In this study, the authors characterize mRNA and protein expression of RBPMS in the retina and present qualitative and quantitative evidence for its potential application as a specific marker for RGCs independent of their connectivity to their central target areas. The data show that this marker could be used as an alternative to retrograde labeling for quantitative analysis of RGCs in normal retina and in retinas with RGC degeneration in glaucoma or other neuropathies.
Abstract
Purpose.
To characterize expression of the RNA binding protein (RBPMS) in the retina as a specific marker for retinal ganglion cells (RGCs).
Methods.
Optic nerve transection (ONT) was performed on adult male Wistar rats. Retrograde RGC labeling was performed with FluoroGold (FG) applied to the cut surface of the optic nerve. RBPMS mRNA and protein expression in the retina was analyzed by in situ hybridization and immunohistochemistry, respectively. The expression of RBPMS in various rat tissues was analyzed with semiquantitative RT-PCR.
Results.
RBPMS mRNA and protein expression was localized primarily to irregularly shaped cells in the ganglion cell layer of the retina. Quantitative analysis showed that almost 100% of RGCs labeled by FG were also RBPMS-positive, irrespective of their location relative to the optic nerve head. Approximately 94% to 97% of RBPMS-positive cells were also positive for Thy-1, neurofilament H, and III β-tubulin. In 2-week ONT retinas, the remaining few RGCs were weakly stained with RBPMS compared with intact RGCs in control retinas. Outside the retina, expression of RBPMS was observed in the heart, kidney, liver, and lungs. No expression was detected in any neuronal tissues except the retina.
Conclusions.
The data indicate that in the retina RBPMS is selectively expressed in RGCs and therefore could serve as a marker for RGC quantification in normal retinas and for estimation of RGC loss in ocular neuropathies.
Ganglion cells, which carry the final neuronal output of the vertebrate retina, collect visual signals from the two preceding layers of nerve cells, bipolar, and amacrine cells, and transmit this information to the brain. The death of retinal ganglion cells (RGCs) and degeneration of their axons in the optic nerve is the cause of vision loss in various optic neuropathies including glaucoma. In experimental rodent models of glaucoma, to evaluate the RGC loss, these cells are commonly retrogradely labeled by injection of tracers such as FluoroGold (FG), DTMR, or DiI into areas of the brain that are targeted by RGCs, primarily superior colliculus (SC), or by exposing the axons in the optic nerve to these dyes. However, both procedures have limitations. Since RGC retrograde labeling with these tracers depends on active axonal transport,1 which has been shown to be affected in animal models of glaucoma,2–6 these labeling techniques do not differentiate between cell loss, axon degeneration and failure of transport. Furthermore, labeling via SC leaves uncounted RGCs projecting to other brain areas. Nevertheless, despite the availability of several antigenic RGC markers, including Thy-1, Brn3, neurofilament and others, retrograde labeling is commonly viewed as the most reliable and accurate way of identifying RGCs.
In the present study, we characterized expression of RNA-binding protein with multiple splicing, RBPMS, or hermes in the retina. We present data supporting the use of anti-RBPMS antibodies for quantitative analysis of the number of RGCs, independent of their connectivity to their central target areas. RBPMS was recently identified in a study designed to analyze gene expression profiles in RGCs.7 RBPMS genes (RBPMS and its paralogue RBPMS2) are members of the RRM (RNA recognition motif) family of RNA-binding proteins. Members of the RRM family are involved in the regulation of gene expression at the posttranscriptional level, including pre-mRNA-processing (splicing, capping, and polyadenylation), RNA stability, transport, localization, and translational regulation. The hermes RNA-binding domain is similar to that of the Drosophila couch potato (cpo) and Caenorhabditis elegans Mec-8 genes (see Fig. 1).8 Mutations in cpo lead to several neurologic phenotypes, including bang-sensitive paralysis, seizure susceptibility, and synaptic transmission defects, indicating an important role for cpo in regulating normal function of the nervous system,9 whereas mutations in Mec-8 affect mechanosensory and chemosensory neuronal function.10 Although the exact functions of hermes genes are unknown, it has been reported that RBPMS could be involved in regulation of mRNA translation and localization during Xenopus laevis development.11 RBPMS has also been shown to physically interact with Smad2, -3, and -4,12 which regulate TGF-β signaling13 and with ataxin 1, a protein responsible for spinocerebellar ataxia type 1 due to expansion of a polyglutamine repeat.14
Figure 1.

RBPMS amino acid sequences and their homology to Drosophila couch potato (cpo) and RBPMS2 proteins. High consensus, red; low consensus, blue; and neutral, black. The RRM, yellow box; and GGKAEKENTPSEANLQEEEVR, used for antibody production, green box. Sequence alignment was performed with MultAlin (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html).53
Methods
Animals
The use of animals for this study was approved by the Animal Research Committee of the University of California, Los Angeles, and was performed in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Adult male Wistar rats (250–300 g) were housed with standard food and water provided ad libitum. The animal room was lit with fluorescent lights (330 lux) automatically turned on at 3 AM and off at 3 PM and was maintained at 21°C. The animals were maintained for at least 1 week in this environment before surgical procedures.
Optic Nerve Transection
The animals were anesthetized by inhalation of isoflurane (1.5%–3.5%) in oxygen, and the optic nerve sheath was incised 2 mm longitudinally, starting 3 mm behind the globe to expose the optic nerve. A cross section of the optic nerve was made with a needle knife without damaging the adjacent blood supply.7 The conjunctival incision was sutured and ophthalmic ointment (tobramycin, Tobrex; Alcon, Fort Worth, TX) was applied topically. ONT was performed on one eye of each rat, while the contralateral eye was used as an untreated control.
