Abstract
Background
MUC13 is over-expressed and aberrantly localized in colon cancer tissue; however, the specific functions and regulation of MUC13 expression are unknown.
Methods
Stable cell lines with either over-expressed or suppressed MUC13 levels were analyzed to determine cell growth, colony formation, cell migration, and cell invasion assays. The molecular mechanisms involved in MUC13 regulation were elucidated via chromatin immunoprecipitation (ChIP) and analysis of interleukin 6 (IL6) treatments. Colon cancer tissues were analyzed by immunohistochemistry (IHC) for the protein levels of MUC13 and P-STAT5 in colon cancer cells.
Results
Over-expression of MUC13 increased cell growth, colony formation, cell migration, and invasion. In concordance, MUC13 silencing decreased these tumorigenic features. Over-expression of MUC13 also modulated various cancer-associated proteins, including telomerase reverse transcriptase (TERT), sonic hedgehog (SHH), B cell lymphoma murine like site 1 (BMI-1), and GATA like transcription factor 1 (GATA1). Additionally, MUC13 over-expressing cells showed increased HER2 and P-ERK expression. ChIP analysis revealed binding of STAT5 to the predicted MUC13 promoter. IL6 treatment of colon cancer cells increased the expression of MUC13 via activation of JAK2/STAT5 signaling pathway. Suppression of JAK2 and STAT5 signaling by chemical inhibitors abolished IL6 induced MUC13 expression. IHC analysis showed increased expression of both P-STAT5 and MUC13 in colon cancer as compared to adjacent normal tissue
Conclusions
The results of this study, for the first time, suggest functional roles of MUC13 in colon cancer progression and provide information regarding the regulation of MUC13 expression via JAK2/STAT5 which may reveal promising therapeutic approaches for colon cancer treatment.
Keywords: IL6, MUC13 regulation, Transcription factors, JAK2/ STAT5
Introduction
Colon cancer is the second most lethal malignancy in the USA and affects over a million people every year [1]. The five year survival rate of patients diagnosed with advanced stage (stage IV) colon carcinoma is less than 10% [2, 3] indicating the need for further research to elucidate molecular pathogenesis and develop novel therapeutic strategies.
Mucins are high molecular weight glycoproteins normally expressed at mucosal surfaces to provide protection from the external environment. However, the over and aberrant expression of mucins has been reported in many cancers, including colon cancer [4-6]. MUC13 is a membrane bound mucin that is normally expressed at low levels in the epithelial surface of gastrointestinal, respiratory and reproductive tracts [7-12]. Dysregulated expression of MUC13 has been shown in ovarian, pancreatic, gastric and colon cancers [7, 8, 12-14]. MUC13 is predicted to contain an extracellular alpha subunit that consists of the tandem repeat (TR) domain, three epidermal growth factor like (EGFI-III) domains, and a Sea urchin sperm protein enterokinase arginine (SEA) domain. The intracellular beta subunit consists of the transmembrane domain (TM) and a cytoplasmic tail (CT). The TR domain, a hallmark of the mucin family, provides a scaffold to build oligosaccharide structures in which extensive O and N glycosylation occurs. The 69 amino acid cytoplasmic tail of MUC13 consists of two tyrosine and eight serine/threonine phosphorylation residues, and a PKC phosphorylation consensus motif. These phosphorylation motifs are predicted to play a critical role in tumorigenesis via signal transduction mechanisms [12]. MUC13 over-expression has been shown to enhance tumorigenic features in ovarian and pancreatic cancers, in both in vitro and in vivo models [8, 13]. As shown by us and others, MUC13 is known to be over-expressed and aberrantly localized in colon cancer tissues [7, 10]; in the present study we provide information pertaining to the functional roles and regulation of MUC13 in colon cancer cells.
Over the last decade it has become evident that cytokines are critical players in cancer pathogenesis [15, 16]. Many cancers, including gastric, colon, breast and prostate cancers, over- express interleukin 6 (IL6) [17-20]. IL6, a regulatory cytokine, uses the gp130 family of receptors which activates the JAK/STAT signaling pathway to affect downstream cellular events, such as cell growth, differentiation, survival and apoptosis [21]. Binding of IL6 to its receptor activates the gp130 subunits, causing phosphorylation of JAK and subsequent phosphorylation of STATs. Once phosphorylated, STATs translocate to the nucleus and regulate transcription of various oncogenes [22].
