, 646–649 [DOI] [PubMed] [Google Scholar]
- 5. Sudol M. (1996) Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65, 113–132 [DOI] [PubMed] [Google Scholar]
- 6. Sudol M., Recinos C. C., Abraczinskas J., Humbert J., Farooq A. (2005) WW or WoW: the WW domains in a union of bliss. IUBMB Life 57, 773–778 [DOI] [PubMed] [Google Scholar]
- 7. Salah Z., Alian A., Aqeilan R. I. (2012) WW domain-containing proteins: retrospectives and the future. Front. Biosci. 17, 331–348 [DOI] [PubMed] [Google Scholar]
- 8. Chen H. I., Sudol M. (1995) The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H. I., Einbond A., Kwak S. J., Linn H., Koepf E., Peterson S., Kelly J. W., Sudol M. (1997) Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J. Biol. Chem. 272, 17070–17077 [DOI] [PubMed] [Google Scholar]
- 10. Kang H. S., Beak J. Y., Kim Y. S., Herbert R., Jetten A. M. (2009) Glis3 is associated with primary cilia and Wwtr1/TAZ and implicated in polycystic kidney disease. Mol. Cell. Biol. 29, 2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M. S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., Meyerson M. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang N. S., Hsu L. J., Lin Y. S., Lai F. J., Sheu H. M. (2007) WW domain-containing oxidoreductase: a candidate tumor suppressor. Trends Mol. Med. 13, 12–22 [DOI] [PubMed] [Google Scholar]
- 13. Del Mare S., Salah Z., Aqeilan R. I. (2009) WWOX: its genomics, partners, and functions. J. Cell. Biochem. 108, 737–745 [DOI] [PubMed] [Google Scholar]
- 14. Gardenswartz A., Aqeilan R. I. (2014) WW domain-containing oxidoreductase's role in myriad cancers: clinical significance and future implications. Exp. Biol. Med. 239, 253–263 [DOI] [PubMed] [Google Scholar]
- 15. Paige A. J., Taylor K. J., Taylor C., Hillier S. G., Farrington S., Scott D., Porteous D. J., Smyth J. F., Gabra H., Watson J. E. (2001) WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc. Natl. Acad. Sci. U.S.A. 98, 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salah Z., Aqeilan R., Huebner K. (2010) WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 6, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aqeilan R. I., Pekarsky Y., Herrero J. J., Palamarchuk A., Letofsky J., Druck T., Trapasso F., Han S. Y., Melino G., Huebner K., Croce C. M. (2004) Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc. Natl. Acad. Sci. U.S.A. 101, 4401–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ludes-Meyers J. H., Kil H., Bednarek A. K., Drake J., Bedford M. T., Aldaz C. M. (2004) WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene 23, 5049–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salah Z., Bar-mag T., Kohn Y., Pichiorri F., Palumbo T., Melino G., Aqeilan R. I. (2013) Tumor suppressor WWOX binds to ΔNp63α and sensitizes cancer cells to chemotherapy. Cell Death Dis. 4, e480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aqeilan R. I., Donati V., Palamarchuk A., Trapasso F., Kaou M., Pekarsky Y., Sudol M., Croce C. M. (2005) WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 65, 6764–6772 [DOI] [PubMed] [Google Scholar]
- 21. Aqeilan R. I., Hassan M. Q., de Bruin A., Hagan J. P., Volinia S., Palumbo T., Hussain S., Lee S. H., Gaur T., Stein G. S., Lian J. B., Croce C. M. (2008) The WWOX tumor suppressor is essential for post-natal survival and normal bone metabolism. J. Biol. Chem. 283, 21629–21639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aqeilan R. I., Palamarchuk A., Weigel R. J., Herrero J. J., Pekarsky Y., Croce C. M. (2004) Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2γ transcription factor. Cancer Res. 64, 8256–8261 [DOI] [PubMed] [Google Scholar]
- 23. Gaudio E., Palamarchuk A., Palumbo T., Trapasso F., Pekarsky Y., Croce C. M., Aqeilan R. I. (2006) Physical association with WWOX suppresses c-Jun transcriptional activity. Cancer Res. 66, 11585–11589 [DOI] [PubMed] [Google Scholar]
- 24. Jin C., Ge L., Ding X., Chen Y., Zhu H., Ward T., Wu F., Cao X., Wang Q., Yao X. (2006) PKA-mediated protein phosphorylation regulates ezrin-WWOX interaction. Biochem. Biophys. Res. Commun. 341, 784–791 [DOI] [PubMed] [Google Scholar]
