Summary
Analysis of hematopoietic stem cell function in non-human primates provides insights that are relevant for human biology and therapeutic strategies. In this study, we applied quantitative genetic barcoding to track the clonal output of transplanted autologous rhesus macaque hematopoietic stem and progenitor cells over a time period of up to 9.5 months. We found that uni-lineage short-term progenitors reconstituted myeloid and lymphoid lineages at one month, but were supplanted over time by multi-lineage clones, initially myeloid-restricted, then myeloid-B clones, and then stable myeloid-B-T multi-lineage long-term repopulating clones. Surprisingly, reconstitution of the natural killer cell lineage, and particularly the major CD16+/CD56− peripheral blood NK compartment, showed limited clonal overlap with T, B or myeloid lineages, and therefore appears to be ontologically distinct. Thus, in addition to providing insights into clonal behavior over time, our analysis suggests an unexpected paradigm for the relationship between NK cells and other hematopoietic lineages in primates.
Introduction
Understanding how hematopoietic stem and progenitor cells (HSPCs) produce a diversity of blood and tissue cell lineages at a high rate for the life of an individual, under homeostatic control, without exhaustion and with only rare catastrophic neoplastic dysregulation, has been a major investigative focus. Insights have been translated into a myriad of clinical advances, including hematopoietic stem cell transplantation, cytokine treatment of cytopenias, and targeted therapies for hematopoietic neoplasms. Despite progress, many questions remain, including the degree of homogeneity of long-term self-renewing hematopoietic stem cells (HSC), hematopoietic lineage ontogenies difficult to model in vitro, control of fate decisions, patterns of contributions of individual cells in vivo over time, and number and life-span of true HSCs (Copley et al., 2012). Murine HSCs have been characterized at close to a single cell level in gold-standard transplantation assays, via cell-surface phenotype and gene expression profiling (Osawa et al., 1996; Smith et al., 1991; Spangrude et al., 1988; Weissman and Shizuru, 2008). Limit-dilution transplantation experiments define what one or small numbers of stem cells can do, but whether HSC behavior in this setting of extraordinary replicative stress fully mimics normal steady-state hematopoiesis or even non-limit dilution transplant is questionable.
Transduction of HSPCs with integrating viral vectors results in passage of the integrated provirus to every daughter cell. Each semi-random pro-viral integration site (VIS) serves as a unique clonal “tag” for individual HSPCs and their progeny. Pioneering studies in mice identified VIS via Southern blot and provided evidence for multi-lineage clonal repopulation by single cells (Jordan and Lemischka, 1990), however this insensitive approach detects only dominant clones, and cannot resolve highly polyclonal patterns or investigate contributions to cells from less abundant lineages such as natural killer (NK) cells. A number of more sensitive PCR-based VIS retrieval methods have been devised, but all have limitations regarding both efficiency and quantitation (Berry et al., 2012; Bushman et al., 2005; Schmidt et al., 2002; Wu et al., 2013). An alternative is generation of high-diversity lentiviral libraries to deliver a unique “barcode” to individual HSPCs, allowing quantitative clonal tracking. This concept was developed to study murine antigen-specific T cell dynamics and extended to proof-of-concept studies in murine HSPCs (Gerrits et al., 2010; Schepers et al., 2008). We combined lentiviral barcoding with PCR retrieval and high throughput sequencing to elucidate the behavior of HSPCs in mice, showing that clonal output from individual cells could be assessed in a sensitive and quantitative manner and yielding important insights regarding murine hematopoietic lineage bias (Lu et al., 2011).
Studying human HSPC clonal behavior is more challenging (Baum et al., 1992; Doulatov et al., 2012). Limit dilution transplantation or vector tagging of human HSPCs in immunodeficient mice have provided valuable information regarding the phenotype of human engrafting cells and some information on their in vivo behavior (Doulatov et al., 2012; Guenechea et al., 2001; Larochelle et al., 1996). However, the behavior of human cells in mice is far from physiologic, given the reliance on a xenogeneic niche, with little release of fully differentiated progeny into the blood, and very limited development of T or NK cells (Coulombel, 2004). There are marked differences between rodent and large animal or human hematopoiesis regarding cytokine utilization, location of stress hematopoiesis, HSPC phenotype and frequency, and lifetime hematopoietic demand (Abkowitz et al., 1996; Abkowitz et al., 2002; Catlin et al., 2011; Larochelle et al., 2011; Shepherd et al., 2007).
We have utilized rhesus macaque autologous transplantation to investigate HSPC behavior in a model with direct relevance to humans (Donahue and Dunbar, 2001). Humans and macaques are phylogenetically close, with comparable lifespans, hematopoietic demand, and HSPC frequencies (Shepherd et al., 2007). We can efficiently mark rhesus HSPCs with lentiviral vectors and track marked progeny cells for many years (Kim et al., 2009). We now report the use of lentiviral HSPC barcoding to track and quantitate clonal contributions following autologous rhesus HSPC transplantation, and provide multiple insights into hematopoietic reconstitution and lineage hierarchies, most notably of NK cells.
Results
Experimental design and validation
In order to track in vivo output from individual HSPCs in the rhesus macaque, we adapted our lentiviral cellular barcoding technology to rhesus CD34+ HSPCs and followed clonal output following myeloablative autologous transplantation (Fig 1A). Highly diverse DNA barcode libraries were cloned into lentiviral vectors also containing a green fluorescent protein (GFP) marker. Using mixtures of single-copy barcoded K562 cell clones we showed that retrieved barcode frequency quantitatively reflects the contribution of an individual clone within a polyclonal mixture, over a range of ratios (Fig S1).
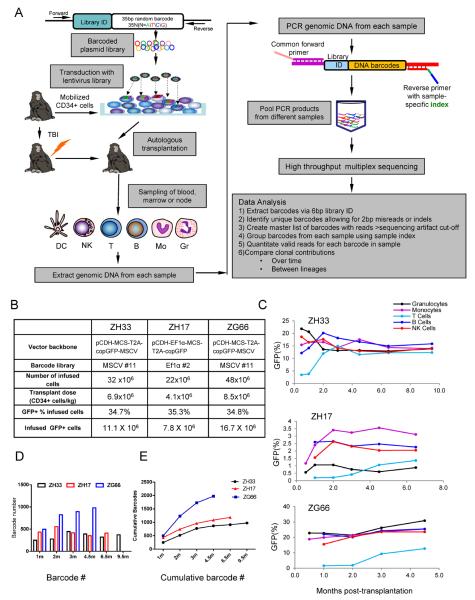
Figure 1. Experimental design and clonal diversity.
(A) Oligonucleotides consisting of a library ID followed by a random barcode were cloned into a lentiviral vector flanked by PCR primer sites. Mobilized PB autologous CD34+ cells were transduced and infused back into the irradiated autologous macaque, and PB, BM and lymph node samples were collected, purified by FACS, and barcode retrieval was performed via PCR and high throughput sequencing. Information relevant to methodologies presented in in Fig S1–S4 and Tables S1–S3. (B) Transduction and transplantation parameters for rhesus macaques ZH33, ZH17 and ZG66. (C) Percentage of GFP+ cells in PB lineages over time following transplantation. (D) Number of independent barcoded clones detected in the PB (combined B, T, Gr, Mono, NK) at each time point (m=months). (E) Cumulative number of independent barcoded clones detected in PB (combined B, T, Gr, Mono, NK) over time.