STAT5, a member of the STAT family of transcription factors, regulates a wide range of cellular processes that are involved in tumorigenesis and metastasis through triggering cell growth and preventing cell apoptosis [23-25]. IL6 has been shown to activate STAT5 in human epithelial cells, M1 myeloid leukemia and T-cells [26-28]. An increased level of STAT5 has been detected in colon cancer patients tissues [29] and the over-expression of P-STAT5 is a poor prognostic indicator for colon cancer [30]. Therefore, we sought to determine the involvement of these inflammatory mediators in the regulation of MUC13 expression.
In this study, we show that exogenous expression of MUC13 enhances tumorigenic features such as cell growth, colony formation, cell migration and invasion of colon cancer cells. In contrast, these tumorigenic features are reduced by suppression of MUC13. Additionally, these phenotypic changes correlate with the modulation of SHH, BMI-I, TERT, GATA1, HER2, P-ERK2 and p53 protein expression. Moreover, we show MUC13 expression is increased via the JAK2/STAT5 signaling pathway. Our results, for the first time, elucidate the regulation of MUC13 and suggest important roles of MUC13 in the progression of colon cancer. Moreover, we show the regulation of MUC13 by IL6 via JAK2/STAT5 signaling pathway.
Experimental Procedures
Cell cultures
Colon cancer cell lines (SW48, SW480, SW620, T84, and HT29), pancreatic cancer cell lines (HPAFII and MiaPaca) and ovarian cancer cell lines (CaOV-3, SKOV-3) were purchased from American tissue culture collection (ATCC). The cells were propagated as follows: CaOV-3, HPAFII, and MiaPaca 2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM). SKOV-3 cells were cultured in RPMI 1640 medium. SW48, SW480, and SW620 cells were cultured in Leivobitz's L15 medium and T84 cells were cultured in a mixture of Ham's F12 and DMEM. Media was supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 2mM L-glutamine, and 5% sodium pyruvate. Cells were cultured in a 5% CO2 humidified incubator at 37°C.
Generation of stable exogenous MUC13 expressing and MUC13 knock-down cell lines
Exogenous MUC13 expressing cell lines (pooled cell populations) were generated using lipofectamine 2000 as per manufacturer's directions (Life technologies, Grand Island, NY). Briefly, SW480 cells were transfected with either empty plasmid vector (pcDNA3.1 V5) (SW480 Vector) and/or green fluorescent protein (GFP) tagged full length human MUC13 construct (SW480 M13OE). Transfected cells were selected in medium containing G418 (400μg/ml) to obtain populations of stable pooled cells transfected either with empty vector and/or MUC13 plasmids.
Stable MUC13 knock-down cell lines (pooled cell populations) were created using MUC13 specific shRNA lentiviral particles as per manufacturer instructions (Sigma Aldrich, Saint Lois, MO). SW620 cells were transduced with scrambled shRNA (SW620 Vector) and MUC13 specific lentiviral shRNA (SW620 M13KD) for 24 hrs. The transduced cells were selected by puromycin (4μg/ml) containing media. Quantitative reverse transcriptase PCR (Q-RT-PCR) and immunofluorescence were performed to validate the expression of MUC13 in stable MUC13 exogenous and MUC13 knock-down pooled cell lines.
Cell growth assay
Cells (×105) were seeded in 6 well plates and incubated for 24, 48, 72, and 96 hours. At the end of each time point, cells were trypsinized and the total number of cells in each well was counted using an automated cell counter (Z1 Automated Particle Coulter Counter, Beckman Coulter). Cell doubling time (Td) was calculated from the growth curve during the exponential growth phase with the formula, Td = 0.693t / ln(Nt/N0), where t is time (days), Nt is cell number at time t, and N0 is cell number at initial time [8, 13].
Clonogenic assay
Cells (1×103) were seeded in 10cm diameter dish in 10 ml of complete media. Cells were allowed to grow for two weeks. The media was replaced on day 7. On 14th day, colonies were washed with phosphate buffer saline (PBS), fixed with ice-cold methanol, stained with 0.05% Hematoxylin, and air dried. Colonies were viewed under Alpha imager. Visible colonies (with >50 cells) were counted to determine clonogenic potential of these cells as described earlier [8, 13].