Retrograde Labeling
Retrograde labeling to identify RGCs was performed by placing a small piece of foam (Gelfoam; Pharmacia & Upjohn-Pfizer, New York, NY) soaked with 6% FG (Fluorochrome, Denver, CO) to the proximal cut surface of the optic nerve after ONT. The rats were euthanatized 1 day or 14 days after ONT. There was no RGC loss on 1 day after ONT, whereas more than 95% of the RGC population was lost 14 days afterward.
RGC Quantification in Wholemounted Retina
For counting RGCs, the eyes were enucleated and immersed in 4% paraformaldehyde in 0.1 M phosphate buffer for 1 hour. The retinas were dissected from the eyeballs and mounted on glass slides with the ganglion cell layer (GCL) facing upward. With several radial cuts, the retina was divided into four quadrants: superior, inferior, nasal, and temporal. Three sampling fields (0.32 × 0.24 mm each) were imaged at each region of 1, 2, 3, and 4 mm from the center of the optic nerve in each retinal quadrant by fluorescence microscopy (LSM410; Carl Zeiss, Oberkochen, Germany) at 200× magnification. The number of RGCs in 48 sampling fields from each retina were counted and analyzed. Morphologically distinguishable glial cells (bright and small cells) were not counted in the retina after ONT. Quantification was performed in a masked manner.
Semiquantitative Polymerase Chain Reaction
Total RNA was extracted from various rat tissues (RNAzol B; Tel-Test, Friendswood, TX) and further purified (RNeasy MinElute Cleanup kit; Qiagen, Valencia, CA). Five micrograms of the total RNA was reverse transcribed to the first-strand cDNA (RETROscript First Strand Synthesis Kit; Applied Biosystems/Ambion, Austin, TX). The cDNA was amplified by PCR with primers specific to the target sequence: RBPMS, 5′-ATTTGTCAGCGGTCTGCCTCT (forward) and 5′-CACCAGTGAAGACCGCATTAATA (reverse); and β-actin, 5′-CTAGACTTCGAGCAAGAGATGGCCACT (forward) and 5′-TAGGAGCCAGGGCAGTAATCTCCTTCT (reverse). Amplification of β-actin was used as a standard to ensure that equivalent amounts of RNA were included in each assay. Amplification conditions were as follows: hot start of 2 minutes at 95°C; 25 cycles of denaturing (95°C for 30 seconds), annealing (60°C for 15 seconds), and extension (72°C for 1 minute); and a final extension of 7 minutes at 72°C. Dilutions of cDNA in the PCR were adjusted with the purpose of staying within the linear range of amplification. The PCR products were quantified using the spot densitometry function of the imager (Alpha Imager 2000; Alpha Innotech, San Leandro, CA) after electrophoresis in a 1.5% agarose gel and staining with 0.5 mg/mL ethidium bromide.
Combination of Retrograde Labeling and In Situ Hybridization
Sequence-specific primers, 5′-ATTTGTCAGCGGTCTGCCTCT (forward) and 5′-ACACCAGTGAAGACCGCATTAATA (reverse), were used in the PCR to generate RBPMS cDNA fragment. This fragment was subcloned into the PCR II TOPO vector (Invitrogen, Carlsbad, CA) containing T7 and Sp6 RNA polymerase promoter sequences. Digoxigenin (DIG)-labeled antisense and sense cRNA probes were synthesized by in vitro transcription with either T7 or Sp6 RNA polymerase according to the manufacturer's protocol (Roche Applied Science, Indianapolis, IN). Sense RNA probes were used in these experiments as the negative control. According to the standard protocol (Roche Applied Science) with minor modifications, in situ hybridizations were performed on 10-μm-thick frozen retinal sections in which RGCs were retrogradely labeled with FG as previously described. Briefly, the sections were washed with PBS for 30 minutes, and equilibrated for 15 minutes in 5× SSC (0.75 M NaCl, 0.075 M Na-Citrate). Prehybridization was performed for 2 hours in a solution containing 50% formamide, 5× SSC, and 40 μg/mL salmon sperm DNA. Sections were then hybridized for 12 to 24 hours in a humid chamber at 58°C in a prehybridization solution, with the addition of the digoxigenin-labeled RNA probe at a concentration of 400 ng/mL. After washing twice with 2× SSC and twice with 0.1× SSC at 65°C for 1 hour, the sections were incubated with an alkaline phosphatase conjugated anti-digoxigenin antibody. Color staining was obtained by incubating sections with NBT/BCIP (nitroblue tetrazolium salt/5-bromo-4-chloro-3-indolyl-phosphate; Roche Applied Science).
After the in situ hybridization, primary rabbit anti-FG polyclonal (1/400; Fluorochrome) and secondary Alexa Fluor 488 goat anti-rabbit IgG (1/1000) antibodies were used to detect FG-labeled cells.
