Background: The pathogenic mechanisms of aniridia-genitourinary anomalies-mental retardation (AGR) syndrome are obscure.
Results: Depletion of Lgr4 in mouse leads to aniridia, polycystic kidney disease, genitourinary anomalies, and mental retardation, similar to the pathological defects of AGR syndrome.
Conclusion: Lgr4 is a candidate gene for the pathogenesis of AGR syndrome.
Significance: Understanding the mechanisms of the pathogenesis of AGR syndrome.
Keywords: CREB, G-protein-coupled Receptors (GPCR), Histone Methylation, Kidney, Ovary, AGR Syndrome, Fbxl10, Jmjd2a, LGR4, Histone Demethylase
Abstract
AGR syndrome (the clinical triad of aniridia, genitourinary anomalies, and mental retardation, a subgroup of WAGR syndrome for Wilm's tumor, aniridia, genitourinary anomalies, and mental retardation) is a rare syndrome caused by a contiguous gene deletion in the 11p13–14 region. However, the mechanisms of WAGR syndrome pathogenesis are elusive. In this study we provide evidence that LGR4 (also named GPR48), the only G-protein-coupled receptor gene in the human chromosome 11p12–11p14.4 fragment, is the key gene responsible for the diseases of AGR syndrome. Deletion of Lgr4 in mouse led to aniridia, polycystic kidney disease, genitourinary anomalies, and mental retardation, similar to the pathological defects of AGR syndrome. Furthermore, Lgr4 inactivation significantly increased cell apoptosis and decreased the expression of multiple important genes involved in the development of WAGR syndrome related organs. Specifically, deletion of Lgr4 down-regulated the expression of histone demethylases Jmjd2a and Fbxl10 through cAMP-CREB signaling pathways both in mouse embryonic fibroblast cells and in urinary and reproductive system mouse tissues. Our data suggest that Lgr4, which regulates eye, kidney, testis, ovary, and uterine organ development as well as mental development through genetic and epigenetic surveillance, is a novel candidate gene for the pathogenesis of AGR syndrome.
Introduction
Hereditary syndromes are caused by genetic abnormalities, such as chromosome microdeletions. WAGR2 syndrome (also named 11p deletion syndrome, consisting of Wilm's tumor, aniridia, genitourinary anomalies, and mental retardation), is a severe and hereditary genetic disease (1, 2). Previous clinical data show that individuals with WAGR syndrome may have variant syndromes combined with the diseases listed above, and about half of WAGR syndrome patients have Wilm's tumor (3). This syndrome is invariably accompanied by a constitutional deletion of all or part of chromosome 11p12–11p14 (including 11p13 deletion in all reported cases), which harbors dozens of genes (4–6). AGR syndrome is a subgroup of WAGR syndrome in which patients do not develop Wilm's tumor. Deletion of chromosome 11p14.1-p13 associates with AGR syndrome (7). Haploinsufficiency of PAX6 causes aniridia, and deletion of the WT1 gene predisposes toward Wilm's tumor, genital abnormality, and nephropathies. These two genes have been identified to be critical in the pathogenesis of WAGR syndrome (8, 9). Recent studies reported that deletion of BDNF (brain-derived neurotrophic factor), which is present in 11p14.1, is attributable to the obesity found in WAGR syndrome (2). 11p14.1 microdeletions are associated with attention-deficit hyperactivity disorder, autism, developmental delay, and obesity in humans (10). PRRG4 (transmembrane γ-carboxyglutamic acid protein 4) and SLC1A2 (solute carrier family 1 member 2), which have been reported to be deleted in some WAGR syndrome patients, may be implicated in autism (4). The molecular mechanisms of the pathogenesis of WAGR syndrome are largely unknown.
G-protein-coupled receptor-48 (Gpr48, also named Lgr4), is a glycoprotein hormone receptor with leucine-rich-repeat domains at the N terminus (11, 12). The LGRs are present very early in evolution, and members share high homology. Up-regulation of Lgr4 has been found to promote cancer cell invasion and metastasis (13). Lgr4/Gpr48 is broadly expressed in diverse organs, including the eye, kidney, testis, ovary, uterus, and brain and plays multiple physiological roles in these organ systems (14). Lgr4 has been implicated in postnatal development (15), integrity of male reproductive tracts (4, 16, 17), and renal development (18). R-spondin(s) is reported to interact with Lgr4/Gpr48 and mediates a Wnt/β-catenin signaling pathway through associated frizzled Lrp5/6 complexes (19, 20). R-spondin potentiates Wnt/β-catenin signaling through LGR4 and LGR5 (21). Norrin is also a ligand for Lgr4 (22). A nonsense point mutation (c.376C>T (p.R126X)) in the LGR4 gene is associated with low bone mineral density, osteoporotic fractures, electrolyte imbalance, late onset of menarche, reduced testosterone levels, and an increased risk of squamous cell carcinoma of the skin and biliary tract cancer in humans (23). Recently, Lgr4 has been identified to play roles in bone formation and remodeling (24), innate immunity (25), mammary stem cells (26), and prostate development (27). However, whether inactivation of Lgr4/Gpr48 plays a role in AGR syndrome is unknown.
Abnormal gene transcriptional regulation contributes to diverse developmental disorders and diseases (28). Multiple processes, such as histone modification, are involved in developmental gene expression regulation (29). Histone (de)methylation is critical in regulating gene activation, silencing, and epigenetic memory (30, 31). Histone demethylases play important roles in various cellular procedures, including apoptosis and cell proliferation (31). Histone demethylase Jmjd2a (also called JHDM3A) is a JmjC histone demethylase that catalyzes the demethylation of di- and trimethylated Lys-9 and Lys-36 in histone H3 (H3K9me2/3 and H3K36me2/3) (32, 33). Jmjd2a depletion increases the histone methylation of nuclear H3K9me3 and H3K36me3 and triggers germ line apoptosis in Caenorhabditis elegans (34). Histone demethylase Jmjd2a is involved in the repression of transcription factor ASCL2 (achaete scute-like homologue 2, also named Hash2) (35) and is also implicated in the regulation of androgen receptor (36). More recently, Jmjd2a has been reported to function in muscle development, and its mRNA is present in oocytes and throughout bovine embryonic developmental stages in vitro, suggesting multiple roles of Jmjd2a in development (37). Fbxl10 (also named Kdm2b or Jhdm1b) is a histone H3 lysine 36 (H3K36) demethylase that regulates cell proliferation and senescence through p15Ink4b (also named as Cdkn2b) (38). However, the role of histone methylation in mediating Lgr4 signaling or a role for histone demethylases in AGR has not been previously reported.
In this study we investigated the roles of Lgr4 in AGR syndrome related aniridia, reproductive system function, and mental retardation in mice as well as Lgr4 functions in regulating histone demethylase expression. We found that LGR4 is the only GPCR gene in human chromosome 11p and that inactivation of Lgr4 not only associates with defects in eye, kidney, testis, ovary, and uterine organ development but also leads to significant mental retardation in mice. We demonstrated that Lgr4 mediates expression of multiple genes critical for organ development through genetic and epigenetic surveillance. Our data suggest that LGR4 may be a novel candidate gene for the pathogenesis of AGR syndrome.
EXPERIMENTAL PROCEDURES
Generation of Lgr4 Gene Null Mice, Antibodies, Plasmids, Cells, and Reagents
The Lgr4 gene trap ES cell clone (LST020, the gene trapping site is located in the intron between the first and second exons) was obtained from William Skarnes (Bay Genomics), and Lgr4-null ES cell clones were injected into C57BL/6 blastocysts and transferred to ICR female mice. Male chimeric mice were mated with C57BL/6 females, resulting in transmission of the inserted allele to the germ line. Positive mice were interbred and maintained on a mixed 129 × C57BL/6 background. Genotyping analyses and identification of wild type and Lgr4 deletion homozygotes were performed as previously described (39). Anti-p-CREB, anti-CREB, and anti-KI-67 antibodies were ordered from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Jmjd2a antibody was ordered from Abcam® (Cambridge, MA). Anti-Fbxl10 antibodies for immunostaining were ordered from LifeSpan Biosciences (Seattle, WA) and for the Western blot were from Santa Cruz Biotechnology. Lgr4, Lgr4T755I, and CREB overexpression plasmids were made by our laboratory (39). ApopTag® Peroxidase In Situ Apoptosis Detection kit was ordered from Chemicon (Millipore, Billerica, MA). LipofectamineTM was ordered from InvitrogenTM. Histostain-Plus kit (DAB) was ordered from Invitrogen®.
Immunohistochemistry and Immunofluorescence Staining
The immunohistochemistry assays were performed as previously described (40). For the DAPI and anti-Jmjd2a (1:1000 dilution) immunofluorescence assays, most procedures were the same as described above. Images were taken by a ZEISS Axioskop 40 fluorescent microscope.
Apoptosis Assay
Apoptosis was analyzed in 1-month-old tissues using the ApopTag® Peroxidase in Situ Apoptosis Detection kit (Chemicon) according to manufacturer's instructions (39).