Cell migration and invasion assay
Cell migration and cell invasion assays were performed using migration and invasion inserts as per manufacturer protocol (BD Biosciences, San Jose, CA). For the migration assay, cells were suspended in media containing 0.1% serum and seeded into the top chamber of the cell culture insert (BD Falcon™ cell culture insert, 12 well, 8.0 micron pore size). For the invasion assay, cells were suspended in media containing 0.1% serum and seeded into the top chamber of a Matrigel coated cell culture insert (BD BioCoat BD Matrigel ™ invasion chamber, 24 well plate, 8.0 micron pore size). Media containing 10% serum was added to the bottom chamber to supply a chemoattractant. Both SW480 and SW620 cells required an additional chemoattractant, chemokine (C-C motif) ligand 21 (CCL21) (PeproTech, Rocky Hill, NJ) (100 ng/ml), to increase the number of migratory and invasive cells. Following incubation at 37°C for 24 hours, the upper side of the membrane (insert) was wiped off to remove cells that had not migrated. The membrane was then fixed with methanol, stained with 0.005% crystal violet and mounted on a glass slide. The membrane was viewed under an Olympus BX 41 microscope (Olympus Corporation; Center Valley, PA). Cells were counted in 10 random fields of view, at 20× magnification, and are reported as the average number of cells per field.
RNA isolation, reverse transcription PCR and real time PCR
Total RNA isolation and reverse transcription was performed as described earlier [8]. Briefly, total RNA was isolated using the Qiagen RNeasy RNA isolation kit (Qiagen, Valancia, CA). Total RNA (2μg) was reverse transcribed using superscript II RNase H-Reverse transcriptase High Capacity RNA to cDNA kit (Applied Biosystems, Foster City, CA). The resultant cDNA was amplified with appropriate primers (supplementary Table 2) using SYBR Green PCR reaction mixture (Applied Biosystems) in a Stratagene Mx3005P QPCR System (Stratagene, La Jolla, CA).
SDS-PAGE and Western blotting
Western blot was performed as described previously [7, 8, 13] using appropriate primary and secondary antibodies (manufacturer name, dilution factor and source of antibody) as listed in supplementary Table 1.
Immunofluorescence and confocal microscopy
Confocal microscopy was performed as described earlier [8]. Briefly, cells were grown in glass chamber slides (Thermo Scientific, Rockford, IL). After incubation for 24 to 48 hours and treatment as indicated, cells were washed with Hanks buffer containing 0.1M HEPES media and PBS containing 0.05% Tween-20. Cells were then fixed with ice cold methanol and blocked with 10% goat serum for 1 hour at RT. Cells were incubated with anti-MUC13 (ppz0020) or P-STAT5 (Santa Cruz 1:200) antibodies, washed and then incubated in FITC or Alexa-568 conjugated goat anti-mouse or goat anti-rabbit secondary antibody. After washing, cover slips were mounted using Vectashield DAPI containing mounting media (Vector Laboratories, Burlingame, CA). Immunostaining was observed and photographed under an Olympus Fluoview FV1000 confocal laser microscope (Olympus Corporation).
Real Time PCR array analysis
Cancer PathwayFinder RT profiler PCR array (96 well format, SA Biosciences, Valencia, CA). This array contained 84 genes which are involved in various cancers along with 6 constitutively active genes as an internal control. PCR array analysis was performed as per manufacturer's protocol (SA biosciences,). Briefly, the RNA samples from SW480 M13OE and SW480 Vector cells were prepared and cDNA was synthesized as described earlier using superscript II RNAase H (High capacity RNA to cDNA kit). The cDNA was amplified by SYBER Green real time PCR using gene specific primers provided in the 96 well plate of the Q-RT-PCR array. The quantitative analysis of the PCR amplification was performed using the software provided by the manufacturer. The expression of various genes in SW480 M13OE cells was determined relative to SW480 Vector cells. Genes that showed at least a 2 fold relative change in expression were considered to be significantly altered.