Retrograde Labeling and Co-localization of RBPMS on Wholemounted Retina
One day after FG labeling, the animals were euthanatized by inhalation of carbon dioxide. The retinas were immediately dissected and fixed in with 4% paraformaldehyde in 0.1 M phosphate buffer for 1 hour, washed extensively with PBS. and processed for whole retina immunohistochemistry according to a published protocol with minor modifications.15 The retinas were incubated with 10% fetal bovine serum for 1 hour to block nonspecific staining and then primary antibody against RBPMS (1/500) in PBS containing 1% Triton, 0.5% BSA, and 0.9% sodium chloride (PBS-T-BSA) overnight at 4°C. Anti-RBPMS antibodies were generated against N-terminal GGKAEKENTPSEANLQEEEVR polypeptide (Fig. 1; ProSci, Poway, CA). The following day, the retinas were washed in PBS-T-BSA and then incubated with secondary Alexa Fluor 488 goat anti-rabbit IgG antibody (1/1000) overnight at 4°C. They were then washed with PBS and counterstained with DAPI, flatmounted, and prepared for quantification under a fluorescence microscope as described earlier. In each sampling field, RGCs labeled by (1) FG and RBPMS immunoreactivity, (2) FG without RBPMS immunoreactivity, and (3) RBPMS immunoreactivity without FG, were counted.
Retrograde Labeling and Double Fluorescence Immunohistochemistry of RGC Markers
One day after FG labeling, the animals were deeply anesthetized with intramuscular injections of 0.8 mL/kg of a cocktail containing ketamine (100 mg/mL), 2.5 mL xylazine (20 mg/mL), 1.0 mL acepromazine (10 mg/mL), and 1.5 mL normal saline and transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate buffer. The eyes were enucleated, immersed in fixative for 1 hour, bisected, and postfixed for 3 hours. The eye cups were incubated with 30% sucrose at 4°C overnight and embedded in OCT compound (Sakura Finetec, Torrance, CA). Ten-micrometer-thick sections were obtained along the vertical meridian through the optic nerve head. The sections were washed three times with PBS in triton-X (T-PBS) for 5 minutes and incubated with blocking buffer (T-PBS containing 5% bovine serum albumin) at room temperature for 30 minutes. After blocking, they were incubated overnight at 4°C with primary antibodies against RBPMS and Thy-1 (1:200, mouse; Chemicon, Temecula, CA), 200 kDa neurofilament (NF-H; 1:500, mouse; Chemicon) or III β-tubulin (TUJ1, 1:200, mouse; Covance, Emeryville, CA). Thy-1 and NF-H are RGC markers and III β-tubulin is a neuronal marker used to distinguish RGCs from displaced amacrine cells in the GCL. The sections were incubated with FITC-conjugated anti-rabbit secondary antibody and rhodamine-conjugated anti-mouse antibody at room temperature for 1 hour and then mounted (Gel/Mount; Biomeda, Foster City, CA) antifade mounting solution. Photomicrographs of the retinal sections were taken with a fluorescence microscope (LSM510; Carl Zeiss Meditec). Negative control sections were incubated without primary antibodies. At least three sections from each eye and four eyes were included for quantitative analysis. For each retinal section, the number of cells in the GCL labeled by FG with immunoreactivity of (1) RBPMS and NF/Thy-1/III β-tubulin, (2) RBPMS but no NF/Thy-1/III β-tubulin, (3) NF/Thy-1/III β-tubulin but no RBPMS were counted.
Results
RBPMS mRNA Expression Profile
RT-PCR was used to determine whether RBPMS is expressed in the retina and whether its expression is affected after ONT. ONT leads to rapid and specific degeneration of RGC, and 2 weeks after transection more than 95% of RGCs loss has been reported in number of studies. Expression of RBPMS in untreated retinas and absence of the corresponding product in retinas 2 weeks after ONT suggest that RBPMS is expressed in RGCs (Fig. 2A). Amplification of β-actin cDNA fragments indicates no significant differences in the amounts of the first-strand cDNA from control and ONT retinas used in this experiment as PCR templates. In situ hybridization with RBPMS antisense riboprobe showed that RBPMS-positive cells were in the retina predominantly localized in the GCL (Fig. 3). Most if not all RBPMS-positive cells were co-localized with retrogradely labeled RGCs. Expression of RBPMS was also analyzed in several other tissues, including cerebellum, cortex, heart, kidney, liver, muscle, lung, intestine, spinal cord, spleen, testis, and prostate (Fig. 2B). Of interest, no RBPMS transcript was detected in any neuronal tissues tested in this experiment. In addition to the retina, PCR products corresponding to RBPMS mRNA were present in the heart, kidney, liver, and lung. β-Actin cDNA amplification shows equal amount of template used in tissue profiling of RBPMS mRNA expression.
Figure 2.
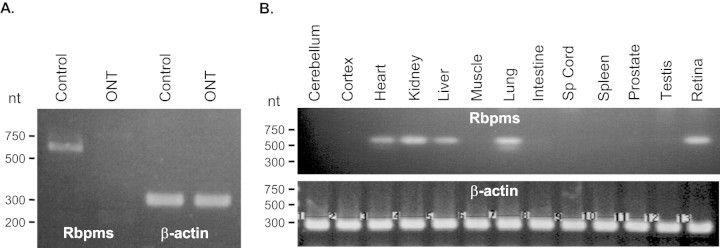
Semiquantitative RT-PCR analysis of RBPMS expression in axotomized retinas (A) and in different rat tissues (B). RBPMS expression was abolished in retinas of optic nerve transection model 2 weeks after surgery (A). Transcription of RBPMS was present in the retina, heart, kidney, liver, and lungs. The amount of the first-strand cDNA used in this experiment was normalized based on the density of the β-actin PCR product.
Figure 3.