Mobilized peripheral blood (PB) CD34+ cells from three macaques were transduced with barcoded vectors and 7.8–16.7 million autologous GFP+ cells were re-infused following ablative total body irradiation (Fig 1B). Vector barcode diversity was assessed following retrieval of barcodes from transduced rhesus cells at the end of transduction and Monte Carlo simulations using this experimental data were performed to ask whether library diversity was sufficient to ensure with high confidence that a single barcode labeled a single cell (Fig S2) (Lu et al., 2011). Based on the best available prior estimates for the frequency of rhesus repopulating cells as 5 per 100,000 CD34+ cells, the barcoded libraries chosen had at least ten-fold higher diversity than required to be more than 95% certain that a single barcode marks only one HSPC clone at these transplanted doses (Kim et al., 2000; Shepherd et al., 2007).
For the most straightforward tracking of individual HSPCs, the presence of only a single barcode per cell is desirable, thus we targeted a maximum CD34+ transduction efficiency of no more than 30–35%, based on prior models (Kustikova et al., 2003; Verovskaya et al., 2013). The level of GFP positivity in circulating blood cells from myeloid and lymphoid lineages following transplant is shown in Fig 1C. As expected, granulocytes (Gr) and monocytes (Mo) counts recovered rapidly, within two week of transplantation, followed by NK, B, and then T cells, reaching normal levels by 3–6 months. 10–30% of hematopoietic cells in all lineages were GFP+ in ZH33 and ZG66, stabilizing by 2–3 months in Gr, Mo, B and NK cells, with transduced T cell engraftment lagging behind (Fig 1C), reflecting slower replacement of endogenous T cells, as we have observed previously (An et al., 2007). ZH17 had a similar pattern, but overall much lower marking, stabilizing at levels of only 1–2%, despite receiving CD34+ cells with the same transduction efficiency as the other two animals, likely reflecting greater competition from endogenous HSPCs surviving radiation, due to heterogeneous radiation sensitivity between individual macaques.
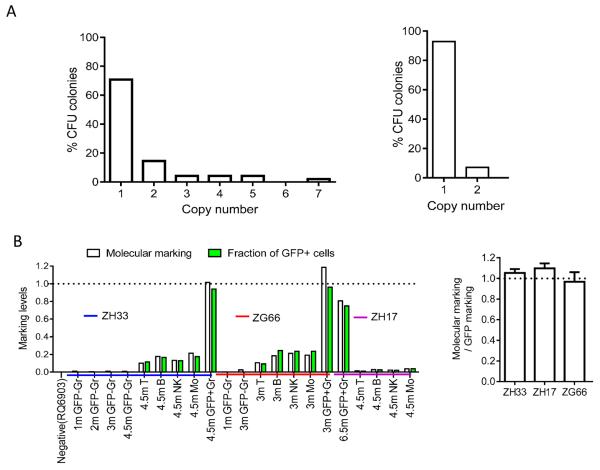
Following engraftment, PB and lymph nodes were collected at various time points, and Gr, Mo, T, B, NK, and plasmacytoid and myeloid dendritic cells (DCs) were flow sorted (purity median 98.8%) (Fig S3, Table S1). The mean barcode copy number per individual transduced HSPC in individual colonies (CFU) derived from CD34+ cells plated at the end of transduction in animal ZH33 was 1.2±0.22 (SEM), and 1.07±0.07 in CFU grown from CD34+ cells obtained from the BM of ZH33 following engraftment, with only 7% of CFU containing 2 copies and none more than 2 (Fig 2A). We also compared vector copy number and %GFP+ cells in PB post-transplantation in all three animals (Fig 2B) and the mean copy number per GFP+ cell was 1.04±0.04. In total, this data confirms that the majority of transduced cells contained only one barcode. A minority of clones containing more than one barcode would skew calculations of the frequency of repopulating clones upwards, but not impact analysis of lineage contributions or kinetics.
Figure 2. Barcode copy number per HSPC clone.
(A) Barcode copy number in individual CFU obtained at the end of transduction (left, n=48) or from marrow CFU post-transplant from ZH33 (right, n=14). (B) Barcode copy number per transduced cell for PB samples. The molecular marking level (vector copy number) was determined by qPCR on sorted GFP negative granulocytes (GFP− Gr), GFP positive granulocytes (GFP+ Gr) and sorted T, B, and NK cells from ZH33, ZH17, and ZG66, and plotted on the left panel (white bars) next to the fraction of GFP+ cells in the same sample as assessed by FACS (green bars). A ratio of 1 implies one vector copy per each GFP+ cell. On the right, the ratio of molecular marking level determined by copy number PCR/GFP marking level for all samples from each of the three animals (mean±SEM) are shown.
Barcode retrieval by PCR, Illumina sequencing, and custom data analysis was performed on purified hematopoietic lineage samples monthly through 9.5m (ZH33), 6.5m (ZH17) and 4.5m (ZG66) (Lu et al., 2011). Our algorithms extract barcodes by library IDs, account for sequencing misreads and indels, quantitate valid reads for each barcode, and create a master list of barcodes for each animal, consisting of barcodes present in at least one sample at a read number over cutoffs determined for each animal to exclude 99.5% of “false” artifactual barcodes (python code in Supplemental Methods, master list of barcodes and contributions to each lineage and time point in Table S4). Very high reproducibility of barcode retrieval and quantitation was demonstrated via sequencing and PCR replicates on the same DNA samples (Fig S4).
We assayed independently-processed replicate blood samples to identify a lower barcode read threshold resulting in 95% barcode retrieval between replicates, designed to exclude false negatives arising from sampling error, related to the number of cells (mean of 70,000 cells processed to yield 200ng DNA) in each purified sample (Fig S4). For highly marked animals ZH33 and ZG66, the majority of barcoded hematopoiesis derived from clones above this threshold, and only clones above the threshold were included in pairwise comparisons between lineages or time points. ZH17, with a much lower overall in vivo marking level, was not used for pairwise comparisons due to a barcode read threshold at least ten-fold higher and therefore limited ability to compare low abundance clones between samples, but individual abundant clones from this animal were tracked, and overall clone numbers scored.
Overall HSPC clonal diversity post-transplantation
Fig 1D and E plot the number of unique barcodes contributing at each time point, and the cumulative number of unique clones identified over time. A total of 976 individual clones in ZH33 (9.5m), 1184 in ZH17, (6.5m), and 1977 in ZG66 (4.5m) were detected at one or more time points, at a read number above the cut-off determined for each animal set to exclude sequencing artifacts. Based on the number of GFP+ cells transplanted (Fig 1B), approximately 1 in 9,000 CD34+ cells contributed at any time point (range 1 in 6,500–11,300). This is a lower limit, given that low-contributing clones were missed due to sampling constraints or excluded by sequencing background cutoffs. The cumulative number of clones detected began to plateau as early as 3–4 months post-transplant, with fewer new clones appearing after that time point. At longest follow-up in each animal (4.5–9.5m), the frequency of clones still contributing was about 2.5-fold lower, 1 in 22,000 CD34+ cells (range 1 in 17,000–30,000).