Chromatin Immunoprecipitation (ChIP) analysis
ChIP assay was performed using the Simple ChIP Enzymatic Chromatin Immunoprecipitation kit according to manufacturer's recommended procedure (Cell Signaling Technology). Briefly, cells were grown in several 150mm plates to 90% confluency (total cell number: 40×106). Cells were cross-linked by adding formaldehyde directly to the media until a 1% formaldehyde concentration was reached. After 10 mins, 10× glycine was added for 5 mins. The media/formaldehyde/gylcine solution was then removed and cells were washed with ice cold PBS. Cells were scraped and centrifuged in PBS containing pregnant mare serum factor. After washing with PBS, cells were subjected to Micrococcal Nulease enzyme digestion at 37°C for 20 mins with frequent mixing. Enzymatic digestion was stopped by 0.5M EDTA, resulting in 100-700 bp cross-linked DNA fragments. The nuclear membrane was broken by sonication (VirTis Versonic 100 ultrasonic homogenizer at setting 6 with a 1/8-inch probe). Cross-linked chromatin samples were further digested by RNAase and proteinase K and purified DNA samples were obtained. The DNA concentration was determined by UV spectrophotometry at 260 nm. Equal amounts of purified DNA were immunoprecipitated by Histone 3 (positive control), non immune IgG (negative control) and ChIP grade STAT5 antibodies (Cell Signaling Technology). The immunoprecipitate was extracted using protein G magnetic beads and proteinase K digestion. Pure DNA was amplified by semi-quantitative PCR and SYBR Green quantitative real time PCR using primers for the STAT5 binding sequence (SA biosciences). For quantitative analysis, gene enrichment was normalized to input and shown as a percentage of the input.
IL6 treatment and inhibition of JAK2 and STAT5
Cells at 60-80% confluent were washed with PBS, serum starved for 6-8 hrs and then treated with either DMSO (vehicle control, Sigma Aldrich, St. Louis, MO) or IL6 (#8904, Cell Signaling Technology, Danvers, MA, dose range: 25-300 ng/ml) for 48-72 hrs and analyzed by Q-RT-PCR, immunoblotting, and confocal microscopy. For the inhibitor assays, serum starved cells were pretreated with JAK2 inhibitor AG490 (EMD Millipore, Calbiochem, Billerica, MA) (dose range 50-200 nM) and/or STAT5 inhibitor (Santa Cruz Biotechnology Inc, Santa Cruz, CA, dose range 25-100 nM) for 6-8 hours. Cells were then treated with IL6 (200 ng/ml) in the presence of JAK2 and/or STAT5 inhibitors for 48-72 hrs. Total RNA and protein lysates were collected from these cells for PCR and immunoblotting analyses.
Tissue specimens and immunohistochemistry
Colon cancer tissue microarrays (TMAs), one containing adjacent normal and colon cancer tissues (non-metastatic, catalog number A303I) and two others containing adjacent normal, metastatic colon cancer and liver metastasis tissues (catalog numbers A203III, A203IV), were procured from AccuMax Arrays (AccuMax Arrays, ISU Abxis Co., Ltd., San Diego, CA). Immunohistochemistry (IHC) was performed as described earlier [7] using MACH4 IHC kit (Biocare Medicals, Concord, CA). The primary antibodies, MUC13 (newly generated monoclonal antibody C18, dilution 1:200), and P-STAT5 (Santa Cruz, dilution 1:200) were used for immunostaining. The semi-quantification of IHC analysis was performed by calculating a composite score (CS, range 0-12) which is a function of the percentage of positively immunostained cancer cells multiplied by the staining intensity as described earlier [7]. The percentage of positively stained cancer cells was scored on a scale of 0 to 4 as follows: 0 for no staining, 1 for 1–25%, 2 for 26–50%, 3 for 51–75%, and 4 for >75% positively stained cancer cells. Immunostaining intensity was graded on a 0 to 3 scale as follows: 0 for no immunostaining, +1 for weak, +2 moderately high, and +3 for very high staining. Based on sub-cellular localization, a CS was calculated for MUC13 immunostaining at the membrane, cytoplasm and nucleus and for P-STAT5 immunostaining at the nucleus by an experienced pathologist (PDS). The mean composite score (MSC) is reported as the average composite score of each group as indicated.
Statistical analysis
Paired Student t-tests were performed to obtain the statistical significance between two groups. P<0.05 was considered significant for all the statistical analyses.