Morris Water Maze Assay
The Morris water maze was performed as described previously with some modifications (41). Mice were trained with three training trials per day for 6 consecutive days with fixed starting and escape platform locations. Then the escape platform was hidden 1 cm under the water surface in the same location of the pool as during the training trials. Mice were released into the pool from a constant starting location. Training trials ended when the mouse was on the platform or 60 s had elapsed, whichever came first. Mice remained for 15 s on the platform before they were removed from the pool. Training trials were given in blocks of two spaced about 90 min apart. Spatial learning was assessed with a probe trial (during which the platform was removed from the pool) given after completion of training. Analysis of variance and t tests were used for analysis of data (quadrant occupancy, target crossings). After initial analysis of variance analysis, specificity of searching was determined by comparing target quadrant measures to average measures of the other quadrants (paired t test).
Open Field Test
The open field test was conducted as previously described with some modifications (41). The apparatus was a square arena (length × width × height = 48 × 35 × 27 cm), and the “center” field was defined as the inner central 20 × 16-cm area (about 19% of the total area). Mice were individually placed in a constant corner and allowed to explore for 10 min, and behavior was video recorded with a Kodak Easyshare z740 Zoom Digital camera. Time spent in the center area was accounted according to the video record.
Elevated Plus Maze Assay
The plus maze assay was performed as described previously with some modifications (41). The plus maze cross was made with X-Acto foam board. The top surfaces of the arms and the top walls of the closed arms were made with dark brown linerboard. A small raised lip (0.5 cm, dark brown linerboard) around the edges of the open arms prevented animals from slipping off. The two open arms (length × width = 67 × 7 cm) and two closed arms (length × width × height = 67 × 7× 18 cm) formed a cross shape with the two open arms opposite to each other. The maze was 41 cm above the floor level. The mouse was placed in the center square, facing a closed arm, and allowed to freely explore the apparatus for 5 min. Behavior was video recorded with a Kodak Easyshare z740 Zoom Digital camera. Time spent in the open arms and the number of entries into the open arms (an open arm entry was defined as all four paws in an open arm) was scored according to the video record.
RT-PCR and Real-time Quantitative-PCR
Total RNAs were extracted from P0 mouse tail or the indicated organs for RT-PCR with Lgr4 primers, 5′-AGTGCTTTGCAGTCTCTACGC-3′ (sense) and 5′-GAAGATGCAGCACTACCAAGC-3′ (antisense), or for real-time quantitative PCR assays with the indicated gene specific primers. Three independent quantitative PCR experiments were performed, and results are shown as mean ± S.D. (n = 3).
Western Blotting and Luciferase Assay
Proteins were extracted from wild type and Lgr4−/− MEF cells, and a Western blotting assay was performed with anti-Jmjd2a and Fbxl10 antibodies (1:1000 dilution). The DNA fragment of −996 ∼ +20 bp of mouse Jmjd2a was amplified and inserted into pGL3 basic vector between KpnI and SacI sites with the primers 5′-ACGTAGGTACCGCGGTGACACTGCAATCCACTA-3′ (sense) and 5′-TACGTGAGCTCAGCCCATAGTCAACCAACCCAA-3′ (antisense). The DNA fragment of −128 ∼ +20 bp of mouse Jmjd2a was amplified and inserted into pGL3 basic vector between KpnI and SacI sites as a control with the primers 5′-ACGTAGGTACCGGAGGCTCAGCGTTTTCTC-3′ (sense) and 5′-TACGTGAGCTCAGCCCATAGTCAACCAACCCAA-3′ (antisense). The DNA fragment of −1 kb ∼ +20 bp of mouse Fbxl10 was amplified and inserted into pGL3 basic vector between MluI and XhoI sites with the primers 5′- ACGTAACGCGTGGTTAGAAGGGCTGCCCAGAGAAT-3′ (sense) and 5′-TACGTCTCGAGGCATAACTTTTAAACTCCCGGGGC-3′ (antisense). The DNA fragment of −136 ∼ +20 bp of mouse Fbxl10 was amplified and inserted into pGL3 basic vector between MluI and XhoI sites as a control with the primers 5′-ACGTAACGCGTGTACTACCGAGGCTATCCGAATG-3′ (sense) and 5′-TACGTCTCGAGGCATAACTTTTAAACTCCCGGGGC-3′ (antisense). Indicated constructs were transfected into HEK293 cells with LipofectamineTM (InvitrogenTM), and luciferase activity was measured using the luciferase assay system (Promega, Madison, WI) with Top Count Microplate Scintillation Counter (Canberra, Meriden, CT). For the fetal bovine serum (FBS) series concentration treatment, plasmid-transfected HEK293 cells were cultured with DMEM without FBS overnight followed by treatment with 0, 0.1, 1, or 10% FBS for 18 h, and luciferase activity was measured.
Statistics
Unless otherwise indicated, results are the mean ± S.D. p values were determined using two-tailed Student's t test. A p value less than 0.05 was considered significant.
Study Approval
Animal studies conformed to the principles for laboratory animal research outlined by the Animal Welfare Act (NIH/Department of Health and Human Services), and the use of animals was approved by the Institutional Animal Care and Use Committee of Texas A&M University System Health Science Center.
RESULTS
Deletion of Lgr4 Leads to Defects in Iris Development and Aniridia in Mouse Eyes
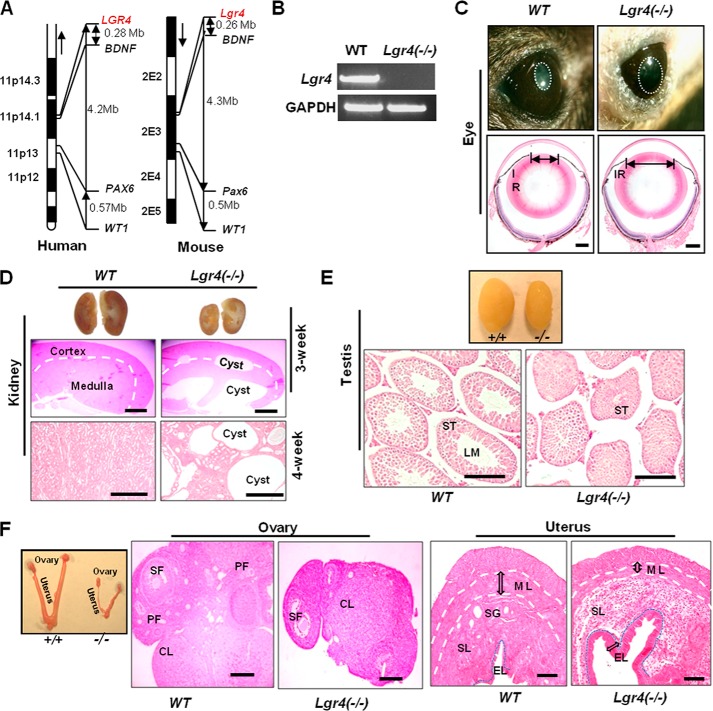
We found that LGR4 is the only GPCR in the “11P deletion” chromosome fragment (11p12–11p14) and positioned ∼4 megabases away from the WAGR syndrome pathogenesis genes WT1 and PAX6 and only ∼0.2 megabases from BDNF (Fig. 1A). To investigate whether Lgr4 is a candidate gene in the pathogenesis of WAGR syndrome, we generated Lgr4-null mice (Fig. 1B). To evaluate the effects of Lgr4 deletion on iris development, we examined pupil formation by examining wild type and Lgr4−/− mouse eyes under strong light (n = 12 in each group). Smaller pupils appeared in the eyes of each wild type mouse in response to strong light, whereas 72% of Lgr4−/− mice did not show pupil constriction under the same conditions (Fig. 1C, top). The remaining 28% of Lgr4−/− mice had cataract diseases and had no iris response to strong light. To confirm these observations, we examined the pupil diameters of these abnormal Lgr4−/− eyes by H&E. We observed significantly wider diameter pupils in Lgr4−/− eyes than in wild type eyes (Fig. 1C, bottom), suggesting that deletion of Lgr4 leads to defects in iris development. The aniridia found in Lgr4−/− mouse eyes suggest that Lgr4 inactivation leads to defects in iris development.
FIGURE 1.
Lgr4 deletion leads to aniridia, polycystic kidney disease, and reproductive defects in mice. A, genetic loci of Lgr4, BDNF, Pax6, and WT1 in synteny chromosome fragments of human 11P and mouse 2E. Mb, megabases. B, RT-PCR results of Lgr4 expression in P0 wild type and Lgr4 depletion mouse tails. C, 1-month-old wild type and Lgr4−/− mouse eyes under strong light (top) demonstrated a significantly bigger pupil (white circle) in Lgr4−/− mouse eyes. H&E-stained samples of indicated mouse eyes showed a bigger pupil diameter in Lgr4−/− eyes (bottom). D, deletion of Lgr4 induces polycystic kidney disease. Cross-section of 3-week-old wild type and Lgr4−/− kidneys, with cysts in the Lgr4−/− kidney (top). The middle panel is a magnified H&E stained section of the top panel. H&E-stained 4-week-old wild type and Lgr4−/− kidney sections showed polycystic disease in the Lgr4−/− kidney (bottom). Representative photos are of n = 9 mice. E, Lgr4−/− mice present testicular development deficits. Representative 4-week-old wild type and Lgr4−/− testes are shown (top). H&E-stained sections (bottom) come from the top panel and show smaller seminiferous tubules and lumen formation defects in Lgr4−/− mice. F, representative 4-week-old wild type and Lgr4−/− ovary and H&E-stained samples are shown, with dramatically smaller size and fewer follicles in Lgr4−/− ovary. Lgr4−/− uterus had a significantly thinner muscle layer, almost no secretory glands, enhanced inner epithelial layer and abnormal stromal layer structure. IR, iris; LM, lumen; ST, seminiferous tubule; PF, primary follicle; SF, secondary follicle; CL, corpus luteum; ML, muscle layer; SG, secretory gland; SL, stroma layer; EL, inner epithelial layer. Scale bar = 20 μm.