Lineage and longitudinal clonal contributions
We assessed fractional contributions of individual clones to different hematopoietic lineages over time. As shown in Fig 3 (ZH33) and Fig S5 (ZG66), the fractional contribution of each individual barcoded clone can be directly compared between lineages and time points, and the distribution of lineage bias in the population of clones assessed. The frequency of highly biased clones, defined as ≥10 fold fractional representation of a barcode in one lineage or time point sample versus another, can be quantified, as can pairwise Pearson correlations between all lineages and time points (Fig 4). Spearman analysis of the dataset gave essentially the same results, although because of the different nature of the two tests, some patterns of biological interest are not as apparent in the Spearman analyses (Fig S6).
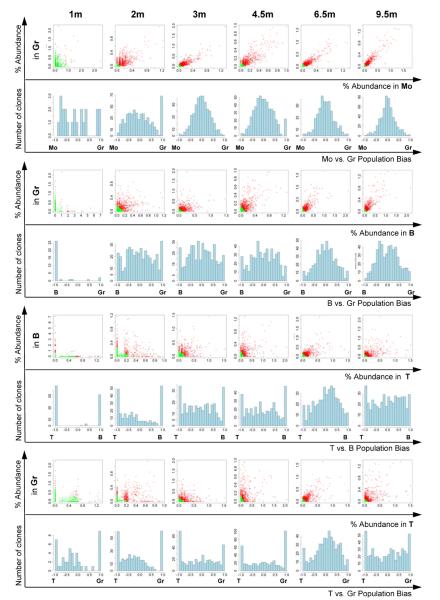
Figure 3. Clonal comparisons between lineages.
Scatter plots compare sequencing read fractions in ZH33 between pairs of lineages over time. Each dot represents an individual barcode/clone from the master list, detected in either lineage. Green dots are barcodes with reads falling below the sampling threshold of 1144 reads combined between the two lineages, red dots are barcodes with reads above the threshold. The bias histograms are generated from the scatter plots for barcodes falling above the threshold by measuring the angle from the axis for each barcode, and renormalizing from -1 to 1 before placing each barcode into a bias bin. The number of clones in each bias bin is on the y and the degree of bias on the x axes, with clones having equal lineage contributions defined as 0. Each clone is counted equally for the bias histograms, without regard to its overall level of contribution. Analogous data from animal ZG66 shown in Fig S5.
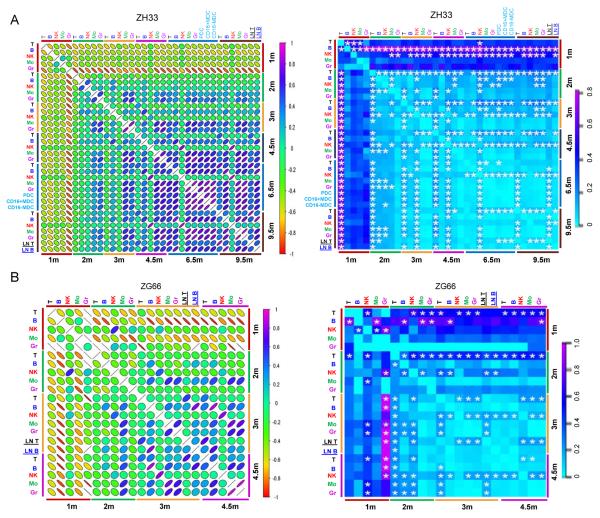
Figure 4. Summary of clonal lineage and kinetic relationships over time.
(A) Pearson correlation coefficients comparing barcode contributions for all barcodes falling above the sampling threshold of 1144 for pairwise comparisons, between all lineages and time points, for animal ZH33 and ZG66 (r values and their associated p values and 95% confidence intervals are given in Table S5). The color scale bar for r values is on the right. Slope indicates negative or positive correlation; the shape and the color signify the strength of the correlation. MDC-myeloid dendritic cells, PDC-plasmacytoid dendritic cells, LN-lymph node. Alternative analysis utilizing Spearman correlation shown in Fig S6 and Table S6 (B) Fraction of highly biased (>10 fold greater fractional representation in one sample versus the other) barcodes between all lineages and time points for ZH33 and ZG66. Bias is towards the sample shown in the row, and away from the sample in the column. Color scale is given on the right. P values <0.05 are starred; the frequency of highly biased clones in these comparisons differed significantly from the total frequency of highly biased clones at the time point combination. Hierarchical clustering analysis for fractional barcode contributions in ZH33 and ZG66 given in Fig S7.
At 1m post-transplantation, there was no correlation between lineages, with uni-lineage or extremely biased low abundance clones predominating. Clones contributing at 1m did not persist in contributing substantially to any lineage at 3m or later (Fig 3,4), defining them as short-term, lineage-restricted progenitors. The first bi-lineage Gr/Mo clones began to emerge at 2m, and rapidly dominated. By 3m, virtually all Gr and Mo clones were shared and there were few highly biased clones, directly demonstrating in vivo output from common Gr/Mo progenitors. By 3m, clones shared between myeloid (Gr/Mo) and B lineages began to dominate, with stronger correlations as well as a decrease in highly biased Gr/Mo or B clones, before clones contributing to T and B lineages appeared (Fig 3,4). By 4.5–6.5m, multi-lineage, less biased Gr/Mo/B/T clones became dominant. CD11c+CD16+ and CD11c+CD16− myeloid DCs, as well as CD11c− CD123+ plasmacytoid DCs collected at 6.5m months from ZH33 were most closely related to each other and to the Gr/Mo myeloid lineages.
To further depict the complexities of clonal contributions to all lineages, we performed hierarchical clustering analysis for ZH33 and ZG66 (Fig S7), which demonstrated grouping of the 1m samples separate from later time points for all lineages, confirmed the close relationship between Gr and Mo, and the lack of a clear grouping of B and T lineages separate from myeloid lineages at any time point.
We also compared the clonal composition of blood versus lymph node T and B cells in ZH33 (9.5m) and ZG66 (3m), and found shared clonal patterns of these lineages in blood versus node (Fig 4), with a single expanded T cell clone outlier in the PB of ZG66 accounting for almost 20% of marked cells not found at a similar level in the one node sampled.