Results
MUC13 enhances tumorigenic features of colon cancer cells
To assess the functional role of MUC13 expression in colon cancer cells, we chose two paired cell lines which express low (SW480) and high (SW620) amounts of MUC13, as we have previously shown [8]. SW480 (non metastatic) cells were isolated from the primary tumor of a colon cancer patient and, a year later, SW620 (metastatic) cells were isolated from a lymph node metastatic lesion from the same patient [31]. Thus, SW480 (low MUC13 expressing and non-metastatic) and SW620 (high MUC13 expressing and metastatic) cells are ideal to examine fundamental changes that may occur during the transition to metastatic colon cancer. To create an over-expressing cell line, the full length MUC13 plasmid construct was stably transfected into SW480 cells. A pool of MUC13 over-expressing cells (SW480 M13OE) and a pool of empty vector cells (SW480 Vector) were selected for further experiments. Additionally, MUC13 knock-down (SW620 M13KD) and scrambled shRNA control (SW620 Vector) cells were generated utilizing lentiviral shRNA specific for MUC13. Immunofluorescence (Fig1. A1, B1, top) and Western blot (Fig. A1, B1, bottom) analysis showed increased expression of MUC13 in SW480 M13OE and decreased expression in SW620 M13KD cells.
Fig. 1. MUC13 expression enhances the tumorigenic features of colon cancer cells.
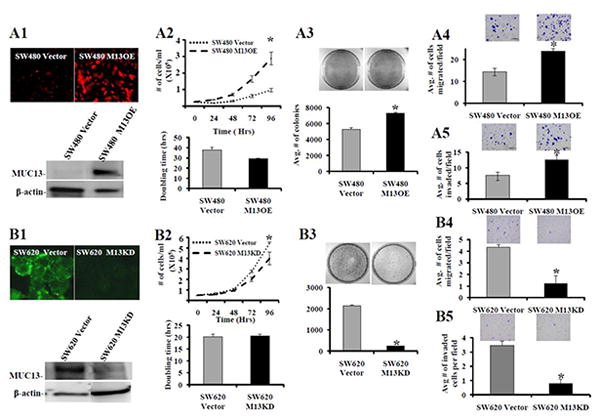
(A) SW480 cells were transfected with GFP tagged full length MUC13 to obtain MUC13 over-expressing (SW480 M13OE) cells. Vector control (SW480 Vector) cells were also selected. (B) SW620 cells were transduced with MUC13 specific shRNA lentiviral particles to obtain MUC13 knock-down (SW620 M13KD) cells. Vector control (SW620 Vector) cells were also obtained. The selected cell populations were screened for MUC13 by immunofluorescence (A1 and B1, top) and confirmed by Western blot (A1 and B1, bottom). Cell growth (A2, B2, top), cell doubling time (A2 and B2, bottom), colony formation (A3 and B3, top and bottom), cell migration (A4 and B4), and cell invasion (A5 and B5) assays were performed with MUC13 over-expressing and MUC13 knock-down cells, respectively, as described in experimental procedures. Representative images are shown above bar graph for the corresponding assays. Bar indicates the mean, error bar indicates the SEM, N=3, * P<0.05.
To determine the effects of MUC13 expression on tumorigenic characteristics, MUC13 over-expressing and MUC13 knock-down cells were used. Significantly higher (P<0.05) cell growth was observed in MUC13 over-expressing cells (SW480 M13OE) compared to SW480 Vector control cells at 96 hrs (Fig. 1, A2, top). Likewise, MUC13 over-expression decreased the cell doubling time in SW480 M13OE (29.4 hrs) compared to SW480 Vector control (38 hrs) cells (Fig. 1, A2, bottom). Conversely, significantly lower (P<0.05) cell growth was observed in MUC13 knock-down cells (SW620 M13KD) compared to SW620 Vector control cells at 96 hrs (Fig. 1, B2, top). Additionally, although not statistically significant, SW620 M13KD cells showed an increase in cell doubling time (20.5 hrs) compared to SW620 Vector control cells (duplication time 20.1 hrs) (Fig. 1, B2, bottom). All together these results suggest that MUC13 expression is associated with increased cell growth and reduced cell doubling time of colon cancer cells. Additionally, MUC13 over-expressing cells (SW480 M13OE) have a significantly (P<0.05) increased ability to form colonies compared to SW480 Vector control cells (Fig. 1, A3, top and bottom). MUC13 knock-down cells (SW620 M13KD) revealed a significant (P<0.05) decrease in total number of colonies compared to SW620 Vector control cells (Fig. 1, B3, top and bottom).