Deletion of Lgr4 Results in Polycystic Kidney Disease in Mice
To examine the roles of Lgr4 in kidney development, 3-week-old (n = 5 in each group) and 4-week-old (n = 9 in each group) mouse kidneys were examined. We observed that all Lgr4−/− kidneys were much smaller than those of wild type kidneys (Fig. 1D, top). Two of the 3-week-old (Fig. 1D, top and middle) and 4 of the 4-week-old (Fig. 1D, bottom) Lgr4−/− kidneys, but none of the wild type kidneys, presented severe polycystic kidney disease (PKD), suggesting that Lgr4 inactivation results in high risk of polycystic kidney disease (PKD frequency ≈ 44.4% at the 1-month stage). There were no Wilm's tumors in all examined kidneys (data not shown). The wide distribution and high expression level of Lgr4 in kidneys (12), the smaller size of Lgr4−/− mouse kidneys, and the high risk of severe PKD disease in Lgr4−/− mouse kidneys indicates that Lgr4 plays a critical role in the development and function of the kidney.
Lgr4 Inactivation Leads to Defects in Reproductive System Development in Mice
To investigate the roles of Lgr4 in male reproductive system development, 4-week-old wild type and Lgr4−/− testes were examined (n = 12 in each group). All Lgr4−/− testes were smaller than wild type testes (Fig. 1E, top). Each wild type testis had lumens in the seminiferous tubules, whereas Lgr4−/− seminiferous tubule lumens were significantly smaller or even absent (Fig. 1E, bottom). We further tracked 3- and 6-week old testes; smaller seminiferous tubules and lumens were present in all Lgr4−/− testes. Over a study period of two years, we observed that all Lgr4−/− male mice were infertile (n > 60). The lumen supplies a pathway to deliver sperm cells to the collection tubule and finally to the epididymis, which is important for spermatogenesis. The dramatically delayed or arrested lumen formation and seminiferous tubule development in Lgr4−/− testes together with the absolute infertility of Lgr4−/− mice suggest that Lgr4 deletion disrupts male reproductive system development and function.
To examine the roles of Lgr4 in the female reproductive system, the ovaries and uteruses of 4-week-old wild type and Lgr4−/− mice were investigated (n = 12 in each group). All Lgr4−/− ovaries were much smaller than wild type (Fig. 1F), and the number of primary follicles and secondary follicles was significantly reduced in Lgr4−/− ovaries (Fig. 1F). All Lgr4−/− uteruses presented strikingly thinner smooth muscle layers and almost no secretory glands in the dilated stromal layers (Fig. 1F). Within a 2-year-long study period, Lgr4−/− female mice were found to be absolutely infertile (n > 50). These data indicate that Lgr4 deletion associates with defects in female mouse genitalia, and Lgr4 is required for female reproductive system function. Collectively, these results indicate that Lgr4 inactivation associates with reproductive system defects in mice.
Deletion of Lgr4 Decreases Learning Ability and Anti-anxiety Ability in Mice
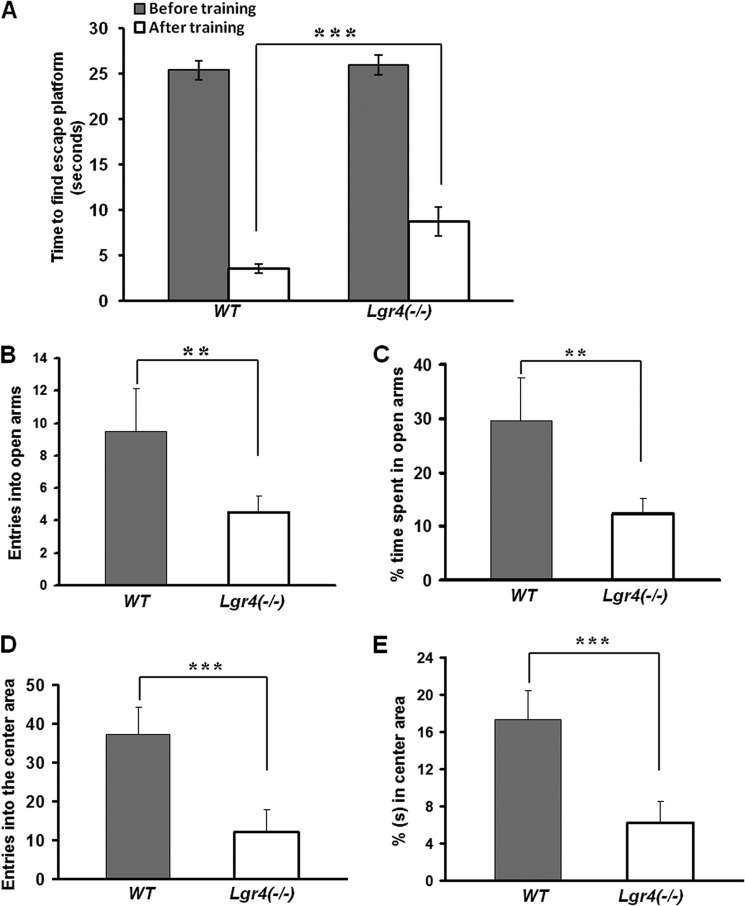
As spatial memory learning ability and anti-anxiety ability are two important aspects in mental development and maturation (41), we then investigated the spatial memory ability and the anti-anxiety ability in 4-week-old wild type and Lgr4−/− mice. In the Morris water maze assays, Lgr4−/− mice took almost the same time to reach the escape platform as wild type mice in the first training trial (n = 6 in each group). After being trained for six consecutive days with three training trials per day, the wild type and Lgr4−/− mice were tested, and the time spent was recorded. The average time of Lgr4−/− mice after training was 8.8 ± 1.6 s, more than that of wild type mice with 3.6 ± 0.5 s (Fig. 2A), indicating that the spatial memory learning ability decreased in Lgr4−/− mice. We further examined the anti-anxiety ability using the elevated plus maze assay (Fig. 2, B and C) and open field assay (Fig. 2, D and E). We observed that the anti-anxiety ability was significantly decreased in Lgr4−/− mice (Fig. 2, B–E). Together, these data suggest that Lgr4 deletion contributes to mental retardation. As obesity is present in some (W)AGR syndrome patients (2), we also examined the potential for obesity in Lgr4−/− mice but found no obesity in Lgr4−/− mice at 4 weeks, 6 weeks, and 6 months of age (n = 6 per group at each age, data not shown). Taken together, Lgr4 deletion leads to aniridia, polycystic kidney disease, reproductive defects, and mental retardation in mice.
FIGURE 2.
Lgr4 deletion results in mental retardation. A, wild type and Lgr4−/− mice that took almost the same amount of time to find and climb the escape platform in the first training trial were tested in the Morris water maze assay (n = 6 in each group). After training trails, wild type mice used 3.6 ± 0.5 s to find the escape platform, whereas Lgr4−/− mice required 8.8 ± 1.6 s. B and C, wild type and Lgr4−/− mice (n = 6 for each group) were tested in the elevated plus maze assay. The average entries into open arms in 5 min of wild type versus Lgr4−/− mice were 9.5 ± 2.6 versus 4.5 ± 1 (B). The average time percentage spent in open arms in 5 min was 29.6 ± 8% for wild type versus 12.3 ± 2.8% for Lgr4−/− mice (C). D and E, wild type and Lgr4−/− mice (n = 6 for each group) were tested in the open field assay. The average entries into the open area in 10 min were 37.3 ± 7.8 for wild type versus 12 ± 5.8 for Lgr4−/− mice (D). The average time percentage spent in open arms in 10 min was 17.3 ± 3.2% for wild type versus 6.2 ± 2.6% for Lgr4−/− mice (E) (mean ± S.D., n = 6). **, p < 0.05; ***, p < 0.01.
Lgr4 Inactivation Increases Cell Apoptosis in Genitourinary Systems
Previous studies reported that Lgr4 regulates epithelial cell proliferation (42). To understand the underlying molecular mechanisms of the above multiple defects in WAGR syndrome-related organs, we examined the roles of Lgr4 in cell apoptosis in the tissues of these organs. We performed apoptosis assays in the cross-sections of 4-week-old kidney, testis, ovary, and uterus. We observed that apoptosis significantly increased in Lgr4−/− kidney (Fig. 3, A and B), testis (Fig. 3, C and D), ovary (Fig. 3, E and F), and uterus (Fig. 3, G and H) compared with that in wild type. Together, these data suggest that Lgr4 deletion suppresses cell proliferation and increases cell apoptosis in mouse genitourinary systems.
FIGURE 3.