Natural killer cell ontogeny
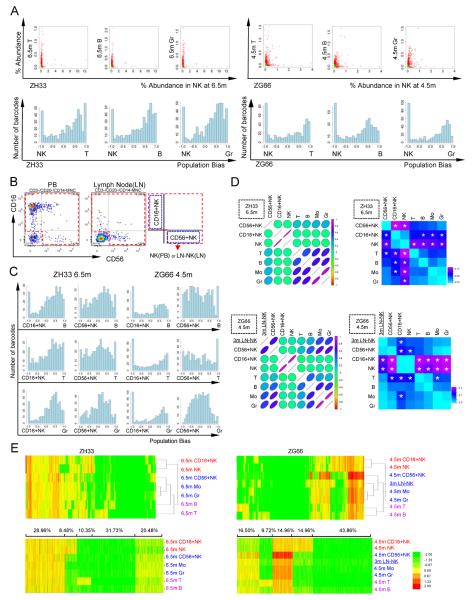
We were surprised to find a very different pattern of clonal contributions in NK cells (Fig 4,5,S6). Between 3 and 6.5m, when many shared Gr/Mo/T/B clones were contributing, NK clonal composition remained distinct, before beginning to converge towards the other lineages by 9.5m (Fig 4,5). Highly biased clones predominated, compared to the other 4 lineages at the same time points. Triangle plots (Fig 6) confirmed the presence of highly skewed NK contributing clones, distinct from shared T and B contributing clones at 6.5m (ZH33) or 4.5m (ZG66), versus clusters of clones contributing to T, B and Gr at the same time points. The extent of NK clonal diversity was similar to other lineages in terms of total number of clones, however a number of very high contributing clones dominated the PB at individual time points.
Figure 5. Clonal contributions to NK cells.
(A) Scatter and bias plots for NK versus T, B, and Gr from ZH33(6.5m) and ZG66(4.5m). (B) Example of FACS gating for sorting of NK subsets in blood (ZH33, 6.5m) and node (ZG66, 3m). (C) Bias plots comparing CD16+ and CD56+ NK with B, T, and Gr from ZH33 at 6.5m (left) and ZG66 at 4.5m (right). (D) Graphical summary of Pearson correlation coefficients (left) (Table S7 gives r values, p values and 95% confidence intervals) and fraction of highly biased (>10 fold difference in fractional representation) clones (right) (p values <0.05 are starred), comparing NK cells and subsets from the peripheral blood (ZH33 6.5m, upper panel) and from ZG66 blood (4.5m) and node (3m) (lower panel). Alternative analysis utilizing Spearman correlation shown in Fig S6 and Table S7 (E) Euclidian distance hierarchical clustering (upper panel) and K-mean clustering with k=5 (lower panel), ZH33 (6.5m) and ZG66 (4.5m blood, 3m node). For each animal, all barcodes on the master clone list are included. For K-mean clustering, the % of clones in each cluster is shown above the plots. NK:NK cells in PB; LN-NK: lymph node NK; CD16+NK: CD16+CD56−; 56+NK: CD16−CD56+
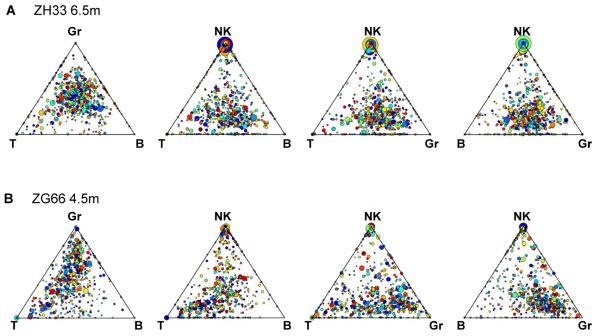
Figure 6. Triangle plots of clonal contributions to T, B, NK and Gr lineages.
Triangle plot of clonal contributions to B, T, NK and Gr in ZH33 at 6.5m (A), or in ZG66 at 4.5m (B). Each dot represents an individual barcoded clone, with the size of the dots corresponding to the total fractional contribution of the individual barcode to the total barcodes retrieved at the time point. The color of the dot is assigned randomly to help distinguish adjacent dots. The location of the barcode represents the ratio of barcode reads in each of the three cell lineages. The presence of large, dots clustering at the vertex of plots including NK cells suggests a unique population of highly biased NK clones in both animals, in contrast to clusters of dots in the center of B/T/Gr triangles, suggesting common clones contributing in a balanced manner to these lineages, particularly by 6.5m in ZH33.
We fractionated NK cells into distinct populations at 6.5m (ZH33) or 4.5m (ZG66): the major CD16+/CD56− PB fraction and the minor blood CD16−/CD56+ fraction (Fig 5B). We also obtained lymph node NK cells from ZG66, which in contrast to PB NK cells are primarily CD16−/CD56+. The CD16+/CD56− PB NK cells accounted for the clones that were distinct from T/B/Gr/Mo lineages, as shown by bias plots, Pearson correlation coefficients, the fraction of highly biased (≥10 fold) clones, and both K-mean and hierarchical clustering (Fig 5). Spearman correlation coefficients also supported this result (Fig S6), though Pearson correlation is more biologically relevant. In contrast, the clonal patterns in the CD16−/CD56+ subsets of NK cells from blood and lymph nodes strongly correlated with T/B/Gr/Mo lineages.
Analysis of abundant clones
In highly polyclonal samples, it is likely that some clones may still intermittently fall below sampling detection limits despite use of thresholds set to exclude false negatives, complicating analysis of lineage contributions. We did not have sufficient DNA to run duplicate samples on every lineage at every time point, thus thresholds set to exclude false negative bias due to sampling error were based on limited information. Thus we also individually mapped the 10 most abundant clones identified in each lineage at 1m, 3m and at longest follow-up (9.5m ZH33, 6.5m ZH17, 4.5m ZG66), tracking contributions from each clone over all time points and in all other lineages (Fig 7). These abundant clones are inherently interesting, because they account for a disproportionate fraction of ongoing hematopoiesis, and are less likely to be misinterpreted due to sampling error. Even the most dominant clones at 1m were primarily uni-lineage, and ceased or greatly decreased their relative contributions by 2–3m. T and B cells produced during initial lineage recovery even from these highly productive clones disappeared or greatly decreased at later time points.
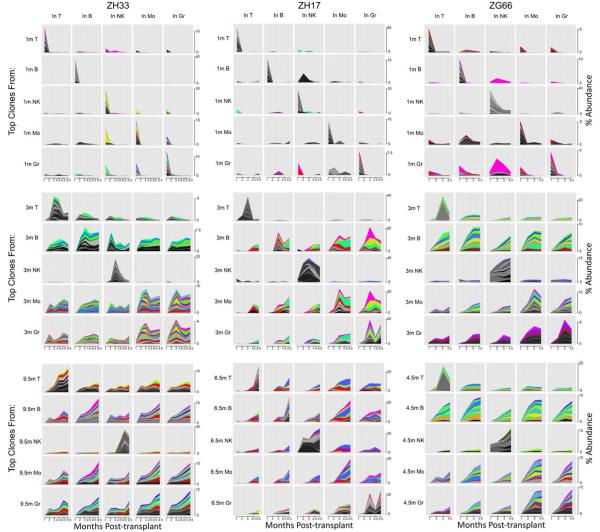
Figure 7. Tracking of individual high-contributing clones.