- 25. Hu H., Columbus J., Zhang Y., Wu D., Lian L., Yang S., Goodwin J., Luczak C., Carter M., Chen L., James M., Davis R., Sudol M., Rodwell J., Herrero J. J. (2004) A map of WW domain family interactions. Proteomics 4, 643–655 [DOI] [PubMed] [Google Scholar]
- 26. O'Keefe L. V., Colella A., Dayan S., Chen Q., Choo A., Jacob R., Price G., Venter D., Richards R. I. (2011) Drosophila orthologue of WWOX, the chromosomal fragile site FRA16D tumour suppressor gene, functions in aerobic metabolism and regulates reactive oxygen species. Hum. Mol. Genet. 20, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossi M., De Laurenzi V., Munarriz E., Green D. R., Liu Y. C., Vousden K. H., Cesareni G., Melino G. (2005) The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 24, 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oka T., Schmitt A. P., Sudol M. (2012) Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene 31, 128–134 [DOI] [PubMed] [Google Scholar]
- 29. Rossi M., Aqeilan R. I., Neale M., Candi E., Salomoni P., Knight R. A., Croce C. M., Melino G. (2006) The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. U.S.A. 103, 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim T., Tyndel M. S., Huang H., Sidhu S. S., Bader G. D., Gfeller D., Kim P. M. (2012) MUSI: an integrated system for identifying multiple specificity from very large peptide or nucleic acid data sets. Nucleic Acids Res. 40, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 33. Tonikian R., Zhang Y., Boone C., Sidhu S. S. (2007) Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2, 1368–1386 [DOI] [PubMed] [Google Scholar]
- 34. Aqeilan R. I., Donati V., Gaudio E., Nicoloso M. S., Sundvall M., Korhonen A., Lundin J., Isola J., Sudol M., Joensuu H., Croce C. M., Elenius K. (2007) Association of Wwox with ErbB4 in breast cancer. Cancer Res. 67, 9330–9336 [DOI] [PubMed] [Google Scholar]
- 35. Tong A. H., Drees B., Nardelli G., Bader G. D., Brannetti B., Castagnoli L., Evangelista M., Ferracuti S., Nelson B., Paoluzi S., Quondam M., Zucconi A., Hogue C. W., Fields S., Boone C., Cesareni G. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295, 321–324 [DOI] [PubMed] [Google Scholar]
- 36. Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., Boone C., Bader G. D., Sidhu S. S. (2008) A specificity map for the PDZ domain family. PLoS Biol. 6, e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDonald C. B., Buffa L., Bar-Mag T., Salah Z., Bhat V., Mikles D. C., Deegan B. J., Seldeen K. L., Malhotra A., Sudol M., Aqeilan R. I., Nawaz Z., Farooq A. (2012) Biophysical basis of the binding of WWOX tumor suppressor to WBP1 and WBP2 adaptors. J. Mol. Biol. 422, 58–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouteille N., Driouch K., Hage P. E., Sin S., Formstecher E., Camonis J., Lidereau R., Lallemand F. (2009) Inhibition of the Wnt/β-catenin pathway by the WWOX tumor suppressor protein. Oncogene 28, 2569–2580 [DOI] [PubMed] [Google Scholar]
- 39. Troyanovsky B., Levchenko T., Månsson G., Matvijenko O., Holmgren L. (2001) Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J. Cell Biol. 152, 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bratt A., Wilson W. J., Troyanovsky B., Aase K., Kessler R., Van Meir E. G., Holmgren L. (2002) Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene 298, 69–77 [DOI] [PubMed] [Google Scholar]
- 41. Salah Z., Aqeilan R. I. (2011) WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan S. W., Lim C. J., Chong Y. F., Pobbati A. V., Huang C., Hong W. (2011) Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 286, 7018–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bernassola F., Karin M., Ciechanover A., Melino G. (2008) The HECT family of E3 ubiquitin ligases. Multiple players in cancer development. Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 45. Melino G., Gallagher E., Aqeilan R. I., Knight R., Peschiaroli A., Rossi M., Scialpi F., Malatesta M., Zocchi L., Browne G., Ciechanover A., Bernassola F. (2008) Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 15, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 46. Sundvall M., Korhonen A., Paatero I., Gaudio E., Melino G., Croce C. M., Aqeilan R. I., Elenius K. (2008) Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc. Natl. Acad. Sci. U.S.A. 105, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Di Marcotullio L., Greco A., Mazzà D., Canettieri G., Pietrosanti L., Infante P., Coni S., Moretti M., De Smaele E., Ferretti E., Screpanti I., Gulino A. (2011) Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene 30, 65–76 [DOI] [PubMed] [Google Scholar]