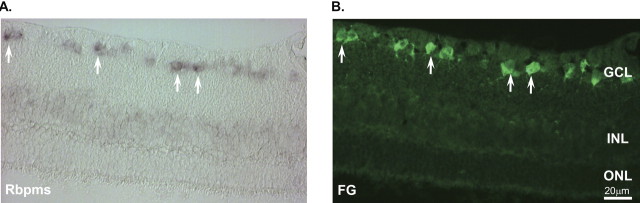
Co-localization of the RBPMS in situ hybridization signals (A) and RGCs retrogradely labeled with FG (B). Arrows: RBPMS- and FG-positive cells. ONL, outer nuclear layer; INL, inner nuclear layer.
RBPMS Immunostaining in the Retina
RBPMS specific antibodies recognizing N-terminal end of the protein (Fig. 1) were used to analyze expression of RBPMS in the retina. This protein is abundantly expressed in the GCL (Fig. 4A). RBPMS staining is present in cells with various soma sizes as well as in dendrites extending into the inner plexiform layer (IPL) and nerve fiber layer (NFL). RBPMS-positive cells were co-localized with FG-labeled RGCs (Fig. 4B). Very few FG-labeled cells appeared to be negative for RBPMS (Fig. 4C, arrows). In the ONT model, which, as described above, is characterized by rapid and specific degeneration of RGCs, very few RGCs were present (Figs. 4D, 4E). Compared with intact RGCs, most of these cells were faintly labeled with FG and weakly, almost at the level of the background, stained with RBPMS. The bright FG-positive and RBPMS-negative cell on transverse sections of ONT retina in Figure 4E (arrow) appears to be a glial cell that has engulfed dead RGCs.16,17 No immunostaining was observed in untreated or ONT retinas when RBPMS primary antibodies were omitted in the reaction.
Figure 4.
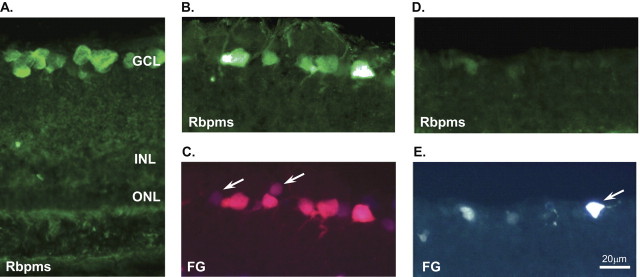
RBPMS immunohistochemistry and its co-localization with retrogradely labeled RGCs in the control retina (A–C) and in the retina 2 weeks after ONT (D, E). Most cells reacting with antibodies against RBPMS (B) and FG (C) were co-localized. (C) Arrows: RGCs that appeared to be negative for RBPMS staining. Very few RBPMS- and FG-positive cells were present in the retinas 2 weeks after ONT (D, E). Arrow: A brightly FG-positive and RBPMS-negative cell (E) appears to be a glial cell that has engulfed dead RGCs. ONL, outer nuclear layer; INL, inner nuclear layer.
Quantitative Analysis of RBPMS-Positive Cells in the Retina
To determine whether RBPMS expression is restricted to certain types of these cells or if it is expressed in all RGCs, we analyzed the distribution of RBPMS- and FG-positive cells at four distances from the center of the optic nerve head. Almost 100% (over 99.5%) of cells labeled by FG were also RBPMS positive irrespective of their location relative to the optic nerve head (Fig. 5, Table 1). Approximately 97% of RBPMS-stained cells were back labeled by FG. Nuclear labeling with DAPI demonstrated selective labeling of RGCs with anti-RBPMS antibodies, whereas non-RGCs remained unlabeled. Large, irregularly shaped cells, as well as cells with smaller somas were among the labeled cells. Although the nuclear localization of the RBPMS is visible, it is clear that this protein is abundantly present in the cytoplasm. Many RBPMS-positive cells, especially those with medium size somas, showed prominent perinuclear staining in combination with a cytoplasmic staining pattern. Staining in large cells was present more evenly throughout the cytoplasm, with less perinuclear accumulation.
Figure 5.
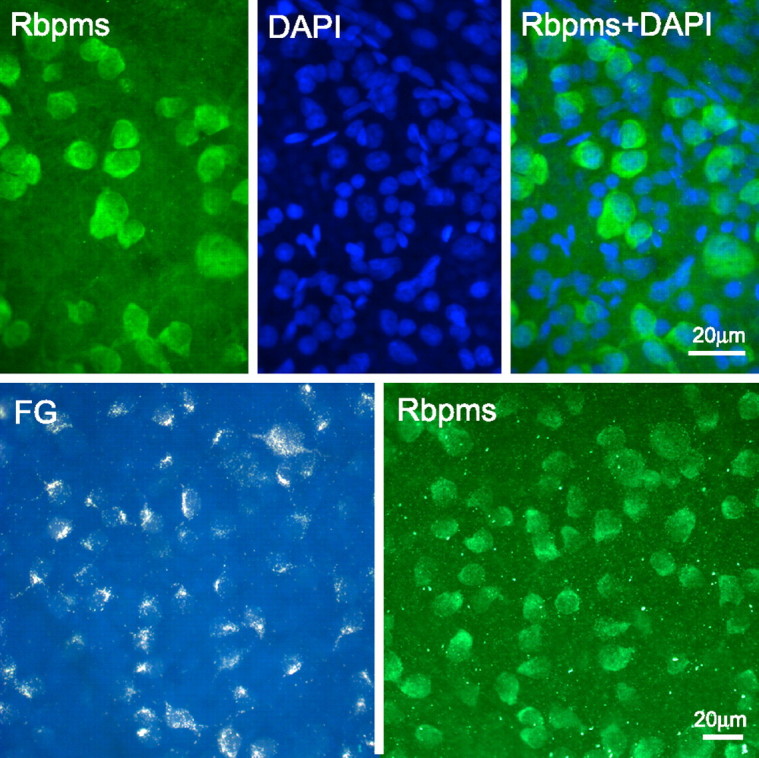
RBPMS immunohistochemistry in the wholemount retina and its co-localization with retrogradely labeled RGCs. Large, irregularly shaped cells, as well as cells with smaller somas were among the labeled cells. RBPMS staining was present in the nucleus but was predominantly localized in the cytoplasm. Many RBPMS-positive cells showed prominent perinuclear staining in combination with cytoplasmic staining pattern. An almost complete overlap of RBPMS-positive cells with FG-labeled RGCs was observed. DAPI was used for nuclear staining.