Lgr4 inactivation increases cell apoptosis in multiple organs. A and B, apoptosis was dramatically increased in Lgr4−/− kidney. Representative 4-week-old wild type and Lgr4−/− kidney samples are shown. C and D, increased apoptosis was found in Lgr4−/− testis. Representative 4-week-old wild type and Lgr4−/− testis samples are shown. E and F, significantly increased apoptosis was present in Lgr4−/− ovary corpus luteum. Representative 4-week-old wild type and Lgr4−/− ovary samples are shown. G and H, increased apoptosis appeared in the Lgr4−/− uterus. Representative 4-week-old wild type and Lgr4−/− uterus samples are shown. Cell proliferation was decreased in spermatogonia and Leydig cells in Lgr4−/− testis. Scale bar = 10 μm. ST, seminiferous tubule; GC, granulosa cell; CL, corpus luteum; L, stroma layer.
Lgr4 Regulates the Expression of Multiple Key Genes in the Eye and the Genitourinary Systems
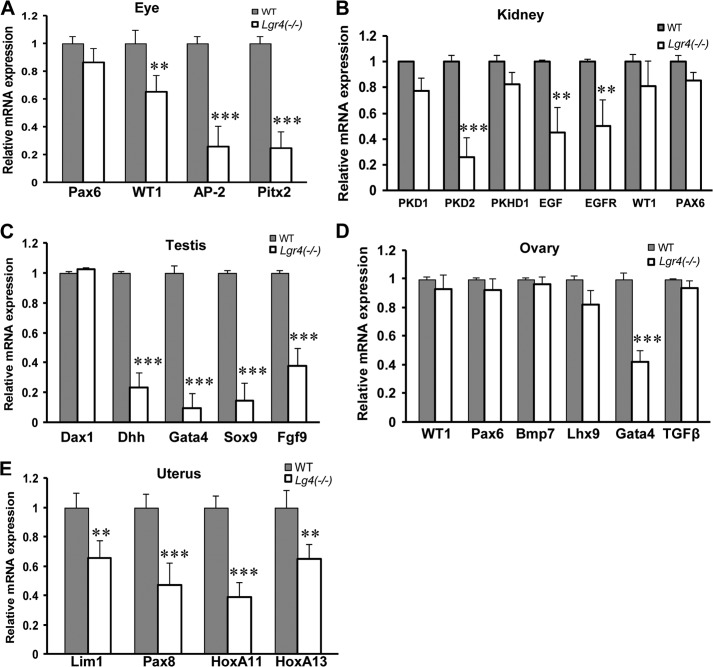
To explore the underlying molecular mechanisms of the above observations, we employed real-time PCR to investigate the transcription changes of multiple genes important in the development of the eye, kidney, and genitourinary system, including Pax6 and WT1. In accord with our previous findings (39), we observed that deletion of Lgr4 decreased the expression levels of Pitx2 in the eye (Fig. 4A). In addition, Lgr4−/− mice had decreased expression of AP-2 (43, 44) in the eye, PKD2 (45), EGF (46), EGFR (47) in the kidney (Fig. 4B), Dhh (48), Gata4 (49, 50), Sox9 (51), and Fgf9 (52) in the testis (Fig. 4C), Gata4 (53, 54) in the ovary (Fig. 4D), and Lim1 (55), Pax8 (56), and HoxA11 (57) in the uterus (Fig. 4E). Together, these data indicate that Lgr4 is broadly implicated in the transcriptional regulation of a wide range of key genes in WAGR syndrome-related organs.
FIGURE 4.
Lgr4 depletion reduces mRNA expression of multiple genes in eye, kidney, testis, ovary, and uterus. Total mRNAs were extracted from 4-week-old wild type and Lgr4−/− mouse eye (A, Lgr4−/− eye with aniridia), kidney (B, Lgr4−/− kidney with polycystic kidney disease), testis (C), ovary (D), and uterus (E). The average results shown come from three independent real time PCR analyses of three individual mice (mean ± S.D., n = 3). **, p < 0.05; ***, p < 0.01.
Lgr4 Is Implicated in Transcriptional Regulation of Multiple Histone Demethylases in Vitro
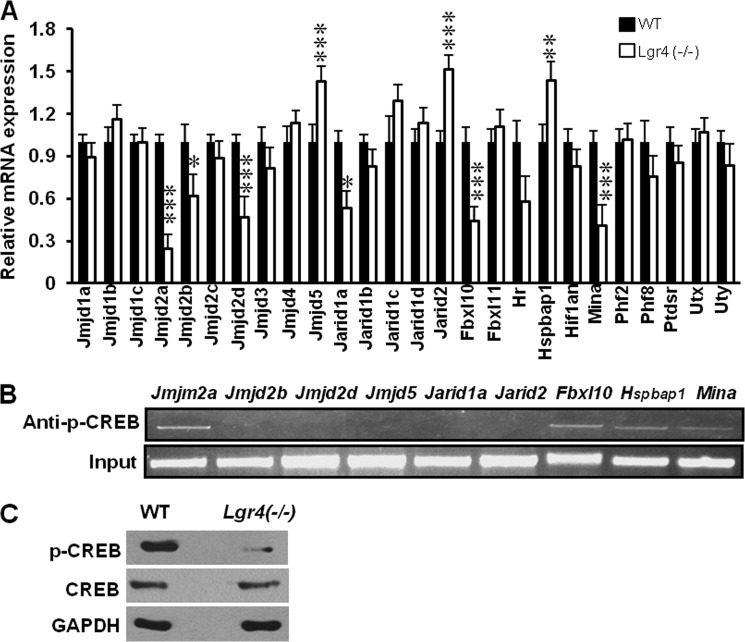
Histone demethylases demethylate nuclear histones and have been implicated in regulating transcription of a vast spectrum of genes (31). G-protein-coupled receptors regulate a wide range of cellular responses through genetic and epigenetic means including histone (de)methylation surveillance (58–60). To examine whether Lgr4 plays a role in the regulation of histone demethylases and, therefore, modulates the transcription of multiple genes in (W)AGR syndrome-related organs, we screened the mRNA levels of all 26 mouse Jumonji C (JMJC)-family histone demethylases in both wild type and Lgr4−/− mouse embryonic fibroblast (MEF) cells. We observed that Lgr4 deletion led to a significant decrease in mRNA expression of Jmjd2a, Jmjd2d, Fbxl10, and Mina and significantly increased mRNA expression of Jmjd5, Jarid2, and Hspbap1 (Fig. 5A). Previous studies have reported that Lgr4 signals through the adenylate cyclase-cAMP-CREB signaling pathway (24). We then analyzed the promoter regions of the above down-regulated histone demethylase genes in Lgr4 deletion mutant mice using the Genomatix web site and found that all of these genes have a potential CREB binding site at −1 kb in their promoter regions (data not shown). Using the chromatin immunoprecipitation (CHIP) assay, we screened for phospho-CREB binding to the promoter regions. We found that p-CREB bound to the promoter regions of the histone demethylases Jmjd2a, Fbxl10, Hspbap1, and Mina (Fig. 5B). In addition, Lgr4 depletion decreased CREB phosphorylation (Fig. 5C). These data suggest that Lgr4 might regulate the transcription of these histone demethylases through the cAMP/CREB pathway.
FIGURE 5.
Lgr4 inactivation affects mRNA expression of multiple histone demethylases. A, total RNA was extracted from wild type and Lgr4−/− MEF cells and analyzed by real-time PCR assays. The average results of three independent real-time PCR assays of histone demethylase genes are shown (mean ± S.D., n = 3). *, p < 0.1, **, p < 0.05, ***, p < 0.01. B, CHIP and quantitative PCR screening were performed with anti-p-CREB antibody and gene promoter-specific primers (about the −1-kb promoter region of each gene). The quantitative PCR products were loaded in an agarose gel and separated by electrophoresis, and the images are shown. C, Western blot analyses of p-CREB in wild type and Lgr4−/− MEF cells.