- 48. Omerovic J., Santangelo L., Puggioni E. M., Marrocco J., Dall'Armi C., Palumbo C., Belleudi F., Di Marcotullio L., Frati L., Torrisi M. R., Cesareni G., Gulino A., Alimandi M. (2007) The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J. 21, 2849–2862 [DOI] [PubMed] [Google Scholar]
- 49. Scialpi F., Malatesta M., Peschiaroli A., Rossi M., Melino G., Bernassola F. (2008) Itch self-polyubiquitylation occurs through lysine-63 linkages. Biochem. Pharmacol. 76, 1515–1521 [DOI] [PubMed] [Google Scholar]
- 50. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 51. Weidemann A., Johnson R. S. (2008) Biology of HIF-1α. Cell Death Differ. 15, 621–627 [DOI] [PubMed] [Google Scholar]
- 52. Chen Z. J., Sun L. J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 53. Sudol M., Harvey K. F. (2010) Modularity in the Hippo signaling pathway. Trends Biochem. Sci. 35, 627–633 [DOI] [PubMed] [Google Scholar]
- 54. Aragón E., Goerner N., Xi Q., Gomes T., Gao S., Massagué J., Macias M. J. (2012) Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-β pathways. Structure 20, 1726–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sudol M. (2012) WW domains in the heart of Smad regulation. Structure 20, 1619–1620 [DOI] [PubMed] [Google Scholar]
- 56. Chang N. S., Pratt N., Heath J., Schultz L., Sleve D., Carey G. B., Zevotek N. (2001) Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J. Biol. Chem. 276, 3361–3370 [DOI] [PubMed] [Google Scholar]
- 57. Chang N. S., Doherty J., Ensign A. (2003) JNK1 physically interacts with WW domain-containing oxidoreductase (WOX1) and inhibits WOX1-mediated apoptosis. J. Biol. Chem. 278, 9195–9202 [DOI] [PubMed] [Google Scholar]
- 58. Sze C. I., Su M., Pugazhenthi S., Jambal P., Hsu L. J., Heath J., Schultz L., Chang N. S. (2004) Down-regulation of WW domain-containing oxidoreductase induces Tau phosphorylation in vitro. A potential role in Alzheimer's disease. J. Biol. Chem. 279, 30498–30506 [DOI] [PubMed] [Google Scholar]
- 59. Wang H. Y., Juo L. I., Lin Y. T., Hsiao M., Lin J. T., Tsai C. H., Tzeng Y. H., Chuang Y. C., Chang N. S., Yang C. N., Lu P. J. (2012) WW domain-containing oxidoreductase promotes neuronal differentiation via negative regulation of glycogen synthase kinase 3β. Cell Death Differ. 19, 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schuchardt B. J., Bhat V., Mikles D. C., McDonald C. B., Sudol M., Farooq A. (2013) Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase. Biochemistry 52, 9223–9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chang N. S., Doherty J., Ensign A., Schultz L., Hsu L. J., Hong Q. (2005) WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J. Biol. Chem. 280, 43100–43108 [DOI] [PubMed] [Google Scholar]
- 62. Mahajan N. P., Whang Y. E., Mohler J. L., Earp H. S. (2005) Activated tyrosine kinase Ack1 promotes prostate tumorigenesis. Role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 65, 10514–10523 [DOI] [PubMed] [Google Scholar]
- 63. Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275 [DOI] [PubMed] [Google Scholar]
- 64. Qiu L., Joazeiro C., Fang N., Wang H. Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y. C. (2000) Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275, 35734–35737 [DOI] [PubMed] [Google Scholar]
- 65. Levy D., Adamovich Y., Reuven N., Shaul Y. (2007) The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 14, 743–751 [DOI] [PubMed] [Google Scholar]
- 66. Conforti F., Sayan A. E., Sreekumar R., Sayan B. S. (2012) Regulation of p73 activity by post-translational modifications. Cell Death Dis. 3, e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hansen T. M., Rossi M., Roperch J. P., Ansell K., Simpson K., Taylor D., Mathon N., Knight R. A., Melino G. (2007) Itch inhibition regulates chemosensitivity in vitro. Biochem. Biophys. Res. Commun. 361, 33–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.