Table 1.
Quantification on Double Labeling of FG and RBPMS in Wholemounted Retinas
| Distance from Center of Optic Nerve Head (mm) |
Average | ||||
|---|---|---|---|---|---|
| 0–1 | 1–2 | 2–3 | >3 | ||
| % of RGCs (FG) are RBPMS-positive | 99.68 ± 0.16 | 99.54 ± 0.50 | 99.63 ± 0.28 | 99.63 ± 0.21 | 99.62 ± 0.06 |
| % of RBPMS-positive cells are RGCs (FG) | 97.75 ± 0.52 | 97.72 ± 1.00 | 96.90 ± 1.43 | 96.92 ± 1.26 | 97.32 ± 0.48 |
n = 4. Three images at each distance per quadrant and 48 images per retina in total.
RBPMS staining in the retina correlated with Thy-1, NF-H, and III β-tubulin immunohistochemical staining (Table 2). These proteins are known to be expressed in RGCs and are used commonly as RGC markers. Representative images of retrogradely labeled RGCs and cells stained with RBPMS, Thy-1, NF-H, or III β-tubulin are presented in Figure 6. As expected, III β-tubulin, NF-H, or Thy-1, and particularly the first two markers, showed predominant expression in the NFL, whereas RBPMS staining, although present in RGC extensions, was concentrated in cell somas. Staining patterns of RBPMS were very similar to the pattern produced by FG back-labeling. Almost complete overlapping in RBPMS staining with each of these three markers was observed. Approximately 97%, 95%, and 96% of RBPMS-positive cells were also stained with III β-tubulin, NF-H, or Thy-1, respectively.
Table 2.
Percentage of FG-Positive Cells Labeled by RBPMS and III β-Tubulin, NF-H, or Thy-1 in Retinal Sections
| III β-Tubulin |
NF-H |
Thy-1 |
||||
|---|---|---|---|---|---|---|
| + | − | + | − | + | − | |
| RBPMS | ||||||
| + | 96.69 ± 2.00 | 1.75 ± 1.30 | 94.37 ± 1.77 | 3.91 ± 1.16 | 95.65 ± 2.57 | 2.50 ± 2.05 |
| − | 1.21 ± 1.17 | 0.35 ± 0.61 | 0.70 ± 0.54 | 1.14 ± 0.96 | 1.52 ± 0.77 | 0.32 ± 0.54 |
n = 6, two retinal sections per animal.
Figure 6.
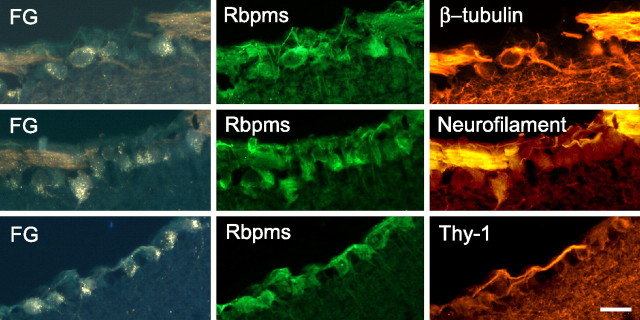
Immunohistochemical co-localization of RBPMS expression with RGC markers Thy-1 and neurofilament H (NF-H), and neuronal marker III β-tubulin. Approximately 97%, 95%, and 96% of RBPMS-positive cells were also stained with III β-tubulin, NF-H, or Thy-1, respectively (Table 2).
Discussion
In the present study, we characterized mRNA and protein expression of the RBPMS in the retina and found qualitative and quantitative evidence for its potential application as a specific RGC marker. RBPMS, or hermes, is a member of the RRM family of RNA-binding proteins and is found in various vertebrate species from fish to human. The RRM, also known as RNA-binding domain (RBD) or ribonucleoprotein domain (RNP), is one of the most abundant protein domains in eukaryotes.18 Members of the RRM family are involved in the regulation of gene expression at the posttranscriptional level, including pre-mRNA-processing (splicing, capping, and polyadenylation), RNA stability, transport, localization, and translational regulation. Two RNP consensus sequences, RNP1 and RNP2, that are involved in direct interaction with RNA have been identified in the RRM domain: Lys/Arg-Gly-Phe/Tyr-Gly/Ala-Phe/Tyr-Val/Ile/Leu-X-Phe/Tyr, where X can be any amino acid, and Ile/Val/Leu-Phe/Tyr-Ile/Val/Leu-X-Asn-Leu.19 The protein encoded by the hermes gene has a single, putative RRM domain in its N terminus, which consists of RNP1 and -2 (Fig. 1). The human RBPMS gene is located on chromosome 8, region p11-12 and spans over 230 kb.20 RBPMS can be expressed as 12 different splice variants, although splice variant 1 (NM_001008710.1) represents the most frequently occurring transcript. RBPMS2, which is considered a RBPMS paralogue, is located on 15q22.31. Only one splice variant, NM_194272, has been reported for this gene.