Distant metastases represent the major cause of death among colon cancer patients [32]. Thus, we performed migration and invasion assays to determine the role of MUC13 expression on metastatic phenotypes (migration/invasion) in colon cancer cells. A significantly higher number of MUC13 over-expressing cells (SW480 M13OE) moved through the membrane compared to SW480 Vector control cells in both migration and invasion assays (P<0.05) (Fig. 1 A4 and A5, respectively). Conversely, significantly fewer MUC13 knock-down (SW620 M13KD) cells moved through the membrane compared to SW620 Vector control cells in both migration and invasion assays (P<0.05) (Fig. 1, B4 and B5, respectively). These data suggest that MUC13 expression might contribute to colon cancer metastasis via influencing the migration and invasive potential of colon cancer cells.
MUC13 modulates multiple cancer pathways
Recently published studies regarding over-expression of MUC13 in ovarian, gastric, pancreatic and colon cancer suggest that MUC13 is implicated in various cancers and may play a critical role in cancer progression [7-10, 12, 14]. Therefore, it is expected that MUC13 may modulate the expression of genes known to be involved in cancer progression and metastasis. Thus, we sought to compare the gene expression profile of several pathways known to be dysregulated in cancer using an RT-PCR array. Q-RT-PCR analysis, comparing MUC13 over-expressing cells (SW480 M13OE) and the SW480 Vector control, showed a significant change in the expression of multiple genes. Genes that were up-regulated more than two fold are listed in Table 1. Among those 6 genes, telomerase reverse transcriptase (TERT) was the most up-regulated gene (4.2 fold). Interestingly, p53 was the most down regulated gene (3.04 fold). To confirm the change in TERT expression, we performed Western blot analysis in MUC13 over-expressing and knock-down cells which showed increased TERT expression in MUC13 over-expressing cells (SW480 M13OE) compared to SW480 Vector cells (Fig. 2A). Conversely, MUC13 knock-down cells (SW620 M13KD) showed decreased TERT expression compared to SW620 Vector cells (Fig. 2B). Similarly, we also investigated the effects of MUC13 expression on p53 expression with Western blot analysis. This analysis confirmed that p53 expression was indeed modulated by MUC13 expression (Fig. 2A and B).
Table 1. Genes up-regulated in RT-PCR array.
| Genes | Genes name | Expression (SW480 M13OE/SW480 Vector) (Fold Change) |
|---|---|---|
| TERT | Telomerase reverse transcriptase | 4.2 |
| IL8 | Interleukin 8 | 2.75 |
| IFNA1 | Interferon, alpha 1 | 2.74 |
| MMP1 | Matrix metalloproteinase 1 | 2.48 |
| FGFR2 | Fibroblast growth factor receptor 2 | 2.4 |
| PDGFB | Platelet-derived growth factor beta polypeptide | 2.13 |
Fig. 2. MUC13 modulates the expression of multiple cancer-associated pathways.
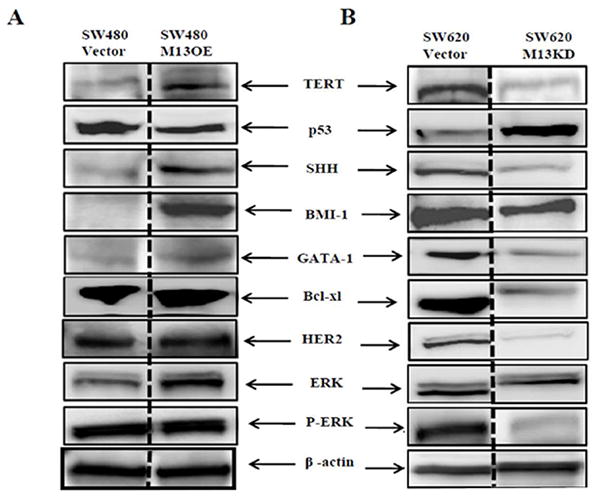
Total cell lysates were collected from MUC13 over-expressing cells (SW480 M13OE) (A), MUC13 knock-down cells (SW620 M13KD) (B) and from corresponding vector controls SW480 Vector and SW620 Vector cells, respectively. The lysates were immunoblotted for multiple proteins, including: TERT, p53, SHH, BMI-1, GATA1, Bcl-xl, HER2, ERK and P-ERK. β-actin was used as the internal loading control for all immunoblot analysis. A representative blot from at least 2 independent lysates is shown.