The fractional contribution of each barcode to the total barcodes retrieved from each lineage at each time point were ranked, and the top ten contributing barcodes in each lineage at 1m, 3m and 9.5m from ZH33 (left column) 1m, 3m and 6.5m from ZH17 (middle column) and 1m, 3m and 4.5m from ZG66 (right column) are tracked for their contributions in all lineages over time. Each graph shows the time post-transplant on the x axis and the percentage of total reads each clone contributes to the lineage as a stacked graph on the y axis. Each y axis scale is linear and identical across each row of graphs, with the scale on the far right of each row, but note that scale is adjusted between rows, in particular when one very dominant clone is being depicted. Clones shown in grey scale are top 10 in only one lineage. Clones shown in color are top 10 in more than one lineage, and each clone has a unique color, allowing comparisons between plots for each animal.
The patterns were very different for the top clones at 3m and the top clones at longest follow-up, with the top Gr, Mo, T and B clones detected in the same lineage tracing back through 2m, but not 1m, and increasing in their relative contributions over time between 2m and 4.5m–9.5m, as short-term progenitor contributions decreased. A number of individual clones fell into the top 10 clones for multiple lineages at the later time points, with the most overlap between B, Gr and Mo, some overlap with T, and least overlap with NK lineages, suggesting that at least some clones were contributing at a relatively balanced and high levels to multiple lineages, confirming the patterns discerned at the population level. Some of these clones also contributed to NK cells, but at much lower levels compared to the relative contributions of the top 10 NK clones. The top NK clones from 3m or latest follow-up were not detected at equivalent or appreciable levels in the other 4 lineages, further support for the unique characteristics of this lineage. In two of the animals, there was transient marked expansion of one or more T cell clones at single time points, suggesting a clonal response to an infection or other environmental insult.
Discussion
The application of HSPC barcoding in the rhesus model has provided a number of new insights into large mammal hematopoiesis. This approach is highly quantitative and requires only small amounts of DNA obtainable from multiple lineages repeatedly over time from the same animal, in contrast to the less efficient and quantitative VIS retrieval-dependent approaches (Bushman et al., 2005). Barcoding relies on vector integration, raising concern of impact on HSPC behavior via activation of adjacent proto-oncogenes (Calmels et al., 2005; Kustikova et al., 2005; Nienhuis et al., 2006). However, lentiviral vectors are much less likely to transform HSPCs compared to murine retroviral vectors, due to lentiviral integration patterns and the deletion of endogenous enhancer sequences in these vectors (Modlich et al., 2009; Montini et al., 2009). Follow-up of 5 to 10 years in rhesus macaques ((Kim et al., 2009) and unpublished) and over 3 years in human clinical trials reveals stable polyclonal engraftment of lentiviral-transduced HSPC, without expansion of dominant clones (Biffi et al., 2011; Cartier et al., 2009).
We believe that non-human primate models provide information highly relevant to human hematopoiesis, difficult to obtain in any other system. Immunodeficient mice lack a physiologic niche for human hematopoiesis, engraftment inefficiently with adult HSPCs, show little maturation of T or NK cells, and do not release human cells normally into the blood, precluding most quantitative analyses of clonal HSPC output or lineage hierarchies. Patients enrolled in clinical gene therapy trials have hematopoiesis tracked long-term via VIS identification, but most have perturbations due to activation of proto-oncogenes by murine retroviral vectors, dysregulated expression of the therapeutic gene, or underlying hematopoietic abnormalities (Deichmann et al., 2011; Deichmann et al., 2007; Gabriel et al., 2009; Schwarzwaelder et al., 2007). Our rhesus approach is not limited by these constraints.
In our studies, CD34+ cells were selected prior to barcoding. Virtually all HSPC engrafting activity in adult BM and mobilized PB in humans and monkeys is within the CD34+ fraction (Andrews et al., 2000; Doulatov et al., 2012). Long-term HSC are a minority of HSPCs, and humans HSCs assayed in xenografts are CD34+38lo90+117+Lin−(Majeti et al., 2007; Park et al., 2008). Human CD34+ subfractions have been placed in a hierarchy of multipotent progenitors (MPP), lymphomyeloid-biased progenitors (LGMP) common lymphoid progenitors (CLP), common myeloid progenitors (CMP), GM progenitors (GMP), megakaryocyte-erythroid progenitors (MEP) then uni-lineage restricted progenitors of the myeloerythroid lineages, as assayed in vitro and in xenografts (Galy et al., 1995; Majeti et al., 2007; Manz et al., 2002). Rigorous identification of cell surface markers delineating each of these subpopulations in the rhesus macaque have not been performed, however our model will allow prospective purification, barcoding and autologous engraftment of putative progenitor populations to define each subset in an autologous, physiologic setting.
We calculate a minimum frequency for rhesus engrafting HSPC as approximately 1 in 9,000 CD34+ cells, corresponding to about 1 per 4.5 million mobilized PBMNCs. At longest follow-up (4.5–9.5m), about 1 in 22,000 CD34+ cells continued to contribute. Limit dilution studies measured “SCID repopulating” HSPC frequency to be 1 in 6 million mobilized human PBMNCs (Wang et al., 1997). The lower frequency of primate or human repopulating cells in xenografts as compared to direct autologous measurement perhaps reflects inefficiency of xenogeneic engraftment. The majority of rhesus clones found as early as 3m were still contributing at 9.5m, with only a small number of new clones appearing for the first time after 3m, suggesting that relatively stable hematopoiesis may be achieved quite rapidly in a physiologic autologous model, perhaps derived from the rhesus equivalent of “intermediate” mouse repopulating HSCs (Benveniste et al., 2010) This rapid clonal stability is in contrast to the much more heterogeneous results recently reported using barcoding of human cord blood cells in a murine xenograft, likely resulting from inefficiencies of engraftment and hematopoietic output in that model (Cheung et al., 2013). The highly polyclonal nature of reconstitution in the barcoded macaques bypasses stochastic variations arising in the setting of limiting stem cell doses, predicted to result in very prolonged periods of hematopoietic instability (Abkowitz et al., 1996). However, hematopoietic tracking for much longer time periods is required to determine the frequency and clonal output of true long-term repopulating cells, and to distinguish models of HSC clonal exhaustion/succession versus clonal stability. A related study from our primate program, published in this issue, has utilized very long-term lentiviral VIS retrieval to provide answers to some of these questions (Kim et al., 2014). Longer follow-up combined with barcoding will allow additional highly quantitative analysis of whether lineage-skewed classes of HSCs exist in non-human primates, analogous to recent findings in murine models regarding lymphoid versus myeloid biased HSC and our companion long-term primate study (Copley et al., 2012)(Kim et al., 2014).