Earlier, we analyzed expression of RBPMS2 at mRNA level in the retinas of wild-type Wistar rats and in retinas 2 weeks after ONT, which leads to specific and almost complete (>90%) degeneration of RGCs.21 Double in situ hybridization with RBPMS2 and Thy-1 riboprobes, the RGC marker in the retina, showed that the majority of the Thy-1-positive cells in the GCL were also RBPMS2 positive. This was also shown by co-localization of hermes in situ hybridization signals in retrogradely labeled RGCs. No in situ signals were detected in the retinal sections after ONT. Dramatic reduction of the RBPMS2 mRNA in axotomized retinas was also observed by RT-PCR.
Similar to the RBPMS2 study, the first evidence of RBPMS expression in RGCs was obtained by analyzing its transcription with RT-PCR and in situ hybridization. Semiquantitative RT-PCR of RBPMS from untreated and ONT retinal RNA showed almost no expression of RBPMS in experimental RGC-deficient retinas, suggesting that the transcription of this gene must be localized in RGCs. However, since gene expression after ONT could be significantly affected in non-RGCs,22 we analyzed RBPMS mRNA localization in the retina with in situ hybridization. The results of this experiment showed predominant expression of RBPMS in the GCL, where RBPMS signals were co-localized with back-labeled RGCs.
Outside of the retina, we observed RBPMS mRNA expression mainly in the heart, liver, kidney, and lung. Two other neuronal tissues, cortex and cerebellum, that were used in this RT-PCR experiment showed no detectable RBPMS expression. Its absence in these tissues is interesting, since RBPMS has been associated with spinocerebellar ataxia type 1 (SCA1) and represents one of the main hubs in the ataxia network, where it interacts with several different ataxia-causing proteins.14 It is possible that the primers used to amplify RBPMS fragment in our experiment were not suitable for the splicing isoform(s) of RBPMS (12 isoforms have been identified for RBPMS) that are expressed in these tissues. A high level of hermes expression in myocardial tissue was detected during the early development of frog, avian, and mammalian embryos.8 RBPMS expression in the Xenopus embryo was also observed in the vegetal cortex of the oocyte and in the kidney, eye, and epiphysis. Expression in the eye was restricted to the GCL. In the chicken embryo, hermes expression was not detected in the retina or eye.23 The abundant expression of RBPMS that we observed in the rat liver and lung was not detected in the corresponding tissues of Xenopus or chicken. These differences in RBPMS expression may be isoform, species, or developmentally related.
Localization of the RBPMS protein in rat retinas was analyzed by immunohistochemistry with antibodies generated against the N-terminal peptide sequence, which is 100% conserved among rat, mouse, and human proteins. As expected from the in situ hybridization data, RBPMS protein was expressed in RGCs. Its presence was demonstrated by co-localization of RBPMS-positive cells with retrogradely labeled RGCs. RBPMS staining was primarily observed in cell somas and also in dendrites extending into the inner plexiform layer and nerve fiber layer. Both large irregularly shaped cells as well as cells with smaller somas were among the labeled cells. RBPMS expression was primarily present in the cytoplasm with weaker nuclear localization. Many RBPMS-positive cells showed prominent perinuclear staining, which suggests its association with the Golgi complex.
Since RGCs include several subtypes of cells, quantitative analysis was performed to demonstrate whether RBPMS expression is restricted to a certain subset of RGCs. Different types of RGCs have been identified based on their morphologic characteristics such as soma size, dendritic field size, and dendritic ramification in a variety of vertebrate species.24–29 Fundamental knowledge has been obtained from cat RGC studies,30–34 which established a correlation between the three main morphologic classes of RGCs—the α (3% of all RGCs), β (45%–50% of all RGCs), and non-α/non-β cells (NAB, 50%–60% of all RGCs)—and three physiological classes—Y, X, and W—classified based on characteristics such as receptive field properties, axonal conduction velocities, and the level of maintained activity.35–39 Similar RGC classifications have been reported in other mammals, including primates.40–49
The number of RBPMS-positive cells in this study correlated with the RGCs retrogradely labeled with FG. To maximize the number of labeled RGCs, we chose to apply FG to the cut surface of the optic nerve, rather than injecting it or applying it to the surface of the SC. In the rat, approximately 98% of RGCs project to the contralateral SC50,51 and up to 95% of RGCs can be labeled via the SC.52 The labeling procedure used in this study targeted all RGCs and was performed 1 day before euthanatization to avoid cellular degeneration. We showed that almost 100% of RGCs were labeled for RBPMS, indicating that this gene is expressed in the entire RGC population. Furthermore, the expression of RBPMS correlated with the expression of the RGC markers, Thy-1, and NF-H and the neuronal marker III β-tubulin. Almost complete overlapping of RBPMS staining with each of these three markers was observed.
In conclusion, the results of this study showed that RBPMS can serve as an RGC marker in the retina and can be used as an alternative to retrograde labeling for quantitative analysis of RGCs in normal retina and in retinas with RGC loss from glaucomatous or other neuropathies.