Lgr4 Regulates Histone Demethylase Jmjd2a in Vitro and in Vivo
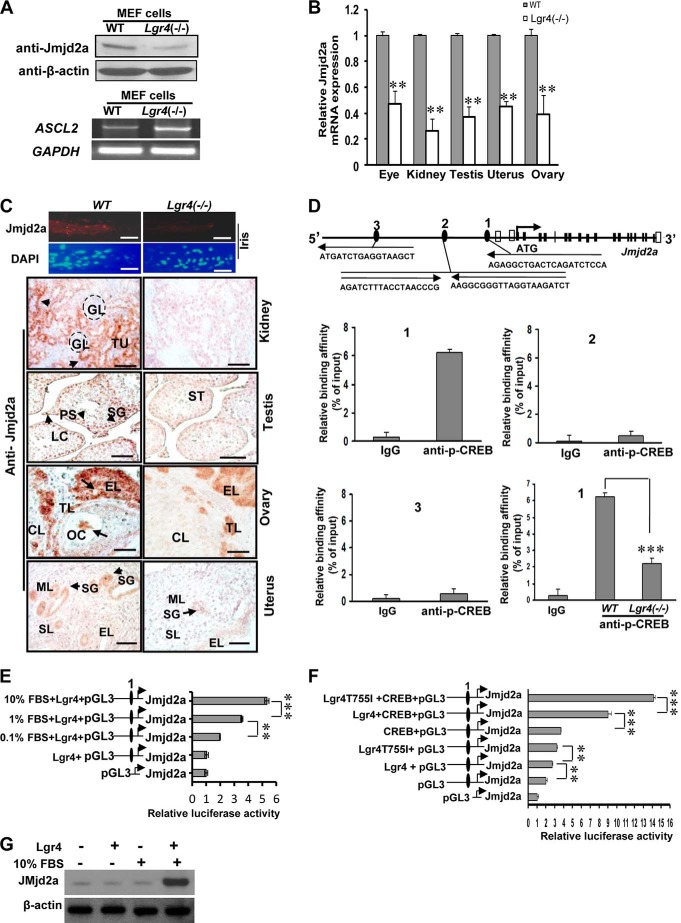
Previous studies show that Jmjd2a (34) and Fbxl10 (38) are important for cell proliferation and apoptosis, whereas the functions of Mina and Hspbap1 are yet to be determined. Here we focused our investigation on the effects of Lgr4 on Jmjd2a and Fbxl10. Western blot analysis of wild type and Lgr4−/− MEF cells indicated that Lgr4 loss down-regulates Jmjd2a in MEF cells (Fig. 6A, top). The transcription factor ASCL2 is a target of the histone demethylase Jmjd2a, and its transcription is repressed by Jmjd2a (36, 38). We then tracked the mRNA level of ASCL2 in wild type and Lgr4−/− MEF cells. As expected, we observed that ASCL2 expression is increased in Lgr4−/− MEF cells (Fig. 6A, bottom). To examine whether Lgr4 deletion reduced the histone demethylase Jmjd2a mRNA level in vivo, tissue-specific total mRNAs were extracted and analyzed by real time PCR. Histone demethylase Jmjd2a mRNA expression is consistently reduced in Lgr4−/− eye, kidney, testis, uterus, and ovary (Fig. 6B). Next, we performed fluorescence immunostaining and immunohistochemistry to evaluate the expression levels of Jmjd2a in wild type and Lgr4−/− tissues. We detected Jmjd2a in the stromal cells and stromal muscle cells of the iris, in the tubule and duct epithelial cells of the kidney, in Leydig cells, spermatogonia and primary spermatocytes in the testes, in the epithelial cells and theca lutein cells in the ovary, and in the epithelial cells of uterine secretory glands of wild type mice (Fig. 6C, left). Meanwhile, we observed a significant decrease of Jmjd2a expression in Lgr4−/− iris, kidney, testis, and ovary (Fig. 6C, right), suggesting that Lgr4 regulates the expression of histone demethylase Jmjd2a in these (W)AGR syndrome-related organs in mice.
FIGURE 6.
Lgr4 regulates histone demethylase Jmjd2a expression through CREB binding. A, Lgr4−/− MEF cells have decreased Jmjd2a protein expression (top) and increased ASCL2 mRNA expression (bottom). B, Jmjd2a mRNA expression was decreased in the indicated tissues of Lgr4−/− mice. C, Lgr4 deletion decreased Jmjd2a protein expression in multiple tissues of 4–6-week-old mice (5 mice for each group). The top four panels show images of the iris tip with anti-Jmjd2a (top) and DAPI (bottom) immunofluorescence. In the bottom eight panels Jmjd2a signals (arrows) were detected in the indicated wild type tissues by immunohistochemistry. Kidney: GL, glomerulus; TU, tubule. Testis: SG, spermatogonia; ST, seminiferous tubule; PS, primary spermatocyte; LC, Leydig cell. Ovary: CL, corpus luteum; GC, granulosa cell; TL, theca lutein cell; EL, epithelial cell; OC, oocyte. Uterus: SG, secretory gland; ML, muscle layer; SL, stromal layer; EL, inner epithelial layer. D, diagram of the mouse Jmjd2a gene (NP_759014) and the −1-kb upstream region. Black boxes, coding exons; empty boxes, non-coding exons; numbered black ellipses, potential CREB sites. Arrows show the direction from 5′ to 3′. The black letters present the minimum CREB site sequences. CHIP and Q- PCR assays were performed in MEF cells. P-CREB bound at the Jmjd2a promoter CREB site 1, and the affinity decreased in Lgr4−/− MEF cells. E, increased FBS concentrations elevate Jmjd2a promoter (containing CREB site 1)-driven luciferase activity. HEK293 cells transfected with plasmids were cultured in DMEM without FBS or with FBS at indicated concentrations. F, Lgr4 enhances CREB-dependent Jmjd2a promoter-induced luciferase activity in HEK293 cells with 10% FBS (mean ± S.D., n = 3). Reporter assay of the indicated Jmjd2a promoter regions driving luciferase expression, co-transfected into HEK293 cells with expression plasmids for CREB, Lgr4, and/or Lgr4(T755I) (a constitutively active Lgr4 mutant (39)). G, overexpression of Lgr4 with FBS elevates cellular Jmjd2a levels in HEK293 cells. After plasmid transfection and culture overnight without FBS, HEK293 cells were cultured with or without 10% FBS for 24 h. Western blot assays were performed using β-actin as a loading control. Scale bar = 20 μm. **, p < 0.05; ***, p < 0.01.
Lgr4 Regulates Histone Demethylase Jmjd2a Transcription via CREB
There are three potential CREB binding sites in the −1-kb region of the histone demethylase Jmjd2a promoter (Fig. 6D). We tested the p-CREB binding affinity of regions 1, 2, and 3 using CHIP and quantitative PCR assays. We found that region 1 showed relatively strong binding affinity for p-CREB (Fig. 6D, 1, top left), indicating that region 1 of the mouse histone demethylase Jmjd2a promoter contains a p-CREB binding site. We then compared the binding affinity of p-CREB on region 1 between wild type and Lgr4−/− MEF cells and observed decreased binding affinity in Lgr4−/− MEF cells compared with that in wild type (Fig. 6D, 1, lower right), suggesting that Lgr4 may regulate histone demethylase Jmjd2a transcription through the PKA/CREB pathway. To verify this observation, we performed luciferase assays using the promoter region of Jmjd2a. Increased concentrations of FBS elevate Lgr4-dependent histone demethylase Jmjd2a promoter activity (Fig. 6E), indicating that FBS contains Lgr4 ligand(s). Furthermore, we investigated the CREB effects on Lgr4-dependent Jmjd2a promoter activity with multiple co-transfection combinations as indicated in Fig. 6F. Overexpression of Lgr4 with FBS promoted CREB-induced histone demethylase Jmjd2a promoter activity and overexpression of the constitutively active-mutant form of Lgr4 further enhanced Jmjd2a promoter activity (Fig. 6F). In addition, overexpressing Lgr4 with FBS elevated cellular Jmjd2a levels (Fig. 6G), indicating that activated Lgr4 up-regulates Jmjd2a expression. Taken together, these data demonstrate that Lgr4 regulates histone demethylase Jmjd2a transcription through the cAMP-CREB signaling pathway.
Lgr4 Regulates Histone Demethylase Fbxl10 in Vitro and in Vivo
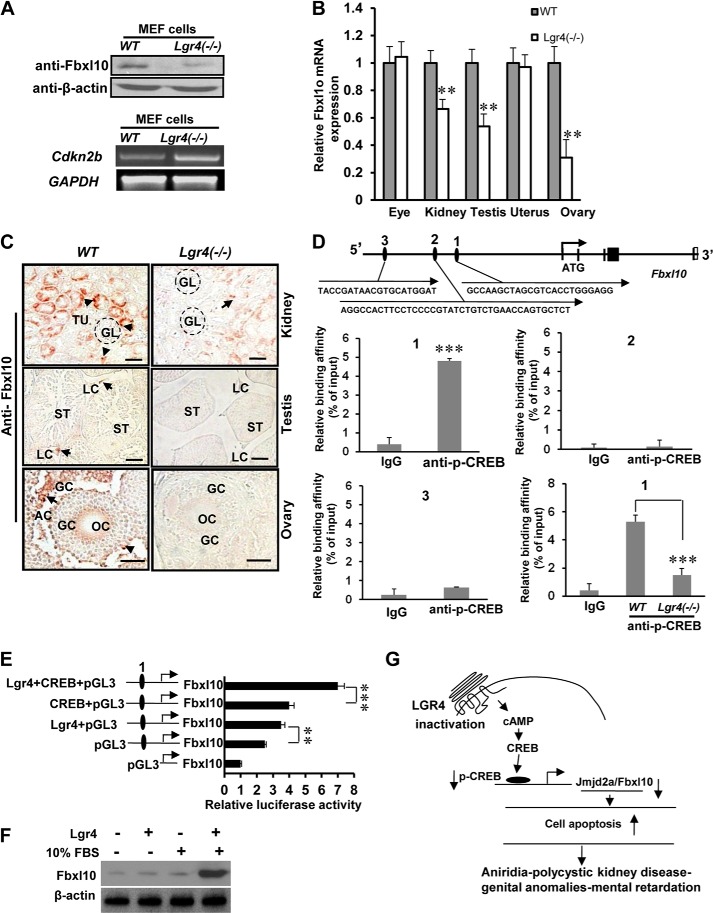
Using Western blot analysis, we examined whether Lgr4 affects the expression level of Fbxl10 and observed that Lgr4 deletion decreased the cellular protein level of Fbxl10 in Lgr4−/− MEF cells (Fig. 7A, top). Cdkn2b is an Fbxl10 target gene and is repressed by Fbxl10 (38). We observed that deletion of Lgr4 increased the mRNA level of Cdkn2b (Fig. 7A, bottom). To examine the effects of Lgr4 on Fbxl10 in vivo, we performed quantitative real-time PCR analysis with total mRNAs from eye, kidney, testis, ovary, and uterus. We observed that Lgr4−/− mice had variable reductions in Fbxl10 mRNA expression levels in kidney, testis, and ovary (Fig. 7B) but not in Lgr4−/− eye and uterus. An explanation of the unaffected Fbxl10 mRNA expression in Lgr4−/− eye and uterus is possible compensatory mechanisms in these organs. In immunostaining assays, detectable Fbxl10 signals decreased in Lgr4−/− tissues compared with those in wild type tissues, including the epithelial cells of tubules of kidney, the Leydig cells of the testis, and the granulosa cells of the ovary (Fig. 7C). These in vitro and in vivo experiments imply that Lgr4 regulates the expression levels of histone demethylase Fbxl10 in the mouse kidney, testis, and ovary.