Our studies demonstrate the presence of uni-lineage engrafting progenitors in all lineages, supporting prior results from murine and xenograft models and a recent paper utilizing barcoding that demonstrated uni-lineage B, granulocytic and dendritic cell output from LMPPs (Guenechea et al., 2001; Jones et al., 1990; Jordan and Lemischka, 1990; Naik et al., 2013). Highly purified ex vivo cultured CD34+ cells do not contain viable mature T, B, or NK cells at the time of reinfusion, thus re-establishment of these lineages must occur from CD34+ progenitor cells, not peripheral expansion of mature barcoded cells. The ability to at least transiently engraft lineage-restricted progenitors is of interest for human transplantation applications, and our model can be applied to identify the phenotype of relevant transplantable progenitor cells. Output from these unilineage short-term repopulating clones largely disappeared from the blood and lymph node by 2–3m, suggesting that the half-life of newly produced mature B and T cells and their direct progenitors is surprisingly short, at least in the post-transplantation setting.
As early as 2 months, Gr and Mo clonal architecture became highly correlated, with shared and unbiased clones contributing, confirming in vivo the close relationship between these lineages revealed by in vitro assays. Beginning at 3m and continuing through 9.5m, myeloid and lymphoid lineages began to share clonal patterns. However, the relationship between B and myeloid cells was closer and faster than between B and T cells. A recent analysis of the lineage potential of putative human HSPC populations suggested that the majority of previously characterized “multi-lymphoid progenitors (MLP)”, the human equivalent of CLP, retain the ability to produce myeloid lineage cells in vitro and in xenografts (Doulatov et al., 2010). A recent murine barcoding analysis did not find a faster or higher correlation between contributions of clones to T and B than myeloid and B cells (Verovskaya et al., 2013). We did not document an appreciable set of clones contributing to T and B but not myeloid lineages at any time during early hematopoietic reconstitution, supporting the existence of an MLP versus a CLP in primates. Multi-lineage clones appeared by 3m and continued to contribute to Gr/Mo/B/T lineages through 9.5m, with faster stabilization than reported in xenografts, perhaps reflecting a more polyclonal and efficient engraftment process in autologous macaques as compared to human cells in murine niches (McKenzie et al., 2006).
Our most interesting findings concern the putative ontogeny of NK cells. NK cells are defined by target cell killing and cytokine production independent of priming or antigen/MHC restriction (Caligiuri, 2008; Vivier et al., 2011). Rhesus NK cells have been well characterized and share most but not all cell surface markers with human NK cells (Mavilio et al., 2005; Webster and Johnson, 2005). The ability of both T and NK cells to lyse targets, as well as some shared cell surface receptors, has suggested that they are ontologically related. In vitro, a human NK-B precursor has been identified, able to produce both lineages under certain culture conditions. However, NK output from this progenitor, or from single “MLP” progenitors able to produce T, B, NK and myeloid lineages in vitro, was not detected in xenografts (Doulatov et al., 2010; Galy et al., 1995). The original studies identifying a mouse CLP able to produce T, B and NK cells in vitro did not show clonal production of all three lineages in vivo (Kondo et al., 1997). In mice, CLP, as a population, do give rise to T, B, and NK cells (Fathman et al., 2011; Karsunky et al., 2008).
In our model, as late as 6.5m, the most abundant NK clones made virtually no contributions to the other four lineages. Analysis of NK subsets demonstrated that blood 16+/56− NK cells accounted for the unique clonal pattern, distinct from the other 4 lineages, and that the less abundant 16−/56+ cells were more closely related to B/T/Gr/Mo lineages. In vitro culture assays and comparisons of node versus blood human NK cells have led to the hypothesis that lymph node 16−/dim/56+ non-cytotoxic cytokine-producing NK cells are precursors to circulating cytotoxic 16+/56−/dim NK cells (Caligiuri, 2008; Reeves et al., 2010). Our results instead suggest separate origins for these two populations. It is of interest that patients with GATA2 mutations and profoundly disordered hematopoiesis characterized by abnormalities of myeloid cells and B cells have an almost complete lack of CD16dim/CD56+ NK cells, but preservation of the CD16+/CD56dim NK population, going against the hypothesis that CD16dim/CD56+ cells are precursors of CD16+/CD56dim cells (Mace et al., 2013).
Alternatively, HSC or other multi-potent progenitors may land in “NK niches”, specifically supporting NK development or proliferation. Self-renewing NK precursors may exist, however, by 9.5m NK cell clonal architecture began to be more closely related to T/B/Gr/Mo, with shared clones contributing to all 5 lineages at appreciable levels. Finally, the most abundant CD16+/CD56− clones in the PB might represent massive expansion of clones to specific environmental stimuli, although lack of contribution of most major NK clones in T/B/Gr/Mo lineages, at any time point, makes this possibility less likely. The ability to track NK clones via barcoding in vivo and in vitro opens new avenues of investigation of this poorly-understood lineage.
Experimental Procedures
Generation of barcoded vectors
DNA barcodes consisting of a 6bp library identifier followed by a 35bp (ZH33, ZG66) or 30bp (ZH17) random barcode were produced from synthesized oligonucleotides and cloned into the non-expressing region of the HIV-derived replication-defective pCDH vector (System Biosciences Mountain View, CA) (Lu et al., 2011) (Fig 1A). High diversity libraries suitable for efficient transduction of rhesus macaque CD34+ cells were produced by calcium phosphate transfection of 293T cells with the barcoded lentiviral vector plasmid and χHIV helper plasmids (Uchida et al., 2012; Uchida et al., 2009).
The diversity of the library was assessed via transduction of the rhesus cell line LCL8864 and CD34+ primary cells, low cycle PCR, Illumina sequencing, custom python code processing, and quantitation of unique barcodes and copy number per barcode (Fig S2). Monte Carlo simulation (Supplemental Methods) utilizing this experimental data was used to determine the number of target cells able to be transduced with this vector barcode library diversity with various probabilities that >95% of retrieved barcodes represent single cells (Lu et al., 2011).
Collection, transduction and transplantation of CD34+ HSPCs
All animal studies were approved by the NHLBI Animal Care and Use Committee. Mobilization with G-CSF and stem cell factor (SCF), CD34+ cell collection and immunoselection, transduction, conditioning, and transplantation were performed as described (Donahue et al., 2005). CD34+ cells were cultured overnight on RetroNectin-coated (Takara, Otsu, Japan) plates in X-vivo 10 media (Lonza, Allendale, NJ) supplemented with 100 ng/ml each human Flt3 ligand, SCF, and thrombopoietin (R&D Systems, Minneapolis, MN), and 1% human albumin, and transduced with barcode-containing lentiviral vectors at a multiplicity of infection (MOI) of 25 in the presence of 4 μg/ml protamine sulfate (Sigma, St Louis, MO). 24 hours later, transduced CD34+ cells were reinfused into the irradiated (500cGy a day for two days) autologous macaque.
Hematopoietic cell purification and analysis
Mononuclear cells from lymph nodes were isolated by gentle mechanical disruption of tissues and collagenase dissociation (Seggewiss et al., 2007). PB cells were separated by Ficoll, stained with antibodies (Table S2) for Gr, T, B, Mo, NK cells, NK subsets and myeloid and plasmacytoid dendritic cells (Gujer et al., 2011) and sorted on a FACSAria II instrument (Fig S3), obtaining purities given in Table S1. 15 ml BM aspirates were obtained from left and right iliac crests. DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) and quantified by NanoDrop (NanoDrop, Wilmington, DE). qPCR was performed to determine vector copy number with an ABI PRISM 7700 (Applied Biosystems, Foster City, CA) using primers and probes given in Table S3.