Footnotes
Supported by Research to Prevent Blindness (JC), and Oppenheimer Family Foundation (JMKK, NP).
Disclosure: J.M.K. Kwong, None; J. Caprioli, None; N. Piri, None
References
- 1. Kobbert C, Apps R, Bechmann I, Lanciego JL, Mey J, Thanos S. Current concepts in neuroanatomical tracing. Prog Neurobiol. 2000; 62: 327–351 [DOI] [PubMed] [Google Scholar]
- 2. Anderson DR, Hendrickson A. Effect of intraocular pressure on rapid axoplasmic transport in monkey optic nerve. Invest Ophthalmol. 1974; 13: 771–783 [PubMed] [Google Scholar]
- 3. Minckler DS, Tso MO, Zimmerman LE. A light microscopic, autoradiographic study of axoplasmic transport in the optic nerve head during ocular hypotony, increased intraocular pressure, and papilledema. Am J Ophthalmol. 1976; 82: 741–757 [DOI] [PubMed] [Google Scholar]
- 4. Quigley HA, Anderson DR. Distribution of axonal transport blockade by acute intraocular pressure elevation in the primate optic nerve head. Invest Ophthalmol Vis Sci. 1977; 16: 640–644 [PubMed] [Google Scholar]
- 5. Quigley HA, Addicks EM, Green WR, Maumenee AE. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch Ophthalmol. 1981; 99: 635–649 [DOI] [PubMed] [Google Scholar]
- 6. Pease ME, McKinnon SJ, Quigley HA, Kerrigan-Baumrind LA, Zack DJ. Obstructed axonal transport of BDNF and its receptor TrkB in experimental glaucoma. Invest Ophthalmol Vis Sci. 2000; 41: 764–774 [PubMed] [Google Scholar]
- 7. Piri N, Kwong JM, Song M, Elashoff D, Caprioli J. Gene expression changes in the retina following optic nerve transection. Mol Vis. 2006; 12: 1660–1673 [PubMed] [Google Scholar]
- 8. Gerber WV, Yatskievych TA, Antin PB, Correia KM, Conlon RA, Krieg PA. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech Dev. 1999; 80: 77–86 [DOI] [PubMed] [Google Scholar]
- 9. Glasscock E, Tanouye MA. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics. 2005; 169: 2137–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lundquist EA, Herman RK, Rogalski TM, Mullen GP, Moerman DG, Shaw JE. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996; 122: 1601–1610 [DOI] [PubMed] [Google Scholar]
- 11. Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006; 124: 521–533 [DOI] [PubMed] [Google Scholar]
- 12. Sun Y, Ding L, Zhang H, et al. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 2006; 34: 6314–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Massague J, Wotton D. Transcriptional control by the TGF-2/Smad signaling system. EMBO J. 2000; 19: 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim J, Hao T, Shaw C, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006; 125: 801–814 [DOI] [PubMed] [Google Scholar]
- 15. Kong WC, Cho EY. Antibodies against neurofilament subunits label retinal ganglion cells but not displaced amacrine cells of hamsters. Life Sci. 1999; 64(19): 1773–1778 [DOI] [PubMed] [Google Scholar]
- 16. Levkovitch-Verbin H, Quigley HA, Martin KR, Zack DJ, Pease ME, Valenta DF. A model to study differences between primary and secondary degeneration of retinal ganglion cells in rats by partial optic nerve transection. Invest Ophthalmol Vis Sci. 2003; 44: 3388–3393 [DOI] [PubMed] [Google Scholar]
- 17. Blair M, Pease ME, Hammond J, et al. Effect of glatiramer acetate on primary and secondary degeneration of retinal ganglion cells in the rat. Invest Ophthalmol Vis Sci. 2005; 46: 884–890 [DOI] [PubMed] [Google Scholar]
- 18. Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS Lett J. 2005; 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- 19. Dreyfuss G, Swanson MS, Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988; 13: 86–91 [DOI] [PubMed] [Google Scholar]
- 20. Shimamoto A, Kitao S, Ichikawa K, et al. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc Natl Acad Sci USA. 1996; 93: 10913–10917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piri N, Kwong JM, Song M, Caprioli J. Expression of hermes gene is restricted to the ganglion cells in the retina. Neurosci Lett. 2006; 405: 40–45 [DOI] [PubMed] [Google Scholar]
- 22. Kielczewski JL, Pease ME, Quigley HA. The effect of experimental glaucoma and optic nerve transection on amacrine cells in the rat retina. Invest Ophthalmol Vis Sci. 2005; 46: 3188–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilmore HP, McClive PJ, Smith CA, Sinclair AH. Expression profile of the RNA-binding protein gene hermes during chicken embryonic development. Dev Dyn. 2005; 233: 1045–1051 [DOI] [PubMed] [Google Scholar]
- 24. Dunn-Meynell AA, Sharma SC. The visual system of the channel catfish (Ictalurus punctatus). I. Retinal ganglion cell morphology. J Comp Neurol. 1986; 247: 32–55 [DOI] [PubMed] [Google Scholar]
- 25. Straznicky C, Straznicky IT. Morphological classification of retinal ganglion cells in adult Xenopus laevis. Anat Embryol. 1988; 178: 143–153 [DOI] [PubMed] [Google Scholar]