FIGURE 7.
Lgr4 regulates histone demethylase Fbxl10 via the CREB signaling pathway. A, Lgr4 deletion reduced Fbxl10 protein expression (top) and increased Cdkn2b mRNA expression (bottom). B, Fbxl10 mRNA expression decreased in Lgr4−/− kidney, testis, and ovary. C, Lgr4 deletion decreased histone demethylase Fbxl10 protein expression in the kidney, testis, and ovary. Fbxl10 immunostaining was performed on tissue sections from kidney (1 month old), testis (1 month old), and ovary (2 months old) of wild type and Lgr4−/− mice (5 mice for each group). Lgr4 deletion decreased Fbxl10 expression in nuclei of epithelial cells in the tubules of kidney, in Leydig cells of testis, and in granulosa cells in ovary. GL, glomerulus; TU, tubule; ST, seminiferous tubule; GC, granulosa cell; LC, Leydig cell; OC, oocyte; AC, antral cavity. D, diagram of the mouse Fbxl10 gene (NP_038938) and the −1-kb upstream region. Black boxes, coding exons; empty boxes, non-coding exons; numbered black ellipses, potential CREB sites. Arrows show the direction from 5′ to 3′. The black letters present the minimum CREB site sequences. Region 1 bound phospho-CREB with relatively high affinity by CHIP and Q-PCR assays. Lgr4 deletion reduced the binding affinity between CREB and Fbxl10 promoter region 1. E, overexpression of Lgr4, CREB, and the combination increased Fbxl10 promoter driven luciferase activity (mean ± S.D., n = 3). HEK293 cells cultured in 10% FBS were transfected with indicated Fbxl10 promoter-driven luciferase constructs as well as expression plasmids for CREB, Lgr4, or empty vector, and luciferase reporter activity was measured. F, overexpression of Lgr4 with FBS increases cellular Fbxl10 in HEK293 cells. After plasmid transfection and culture overnight without FBS, HEK293 cells were cultured with or without 10% FBS for 24 h. Western blot assays were performed with β-actin used as a loading control. G, a schematic presenting the molecular mechanism of Lgr4 inactivation leading to WAGR syndrome. Scale bar = 20 μm. **, p < 0.05; ***, p < 0.01.
Lgr4 Regulates Histone Demethylase Fbxl10 through CREB Transcription Factor
There are three potential CREB binding sites in the −1-kb region of the histone demethylase Fbxl10 promoter (Fig. 7D). We tested the binding affinity between these three regions and p-CREB using CHIP and quantitative PCR assays. We found that region 1 showed relatively high binding affinity compared with region 2 and 3 (Fig. 7D, 1–3), indicating that region 1 of the mouse histone demethylase Fbxl10 promoter contains a p-CREB binding site. We then examined the binding affinity between p-CREB and region 1 of Fbxl10 in wild type and Lgr4 null mutant cells. Our data show that deletion of Lgr4 in MEF cells decreased the binding affinity of p-CREB to region 1 (Fig. 7D, 1, lower right) indicating that Lgr4 regulates Fbxl10 transcription via the PKA/p-CREB pathway. To verify this observation, we performed luciferase assays with Fbx110-luciferase and different combinations of plasmids. We observed that overexpression of Lgr4 and CREB significantly enhanced histone demethylase Fbxl10 promoter-induced luciferase activity (Fig. 7E). In addition, overexpressing Lgr4 with FBS stimulation increased cellular Fbxl10 levels (Fig. 7F). Together, these data demonstrate that Lgr4 regulates histone demethylase Fbxl10 transcription via the phospho-CREB signaling pathway. Collectively, our study indicates that Lgr4 is a (W)AGR syndrome candidate gene and regulates multiple organ development through genetic and epigenetic controls (Fig. 7G).
DISCUSSION
(W)AGR syndrome is caused by constitutional deletion of all or part of chromosome 11p, which harbors dozens of genes; however, the underlying mechanisms of the pathogenesis are still elusive. In this report we identify Lgr4 as a novel candidate gene for (W)AGR syndrome. We demonstrate that inactivation of Lgr4 associates with multiple defects of (W)AGR syndrome, including absolute infertility in both male and female mice, compromised iris development and function (aniridia), smaller kidneys and polycystic kidney disease, decreased learning ability, and anti-anxiety in Lgr4 null mice. Therefore, deletion of Lgr4 leads to aniridia, polycystic kidney disease, reproductive defects, and mental retardation in mice, similar to human (W)AGR syndrome. Because LGR4 is located in the chromosome 11p region deleted in WAGR syndrome, and deletion of Lgr4 reduces the expression levels of key genes in WAGR syndrome related organs in mice, our results strongly suggest that LGR4 is a novel potential candidate gene in the pathogenesis of (W)AGR syndrome.
R-spondins interact with Lgr4 and potentiate the Wnt/β-catenin signaling pathway (20) through the associated frizzled/Lrp Wnt receptor complex (19). It is well established that β-catenin interacts with CREB binding protein (CBP) or a closely related CBP homolog of p300 to form a transcriptionally active complex (61–63). Here we demonstrate that Lgr4 regulates the transcription levels of histone demethylases Jmjd2a and Fbxl10 through the classic cAMP-CBP (CREB) signaling pathway, which is consistent with previous studies that Lgr4 regulates gene transcription via cAMP-CREB signaling (24, 39), and that CBP mutation associates with growth and mental retardation in humans (64). Therefore, our studies suggest that Lgr4/Gpr48 may synergize the expression of key genes through both the Gαs/cAMP-PKA-CBP/CREB pathway and the Wnt/β-catenin signaling pathway.
Interestingly, recent studies reported that several GPCRs employ either CREB signaling or Wnt/β-catenin signaling to regulate expression of histone demethylases. For example, β2-adrenergic receptor/cAMP/CREB signaling regulates transcription of histone demethylases JMJD1A (JHDM2a) (58), vitamin-D receptor/Wnt/β-catenin signaling mediates transcription of histone demethylase JMJD3(KDM6B) (65), and Wnt/β-catenin signaling up-regulates expression of histone demethylase JMJD2C (KDM4C) (66). Here we demonstrated that Lgr4 induces the expression of histone demethylases Jmjd2a and Fbxl10 through cAMP/CREB signaling and regulates transcription of multiple key genes important for AGR syndrome-related organ development. The consistent data shown here and in previous reports link GPCRs to histone demethylases and extend our understanding of the roles of GPCRs to epigenetic regulation.
More recently, a nonsense point mutation in LGR4 is reported to increase the risk of squamous cell carcinoma of the skin and biliary tract cancer in humans (23). Although we did not observe Wilm's tumors in this study, our results do not exclude the possibility of increased risk of Wilms' tumor in Lgr4−/− mice. In this study we found that Lgr4 depletion leads to reduced histone demethylases Jmjd2a and Fbxl10 in MEF cells, mouse kidney, testis, ovary, and uterus. In addition, we observed that Lgr4−/− mice display significantly increased cell apoptosis and decreased cell proliferation in kidney, testis, ovary, and uterus. Our observations are in agreement with previous studies that Jmjd2a and Fbxl10 are important for cell proliferation. These data indicate that Lgr4 not only regulates gene expression via cAMP-CREB and Wnt/β-catenin signaling pathways to modulate cell procedures described here and elsewhere (19, 20, 23, 39) but also employs epigenetic regulation via histone demethylases Jmjd2a and Fbxl10 to play roles in the development of multiple organs.
We demonstrated that Lgr4 depletion leads to decreased learning ability and anti-anxiety, extending our understanding of Lgr4 roles in neuron activity, which has not been previously characterized. The expression of PAX6, an important gene for iris development and located at 11p13, is not significantly affected in Lgr4−/− eyes, which is consistent with previous reports that Lgr4 depletion does not affect PAX6 gene expression in the ocular anterior segment (39). These data suggest that the iris defects in Lgr4−/− mice are not likely to be mediated through down-regulation of PAX6 but are primarily caused by down-regulation of other important genes, such as AP-2 and Pitx-2. We cannot rule out, however, that PAX6 loss contributes to the iris defects observed in some (W)AGR patients.