Barcode extraction, sequencing and analysis
200ng (ZH33 and ZG66) or 500ng (ZH17) DNA underwent a first round of 14 cycle PCR with primers bracketing the barcode, and a second round of 14 cycles with primers adding a sample ID to facilitate multiplex sequencing (Table S3). Equal amounts of gel-purified product from individual samples were pooled together for multiplex sequencing (Illumina HiSeq 2000, San Diego, CA). Sequencing output was processed using custom python code (Supplemental Methods) to extract barcodes via library IDs, combining barcodes to allow for sequencing indels and mismatches up to 2bp, and setting a lower cut-off for read numbers to eliminate 99.5% of background “false barcodes” for each animal (Lu et al., 2011). Barcodes with total read numbers less than these cut-offs (for ZH33-1845; ZH17-1686; and ZG66-541) were eliminated, and a cumulative master list of barcodes with total reads over these cut-offs was generated for each animal (Table S4). The raw sequencing data from all time points and lineages was then reanalyzed versus this master list to pull out sequences representing these barcodes, allowing for mismatches and indels up to 2bps, and determining the final read number for each barcode in each sample. The fractional representation of each barcode in a sample was then calculated from the ratio of an individual barcode's read number divided by the total number of valid barcode reads (Table S4).
For all pairwise comparisons between samples collected from different lineages or at different time points for ZH33 and ZG66, a combined read number “sampling” threshold was set to exclude 95% of clones scoring as falsely “biased” (>10x barcode ratio) between the replicate samples (Fig S4), and was determined to be 1144, based on the median value calculated for all available replicate samples (n=10).
Computational and statistical analysis
Data analysis and plot generation was done using R code, ggplot2, corrplot, and reshape2 software. Pearson and Spearman correlation coefficients were calculated using R (R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0, http://www.R-project.org/). P values for highly biased clones were calculated using Fisher's exact test comparing the frequency of highly biased clones against the overall frequency of highly biased clones at the time point combination being considered.
For global analyses of clonal contributions via hierarchical clustering, three steps of normalization were applied to the total data set consisting of the valid read number for all barcodes: fractional contribution of each barcode to the overall reads in each individual sample, 75th percentile normalization, and log10 transformation. Unsupervised two-way hierarchical clustering by a Euclidean distance algorithm was applied to all samples. K-mean clustering with a k=5 was applied to all lineages at specific time points, filtering out any barcodes not present in any sample prior to K-mean clustering.
Supplementary Material
Highlights
Clonal tracking of individual primate hematopoietic stem and progenitor cell output
Unilineage, bilineage, and multilineage clones can be mapped post-transplant
Primate myeloid and B cells are more closely related than B and T cells
The dominant blood fraction of NK cells forms a unique hematopoietic lineage
Acknowledgments
This research was supported by the Division of Intramural Research of the NHLBI, NHLBI grants NIH-UO1-HL099999 to IW, NIH-K99-HL11304 to RL, and CIRM-TG2-01159 to RL. We thank Naoya Uchida for the χHIV packaging system, Danny Douek for helpful comments, and Ann Williams (NHLBI FACS Core), and Jun Zhu (DNA Sequencing and Genomics Core) for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: CW, BL, AJ, SJK, FK and KL performed and analyzed the experiments; CW, BL, SJK, RL, IW and CED wrote the paper; CED, CW, RL and BL designed the experiments, AEK, MM and RED transplanted the macaques; RL wrote the python code and ran data analyses; COW, SJK, YY, RL and BL performed computational and statistical analyses; ISYC provided important insights.
References
- Abkowitz JL, Catlin SN, Guttorp P. Evidence that hematopoiesis may be a stochastic process in vivo. Nat Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- Abkowitz JL, Catlin SN, McCallie MT, Guttorp P. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood. 2002;100:2665–2667. doi: 10.1182/blood-2002-03-0822. [DOI] [PubMed] [Google Scholar]
- An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao SH, Bonifacino A, Krouse AE, Darlix JL, Baltimore D, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci USA. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RG, Peterson LJ, Morris J, Potter J, Heyward S, Gough M, Bryant E, Kiem HP. Differential engraftment of genetically modified CD34(+) and CD34(−) hematopoietic cell subsets in lethally irradiated baboons. Exp Hematol. 2000;28:508–518. doi: 10.1016/s0301-472x(00)00137-5. [DOI] [PubMed] [Google Scholar]
- Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci U S A. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyan D, Iscove NN. Cell Stem Cell. 8:48–58. doi: 10.1016/j.stem.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28:755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Bartolomae CC, Cesana D, Cartier N, Aubourg P, Ranzani M, Cesani M, Benedicenti F, Plati T, Rubagotti E, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, Sellers S, Hematti P, Schmidt M, von Kalle C, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo L, Caccavelli L, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Catlin SN, Busque L, Gale RE, Guttorp P, Abkowitz JL. The replication rate of human hematopoietic stem cells in vivo. Blood. 2011;117:4460–4466. doi: 10.1182/blood-2010-08-303537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AM, Nguyen LV, Carles A, Beer P, Miller PH, Knapp DJ, Dhillon K, Hirst M, Eaves CJ. Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood. 2013;122:3129–3137. doi: 10.1182/blood-2013-06-508432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;10:690–697. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Coulombel L. Identification of hematopoietic stem/progenitor cells: strength and drawbacks of functional assays. Oncogene. 2004;23:7210–7222. doi: 10.1038/sj.onc.1207941. [DOI] [PubMed] [Google Scholar]
- Deichmann A, Brugman MH, Bartholomae CC, Schwarzwaelder K, Verstegen MM, Howe SJ, Arens A, Ott MG, Hoelzer D, Seger R, et al. Insertion sites in engrafted cells cluster within a limited repertoire of genomic areas after gammaretroviral vector gene therapy. Mol Ther. 2011;19:2031–2039. doi: 10.1038/mt.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, Glimm H, Gyapay G, Prum B, Fraser CC, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue RE, Dunbar CE. Update on the use of nonhuman primate models for preclinical testing of gene therapy approaches targeting hematopoietic cells. Hum Gene Ther. 2001;12:607–617. doi: 10.1089/104303401300057289. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Kuramoto K, Dunbar CE. Large animal models for stem and progenitor cell analysis. Curr Protoc Immunol. 2005;Chapter 22(Unit 22A):21. doi: 10.1002/0471142735.im22a01s69. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 2011;118:5439–5447. doi: 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A, Howe SJ, Recchia A, Cattoglio C, Wang W, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- Gerrits A, Dykstra B, Kalmykowa OJ, Klauke K, Verovskaya E, Broekhuis MJ, de Haan G, Bystrykh LV. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115:2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- Guenechea G, Gan OI, Dorrell C, Dick JE. Distinct classes of human stem cells that differ in proliferative and self-renewal potential. Nat Immunol. 2001;2:75–82. doi: 10.1038/83199. [DOI] [PubMed] [Google Scholar]