- 26. Ammermüller J, Kolb H. The organization of the turtle inner retina: I. ON- and OFF- center pathways. J Comp Neurol. 1995; 358: 1–34 [DOI] [PubMed] [Google Scholar]
- 27. Ammermüller J, Kolb H. The organization of the turtle inner retina: II. Analysis of color-coded and directionally selective cells. J Comp Neurol. 1995; 358: 35–62 [DOI] [PubMed] [Google Scholar]
- 28. Thanos S, Vanselow J, Mey J. Ganglion cells in the juvenile chick retina and their ability to regenerate axons in vitro. Exp Eye Res. 1992; 54: 377–391 [DOI] [PubMed] [Google Scholar]
- 29. Boycott BB, Dowling JE. Organization of the primate retina: light microscopy. Philos Trans R Soc B (Lond). 1969; 255: 109–184 [DOI] [PubMed] [Google Scholar]
- 30. Boycott B, Wässle H. The morphological types of ganglion cells of the domestic cat's retina. J Physiol. 1974; 240: 397–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saito H. Morphology and physiologically identified X-, Y- and W-type retinal ganglion cells of the cat. J Comp Neurol. 1983; 221: 279–288 [DOI] [PubMed] [Google Scholar]
- 32. Rodieck RW, Brening RK. Retinal ganglion cells: properties, types, genera, pathways and trans-species comparisons. Brain Behav Evol. 1983; 23: 121–164 [DOI] [PubMed] [Google Scholar]
- 33. Fukuda Y, Hsiao CH, Watanabe M, Ito H. Morphological correlates of physiologically identified Y-, X-, and W-cells in cat retina. J Neurophysiol. 1984; 52: 999–1013 [DOI] [PubMed] [Google Scholar]
- 34. Rowe MH, Stone J. Naming of neurons: classification and naming of cat retinal ganglion cells. Brain Behav Evol. 1977; 14: 185–216 [DOI] [PubMed] [Google Scholar]
- 35. Enroth-Cugell C, Robson JC. The contrast sensitivity of retinal ganglion cells of the cat. J Physiol. 1966; 187: 517–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cleland BG, Dubin MW, Levick WR. Sustained and transient neurons in the cat's retina and lateral geniculate nucleus. J Physiol. 1971; 217: 473–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fukada Y. Receptive field organization of cat optic nerve fibers with special reference to conduction velocity. Vision Res. 1971; 11: 209–226 [DOI] [PubMed] [Google Scholar]
- 38. Stone J, Hoffmann KP. Very slow-conducting ganglion cells in the cat's retina: a major new functional type. Brain Res. 1972; 43: 610–616 [DOI] [PubMed] [Google Scholar]
- 39. Stone J, Fukuda Y. Properties of cat retinal ganglion cells: a comparison of W-cells with X- and Y-cells. J Neurophysiol. 1974; 37: 722–748 [DOI] [PubMed] [Google Scholar]
- 40. Perry VH. The ganglion cell layer of the retina of the rat: a Golgi study. Proc R Soc Lond B. 1979; 204: 363–375 [DOI] [PubMed] [Google Scholar]
- 41. Doi M, Uji Y, Yamamura H. Morphological classification of retinal ganglion cells in mice. J Comp Neurol. 1995; 356: 368–386 [DOI] [PubMed] [Google Scholar]
- 42. Leventhal AG, Rodieck RW, Dreher B. Retinal ganglion cell classes in the old world monkey: morphology and central projections. Science. 1981; 213: 1139–1142 [DOI] [PubMed] [Google Scholar]
- 43. Ghosh KK, Goodchild AK, Sefton AE, Martin PR. Morphology of retinal ganglion cells in a new world monkey, the marmoset Callithrix jacchus. J Comp Neurol. 1996; 366: 76–92 [DOI] [PubMed] [Google Scholar]
- 44. Dacey DM, Petersen MR. Dendritic field size and morphology of midget and parasol ganglion cells of the human retina. Proc Natl Acad Sci USA. 1992; 89: 9666–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perry VH, Oehler R, Cowey A. Retinal ganglion cells that project to the dorsal lateral geniculate nucleus in the macaque monkey. Neuroscience. 1984; 12: 1101–1123 [DOI] [PubMed] [Google Scholar]
- 46. Perry VH, Cowey A. Retinal ganglion cells that project to the superior colliculus and pretectum in the macaque monkey. Neuroscience. 1984; 12: 1125–1137 [DOI] [PubMed] [Google Scholar]
- 47. Dacey DM. The mosaic of midget ganglion cells in the human retina. J Neurosci. 1993; 13: 5334–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe M, Rodieck RW. Parasol and midget ganglion cells of the primate retina. J Comp Neurol. 1989; 289: 434–454 [DOI] [PubMed] [Google Scholar]
- 49. Hattar S, Kumar M, Park A, et al. Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J Comp Neurol. 2006; 497: 326–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Forrester J, Peters A. Nerve fibres in optic nerve of rat. Nature. 1967; 214: 245–247 [DOI] [PubMed] [Google Scholar]
- 51. Chiu K, Lau WM, Yeung SC, Chang RC, So KF. Retrograde labeling of retinal ganglion cells by application of fluoro-gold on the surface of superior colliculus. J Vis Exp. 2008; 16: 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang W, Hui Y, Zhang M. Retrograde labeling of adult rat retinal ganglion cells with the fluorogold. Yan Ke Xue Bao. 2000; 16: 29–33 [PubMed] [Google Scholar]
- 53. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988; 16(22): 10881–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]