In summary, we identify LGR4 as a candidate gene responsible for many of the (W)AGR syndrome defects and demonstrate that Lgr4 plays a role in epigenetic regulation of gene expression by mediating histone demethylases Jmjd2a and Fbxl10 through the cAMP-CREB signaling pathway.
Acknowledgments
We thank Xin-Hua Feng of Baylor College of Medicine, Fen Wang, and James F. Martin of Texas A&M University Health Science Center for helpful suggestions in the early stages of this project.
This work was supported, in whole or in part, by grants from the State Key Development Programs of China (2012CB910400) and the National Institutes of Health (NCI Grant 1R01CA134731 (to M. L.)).
- (W)AGR
- (Wilm's tumor), aniridia, genitourinary anomalies, and mental retardation
- Lgr4
- leucine-rich repeat-containing G-protein-coupled receptor-4
- Gpr48
- G-protein-coupled receptor-48
- CREB
- cAMP response element-binding protein
- CBP
- CREB binding protein
- GPCR
- G-protein-coupled receptor
- PKD
- polycystic kidney disease
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Fischbach B. V., Trout K. L., Lewis J., Luis C. A., Sika M. (2005) WAGR syndrome. A clinical review of 54 cases. Pediatrics 116, 984–988 [DOI] [PubMed] [Google Scholar]
- 2. Han J. C., Liu Q. R., Jones M., Levinn R. L., Menzie C. M., Jefferson-George K. S., Adler-Wailes D. C., Sanford E. L., Lacbawan F. L., Uhl G. R., Rennert O. M., Yanovski J. A. (2008) Brain-derived neurotrophic factor and obesity in the WAGR syndrome. N. Engl. J. Med. 359, 918–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lennon P. A., Scott D. A., Lonsdorf D., Wargowski D. S., Kirkpatrick S., Patel A., Cheung S. W. (2006) WAGR(O?) syndrome and congenital ptosis caused by an unbalanced t(11;15)(p13;p11.2)dn demonstrating a 7 megabase deletion by FISH. Am. J. Med. Genet A. 140, 1214–1218 [DOI] [PubMed] [Google Scholar]
- 4. Xu S., Han J. C., Morales A., Menzie C. M., Williams K., Fan Y. S. (2008) Characterization of 11p14-p12 deletion in WAGR syndrome by array CGH for identifying genes contributing to mental retardation and autism. Cytogenet. Genome Res. 122, 181–187 [DOI] [PubMed] [Google Scholar]
- 5. Scott D. A., Cooper M. L., Stankiewicz P., Patel A., Potocki L., Cheung S. W. (2005) Congenital diaphragmatic hernia in WAGR syndrome. Am. J. Med. Genet A 134, 430–433 [DOI] [PubMed] [Google Scholar]
- 6. Narahara K., Kikkawa K., Kimira S., Kimoto H., Ogata M., Kasai R., Hamawaki M., Matsuoka K. (1984) Regional mapping of catalase and Wilms tumor–aniridia, genitourinary abnormalities, and mental retardation triad loci to the chromosome segment 11p1305–p1306. Hum. Genet. 66, 181–185 [DOI] [PubMed] [Google Scholar]
- 7. Gessler M., Bruns G. A. (1988) Molecular mapping and cloning of the breakpoints of a chromosome 11p14.1-p13 deletion associated with the AGR syndrome. Genomics 3, 117–123 [DOI] [PubMed] [Google Scholar]
- 8. Pritchard-Jones K., Fleming S., Davidson D., Bickmore W., Porteous D., Gosden C., Bard J., Buckler A., Pelletier J., Housman D. (1990) The candidate Wilms' tumour gene is involved in genitourinary development. Nature 346, 194–197 [DOI] [PubMed] [Google Scholar]
- 9. Jordan T., Hanson I., Zaletayev D., Hodgson S., Prosser J., Seawright A., Hastie N., van Heyningen V. (1992) The human PAX6 gene is mutated in two patients with aniridia. Nat. Genet 1, 328–332 [DOI] [PubMed] [Google Scholar]
- 10. Shinawi M., Sahoo T., Maranda B., Skinner S. A., Skinner C., Chinault C., Zascavage R., Peters S. U., Patel A., Stevenson R. E., Beaudet A. L. (2011) 11p14.1 microdeletions associated with ADHD, autism, developmental delay, and obesity. Am. J. Med. Genet. 155A, 1272–1280 [DOI] [PubMed] [Google Scholar]
- 11. Loh E. D., Broussard S. R., Liu Q., Copeland N. G., Gilbert D. J., Jenkins N. A., Kolakowski L. F., Jr. (2000) Chromosomal localization of GPR48, a novel glycoprotein hormone receptor like GPCR, in human and mouse with radiation hybrid and interspecific backcross mapping. Cytogenet. Cell Genet. 89, 2–5 [DOI] [PubMed] [Google Scholar]
- 12. Mazerbourg S., Bouley D. M., Sudo S., Klein C. A., Zhang J. V., Kawamura K., Goodrich L. V., Rayburn H., Tessier-Lavigne M., Hsueh A. J. (2004) Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Mol. Endocrinol. 18, 2241–2254 [DOI] [PubMed] [Google Scholar]
- 13. Gao Y., Kitagawa K., Hiramatsu Y., Kikuchi H., Isobe T., Shimada M., Uchida C., Hattori T., Oda T., Nakayama K., Nakayama K. I., Tanaka T., Konno H., Kitagawa M. (2006) Up-regulation of GPR48 induced by down-regulation of p27Kip1 enhances carcinoma cell invasiveness and metastasis. Cancer Res. 66, 11623–11631 [DOI] [PubMed] [Google Scholar]
- 14. Van Schoore G., Mendive F., Pochet R., Vassart G. (2005) Expression pattern of the orphan receptor LGR4/GPR48 gene in the mouse. Histochem. Cell Biol. 124, 35–50 [DOI] [PubMed] [Google Scholar]
- 15. Hoshii T., Takeo T., Nakagata N., Takeya M., Araki K., Yamamura K. (2007) LGR4 regulates the postnatal development and integrity of male reproductive tracts in mice. Biol. Reprod. 76, 303–313 [DOI] [PubMed] [Google Scholar]
- 16. Mendive F., Laurent P., Van Schoore G., Skarnes W., Pochet R., Vassart G. (2006) Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Dev. Biol. 290, 421–434 [DOI] [PubMed] [Google Scholar]
- 17. Qian Y., Liu S., Guan Y., Pan H., Guan X., Qiu Z., Li L., Gao N., Zhao Y., Li X., Lu Y., Liu M., Li D. (2013) Lgr4-mediated Wnt/β-catenin signaling in peritubular myoid cells is essential for spermatogenesis. Development 140, 1751–1761 [DOI] [PubMed] [Google Scholar]
- 18. Kato S., Matsubara M., Matsuo T., Mohri Y., Kazama I., Hatano R., Umezawa A., Nishimori K. (2006) Leucine-rich repeat-containing G protein-coupled receptor-4 (LGR4, Gpr48) is essential for renal development in mice. Nephron Exp. Nephrol. 104, e63–e75 [DOI] [PubMed] [Google Scholar]
- 19. de Lau W., Barker N., Low T. Y., Koo B. K., Li V. S., Teunissen H., Kujala P., Haegebarth A., Peters P. J., van de Wetering M., Stange D. E., van Es J. E., Guardavaccaro D., Schasfoort R. B., Mohri Y., Nishimori K., Mohammed S., Heck A. J., Clevers H. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 [DOI] [PubMed] [Google Scholar]
- 20. Carmon K. S., Gong X., Lin Q., Thomas A., Liu Q. (2011) R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 11452–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruffner H., Sprunger J., Charlat O., Leighton-Davies J., Grosshans B., Salathe A., Zietzling S., Beck V., Therier M., Isken A., Xie Y., Zhang Y., Hao H., Shi X., Liu D., Song Q., Clay I., Hintzen G., Tchorz J., Bouchez L. C., Michaud G., Finan P., Myer V. E., Bouwmeester T., Porter J., Hild M., Bassilana F., Parker C. N., Cong F. (2012) R-Spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS ONE 7, e40976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng C., Reddy P., Cheng Y., Luo C. W., Hsiao C. L., Hsueh A. J. (2013) Multi-functional norrin is a ligand for the LGR4 receptor. J. Cell Sci. 126, 2060–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Styrkarsdottir U., Thorleifsson G., Sulem P., Gudbjartsson D. F., Sigurdsson A., Jonasdottir A., Jonasdottir A., Oddsson A., Helgason A., Magnusson O. T., Walters G. B., Frigge M. L., Helgadottir H. T., Johannsdottir H., Bergsteinsdottir K., Ogmundsdottir M. H., Center J. R., Nguyen T. V., Eisman J. A., Christiansen C., Steingrimsson E., Jonasson J. G., Tryggvadottir L., Eyjolfsson G. I., Theodors A., Jonsson T., Ingvarsson T., Olafsson I., Rafnar T., Kong A., Sigurdsson G., Masson G., Thorsteinsdottir U., Stefansson K. (2013) Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497, 517–520 [DOI] [PubMed] [Google Scholar]
- 24. Luo J., Zhou W., Zhou X., Li D., Weng J., Yi Z., Cho S. G., Li C., Yi T., Wu X., Li X. Y., de Crombrugghe B., Höök M., Liu M. (2009) Regulation of bone formation and remodeling by G-protein-coupled receptor 48. Development 136, 2747–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du B., Luo W., Li R., Tan B., Han H., Lu X., Li D., Qian M., Zhang D., Zhao Y., Liu M. (2013) Lgr4/Gpr48 negatively regulates TLR2/4-associated pattern recognition and innate immunity by targeting CD14 expression. J. Biol. Chem. 288, 15131–15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y., Dong J., Li D., Lai L., Siwko S., Li Y., Liu M. (2013) Lgr4 regulates mammary gland development and stem cell activity through the pluripotency transcription factor Sox2. Stem Cells 31, 1921–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo W., Rodriguez M., Valdez J. M., Zhu X., Tan K., Li D., Siwko S., Xin L., Liu M. (2013) Lgr4 is a key regulator of prostate development and prostate stem cell differentiation. Stem Cells 31, 2492–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reik W. (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 29. Dunn R. K., Kingston R. E. (2007) Gene regulation in the postgenomic era. Technology takes the wheel. Mol. Cell 28, 708–714 [DOI] [PubMed] [Google Scholar]