- Gujer C, Sundling C, Seder RA, Karlsson Hedestam GB, Lore K. Human and rhesus plasmacytoid dendritic cell and B-cell responses to Toll-like receptor stimulation. Immunology. 2011;134:257–269. doi: 10.1111/j.1365-2567.2011.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RJ, Wagner JE, Celano P, Zicha MS, Sharkis SJ. Separation of pluripotent haematopoietic stem cells from spleen colony-forming cells. Nature. 1990;347:188–189. doi: 10.1038/347188a0. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Tisdale JF, Wu T, Takatoku M, Sellers SE, Zickler P, Metzger ME, Agricola BA, Malley JD, Kato I, et al. Many multipotential gene-marked progenitor or stem cell clones contribute to hematopoiesis in nonhuman primates. Blood. 2000;96:1–8. [PubMed] [Google Scholar]
- Kim YJ, Kim YS, Larochelle A, Renaud G, Wolfsberg TG, Adler R, Donahue RE, Hematti P, Hong BK, Roayaei J, et al. Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector-transduced CD34+ cells. Blood. 2009;113:5434–5443. doi: 10.1182/blood-2008-10-185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim N, Presson AP, Metzger ME, Bonifacino AC, Sehl M, Chow SA, Crooks GM, Dunbar CE, An DS, et al. Dynamics of HSPC repopulatoin in nono-human primate revealed by a decade-long clonal tracking study. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K, von Neuhoff N, Schlegelberger B, Li Z, Baum C. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Wahlers A, Kuhlcke K, Stahle B, Zander AR, Baum C, Fehse B. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood. 2003;102:3934–3937. doi: 10.1182/blood-2003-05-1424. [DOI] [PubMed] [Google Scholar]
- Larochelle A, Savona M, Wiggins M, Anderson S, Ichwan B, Keyvanfar K, Morrison SJ, Dunbar CE. Human and rhesus macaque hematopoietic stem cells cannot be purified based only on SLAM family markers. Blood. 2011;117:1550–1554. doi: 10.1182/blood-2009-03-212803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle A, Vormoor J, Hanenberg H, Wang JC, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao XL, Kato I, et al. Identification of primitive human hematopoietic cells capable of repopulating NOD/SCID mouse bone marrow: implications for gene therapy. Nat Med. 1996;2:1329–1337. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotech. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace EM, Hsu AP, Monaco-Shawver L, Makedonas G, Rosen JB, Dropulic L, Cohen JI, Frenkel EP, Bagwell JC, Sullivan JL, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood. 2013;121:2669–2677. doi: 10.1182/blood-2012-09-453969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio D, Benjamin J, Kim D, Lombardo G, Daucher M, Kinter A, Nies-Kraske E, Marcenaro E, Moretta A, Fauci AS. Identification of NKG2A and NKp80 as specific natural killer cell markers in rhesus and pigtailed monkeys. Blood. 2005;106:1718–1725. doi: 10.1182/blood-2004-12-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, Schambach A, Charrier S, Galy A, Thrasher AJ, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, Benedicenti F, Sergi LS, Ambrosi A, Ponzoni M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Perie L, Swart E, Gerlach C, van Rooij N, de Boer RJ, Schumacher TN. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Park CY, Majeti R, Weissman IL. In vivo evaluation of human hematopoiesis through xenotransplantation of purified hematopoietic stem cells from umbilical cord blood. Nat Protoc. 2008;3:1932–1940. doi: 10.1038/nprot.2008.194. [DOI] [PubMed] [Google Scholar]
- Reeves RK, Gillis J, Wong FE, Yu Y, Connole M, Johnson RP. CD16- natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood. 2010;115:4439–4446. doi: 10.1182/blood-2010-01-265595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K, Swart E, van Heijst JW, Gerlach C, Castrucci M, Sie D, Heimerikx M, Velds A, Kerkhoven RM, Arens R, et al. Dissecting T cell lineage relationships by cellular barcoding. The J Exp Med. 2008;205:2309–2318. doi: 10.1084/jem.20072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Zickler P, Hoffman G, Haas S, Wissler M, Muessig A, Tisdale JF, Andrews RG, Wu T, Kiem H-P, et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002 doi: 10.1182/blood-2002-02-0407. 2002-2002-0407. [DOI] [PubMed] [Google Scholar]
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, Schmidt S, Prinz C, Wissler M, King DJ, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggewiss R, Lore K, Guenaga FJ, Pittaluga S, Mattapallil J, Chow CK, Koup RA, Camphausen K, Nason MC, Meier-Schellersheim M, et al. Keratinocyte growth factor augments immune reconstitution after autologous hematopoietic progenitor cell transplantation in rhesus macaques. Blood. 2007;110:441–449. doi: 10.1182/blood-2006-12-065623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd BE, Kiem HP, Lansdorp PM, Dunbar CE, Aubert G, LaRochelle A, Seggewiss R, Guttorp P, Abkowitz JL. Hematopoietic stem-cell behavior in nonhuman primates. Blood. 2007;110:1806–1813. doi: 10.1182/blood-2007-02-075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Weissman IL, Heimfeld S. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc Natl Acad Sci U S A. 1991;88:2788–2792. doi: 10.1073/pnas.88.7.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Uchida N, Hargrove PW, Lap CJ, Evans ME, Phang O, Bonifacino AC, Krouse AE, Metzger ME, Nguyen AD, Hsieh MM, et al. High-efficiency transduction of rhesus hematopoietic repopulating cells by a modified HIV1-based lentiviral vector. Mol Ther. 2012;20:1882–1892. doi: 10.1038/mt.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Washington KN, Hayakawa J, Hsieh MM, Bonifacino AC, Krouse AE, Metzger ME, Donahue RE, Tisdale JF. Development of a human immunodeficiency virus type 1-based lentiviral vector that allows efficient transduction of both human and rhesus blood cells. J Virol. 2009;83:9854–9862. doi: 10.1128/JVI.00357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verovskaya E, Broekhuis MJ, Zwart E, Ritsema M, van Os R, de Haan G, Bystrykh LV. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 2013;122:523–532. doi: 10.1182/blood-2013-01-481135. [DOI] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Doedens M, Dick JE. Primitive human hematopoietic cells are enriched in cord blood compared with adult bone marrow or mobilized peripheral blood as measured by the quantitative in vivo SCID-repopulating cell assay. Blood. 1997;89:3919–3924. [PubMed] [Google Scholar]
- Webster RL, Johnson RP. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology. 2005;115:206–214. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Jares A, Winkler T, Xie J, Metais JY, Dunbar CE. High efficiency restriction enzyme-free linear amplification-mediated polymerase chain reaction approach for tracking lentiviral integration sites does not abrogate retrieval bias. Hum Gene Ther. 2013;24:38–47. doi: 10.1089/hum.2012.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.