- 30. Shi Y. (2007) Histone lysine demethylases. Emerging roles in development, physiology, and disease. Nat. Rev. Genet 8, 829–833 [DOI] [PubMed] [Google Scholar]
- 31. Klose R. J., Zhang Y. (2007) Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318 [DOI] [PubMed] [Google Scholar]
- 32. Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 33. Couture J. F., Collazo E., Ortiz-Tello P. A., Brunzelle J. S., Trievel R. C. (2007) Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 14, 689–695 [DOI] [PubMed] [Google Scholar]
- 34. Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 [DOI] [PubMed] [Google Scholar]
- 35. Zhang D., Yoon H. G., Wong J. (2005) JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25, 6404–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin S., Janknecht R. (2007) Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 359, 742–746 [DOI] [PubMed] [Google Scholar]
- 37. Peng Y. B., Yerle M., Liu B. (2009) Mapping and expression analyses during porcine foetal muscle development of 12 genes involved in histone modifications. Anim. Genet. 40, 242–246 [DOI] [PubMed] [Google Scholar]
- 38. He J., Kallin E. M., Tsukada Y., Zhang Y. (2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat. Struct. Mol. Biol. 15, 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weng J., Luo J., Cheng X., Jin C., Zhou X., Qu J., Tu L., Ai D., Li D., Wang J., Martin J. F., Amendt B. A., Liu M. (2008) Deletion of G protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (ASD) through down-regulation of Pitx2. Proc. Natl. Acad. Sci. U.S.A. 105, 6081–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yi T., Tan K., Cho S. G., Wang Y., Luo J., Zhang W., Li D., Liu M. (2010) Regulation of embryonic kidney branching morphogenesis and glomerular development by KISS1 receptor (Gpr54) through NFAT2- and Sp1-mediated Bmp7 expression. J. Biol. Chem. 285, 17811–17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ehninger D., Han S., Shilyansky C., Zhou Y., Li W., Kwiatkowski D. J., Ramesh V., Silva A. J. (2008) Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat. Med. 14, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jin C., Yin F., Lin M., Li H., Wang Z., Weng J., Liu M., Da Dong X., Qu J., Tu L. (2008) GPR48 regulates epithelial cell proliferation and migration by activating EGFR during eyelid development. Invest. Ophthalmol. Vis. Sci. 49, 4245–4253 [DOI] [PubMed] [Google Scholar]
- 43. Chen T. T., Wu R. L., Castro-Munozledo F., Sun T. T. (1997) Regulation of K3 keratin gene transcription by Sp1 and AP-2 in differentiating rabbit corneal epithelial cells. Mol. Cell. Biol. 17, 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nottoli T., Hagopian-Donaldson S., Zhang J., Perkins A., Williams T. (1998) AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proc. Natl. Acad. Sci. U.S.A. 95, 13714–13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mochizuki T., Wu G., Hayashi T., Xenophontos S. L., Veldhuisen B., Saris J. J., Reynolds D. M., Cai Y., Gabow P. A., Pierides A., Kimberling W. J., Breuning M. H., Deltas C. C., Peters D. J., Somlo S. (1996) PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272, 1339–1342 [DOI] [PubMed] [Google Scholar]
- 46. Rall L. B., Scott J., Bell G. I., Crawford R. J., Penschow J. D., Niall H. D., Coghlan J. P. (1985) Mouse prepro-epidermal growth factor synthesis by the kidney and other tissues. Nature 313, 228–231 [DOI] [PubMed] [Google Scholar]
- 47. Richards W. G., Sweeney W. E., Yoder B. K., Wilkinson J. E., Woychik R. P., Avner E. D. (1998) Epidermal growth factor receptor activity mediates renal cyst formation in polycystic kidney disease. J. Clin. Invest. 101, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yao H. H., Whoriskey W., Capel B. (2002) Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 16, 1433–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Viger R. S., Mertineit C., Trasler J. M., Nemer M. (1998) Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Mullerian inhibiting substance promoter. Development 125, 2665–2675 [DOI] [PubMed] [Google Scholar]
- 50. Watanabe K., Clarke T. R., Lane A. H., Wang X., Donahoe P. K. (2000) Endogenous expression of Mullerian inhibiting substance in early postnatal rat sertoli cells requires multiple steroidogenic factor-1 and GATA-4-binding sites. Proc. Natl. Acad. Sci. U.S.A. 97, 1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F. D., Keutel J., Hustert E., Wolf U., Tommerup N., Schempp W., Scherer G. (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 52. Holmes S. D., Spotts G., Smith R. G. (1986) Rat Sertoli cells secrete a growth factor that blocks epidermal growth factor (EGF) binding to its receptor. J. Biol. Chem. 261, 4076–4080 [PubMed] [Google Scholar]
- 53. Heikinheimo M., Ermolaeva M., Bielinska M., Rahman N. A., Narita N., Huhtaniemi I. T., Tapanainen J. S., Wilson D. B. (1997) Expression and hormonal regulation of transcription factors GATA-4 and GATA-6 in the mouse ovary. Endocrinology 138, 3505–3514 [DOI] [PubMed] [Google Scholar]
- 54. Manuylov N. L., Smagulova F. O., Leach L., Tevosian S. G. (2008) Ovarian development in mice requires the GATA4-FOG2 transcription complex. Development 135, 3731–3743 [DOI] [PubMed] [Google Scholar]
- 55. Kobayashi A., Shawlot W., Kania A., Behringer R. R. (2004) Requirement of Lim1 for female reproductive tract development. Development 131, 539–549 [DOI] [PubMed] [Google Scholar]
- 56. Mittag J., Winterhager E., Bauer K., Grümmer R. (2007) Congenital hypothyroid female pax8-deficient mice are infertile despite thyroid hormone replacement therapy. Endocrinology 148, 719–725 [DOI] [PubMed] [Google Scholar]
- 57. Connell K. A., Guess M. K., Chen H., Andikyan V., Bercik R., Taylor H. S. (2008) HOXA11 is critical for development and maintenance of uterosacral ligaments and deficient in pelvic prolapse. J. Clin. Invest. 118, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Y., He J., Sui S., Hu X., Zhao Y., Li N. (2012) Clenbuterol upregulates histone demethylase JHDM2a via the β2-adrenoceptor/cAMP/PKA/p-CREB signaling pathway. Cell. Signal. 24, 2297–2306 [DOI] [PubMed] [Google Scholar]
- 59. Sim C. K., Perry S., Tharadra S. K., Lipsick J. S., Ray A. (2012) Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev. 26, 2483–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nancy P., Tagliani E., Tay C. S., Asp P., Levy D. E., Erlebacher A. (2012) Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science 336, 1317–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Henderson W. R., Jr., Chi E. Y., Ye X., Nguyen C., Tien Y. T., Zhou B., Borok Z., Knight D. A., Kahn M. (2010) Inhibition of Wnt/β-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc. Natl. Acad. Sci. U.S.A. 107, 14309–14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Takemaru K. I., Moon R. T. (2000) The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J. Cell Biol. 149, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) WNT and β-catenin signalling. Diseases and therapies. Nat. Rev. Genet 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 64. Petrij F., Giles R. H., Dauwerse H. G., Saris J. J., Hennekam R. C., Masuno M., Tommerup N., van Ommen G. J., Goodman R. H., Peters D. J. (1995) Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376, 348–351 [DOI] [PubMed] [Google Scholar]
- 65. Pereira F., Barbáchano A., Silva J., Bonilla F., Campbell M. J., Muñoz A., Larriba M. J. (2011) KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum. Mol. Genet 20, 4655–4665 [DOI] [PubMed] [Google Scholar]
- 66. Yamamoto S., Tateishi K., Kudo Y., Yamamoto K., Isagawa T., Nagae G., Nakatsuka T., Asaoka Y., Ijichi H., Hirata Y., Otsuka M., Ikenoue T., Aburatani H., Omata M., Koike K. (2013) Histone demethylase KDM4C regulates sphere formation by mediating the crosstalk between Wnt and Notch pathways in colonic cancer cells. Carcinogenesis 34, 2380–2388 [DOI] [PubMed] [Google Scholar]