To further explore the influence of MUC13 on cell signaling, we examined the effect of MUC13 expression on multiple proteins, including sonic hedgehog (SHH), which is predicted to be upstream of TERT, and known to be involved in cell growth and survival [33]. Western blot analysis suggested that SHH was up-regulated in MUC13 over-expressing cells (SW480 M13OE) compared to SW480 Vector cells. Conversely, down regulation of SHH was observed in MUC13 knock-down cells (SW620 M13KD). B cell lymphoma murine integration site-1 (BMI-1) is a member of the polycomb ring finger oncogene family. Interestingly, BMI-1 is downstream of SHH signaling [33]. Therefore, we investigated the influence of MUC13 on BMI-1 expression. As expected, MUC13 over-expression positively correlated with BMI-1 expression (Fig. 2A). Additionally, suppression of MUC13 decreased BMI-1 expression in MUC13 knock-down (SW620 M13KD) cells compared to SW620 Vector cells (Fig. 2B). GATA binding proteins (GATA) are transcription factors that regulate multiple steps of the cell cycle and are predicted to be downstream targets of BMI-1 [33]. We investigated whether MUC13 influences GATA1 expression. Western blot analysis revealed higher GATA1 expression in MUC13 over-expressing cells (SW480 M13OE) compared to SW480 Vector cells. Conversely, MUC13 knock-down cells (SW620 M13KD) showed reduced GATA1 expression compared to SW620 Vector cells. B cell lymphoma (Bcl-xl) is an anti-apoptotic protein known to be a downstream target of GATA1 [34, 35]. MUC13 expression also influenced the expression of Bcl-xl in our MUC13 over-expressing and knockdown colon cancer cell lines (Fig. 2A and B).
Epidermal growth factor receptors (EGFR) are involved in colon cancer carcinogenesis [36-38]. The up-regulation of HER2 (EGFR-2) has been reported in breast, pancreatic and colon carcinogenesis [8, 39, 40]. MUC13 is predicted to contain 3 EGF-like domains; therefore, we sought to determine if MUC13 increases tumorigenesis via modulation of HER2 expression. Western blot analysis revealed that the total level of HER2 was increased in MUC13 over-expressing cells (SW480 M13OE) compared to SW480 Vector control cells. Conversely, HER2 expression was decreased in MUC13 knock-down cells (SW620 M13KD). We further investigated the involvement of the downstream signaling cascade of HER2, such as ERK. While total ERK expression levels remained unchanged, we found increased levels of phosphorylated ERK (P-ERK) in MUC13 over-expressing cells (SW480 M13OE) compared to SW480 Vector controls cells and decreased levels of P-ERK expression on MUC13 knockdown cells (SW620 M13KD) compared to SW620 Vector control cells. All together this data suggests that MUC13 expression increases tumorigenesis via influencing the expression of multiple oncogenic proteins.
STAT5 regulates MUC13 expression
To date, the regulatory aspects of MUC13 expression are unknown; therefore, we sought to explore the molecular mechanisms affecting MUC13 expression in cancer cells. Initially, we performed in silico analysis to identify potential transcription factors that may regulate MUC13 expression [41, 42] and 11 potential transcription factors were identified (Supplementary Table 3). Subsequently, multiple cell lines were screened by Q-RT-PCR analysis to identify cell lines with high endogenous MUC13 expression. Of these cell lines, the colon cancer cell line T84 and pancreatic cancer cell line HPAFII showed the highest expression levels of MUC13 mRNA transcripts (Fig. 3A). Thus, based on expression level of MUC13, T84 and HPAFII cell lines were selected for further analysis into MUC13 regulation. Q-RT-PCR analysis revealed that STAT5 is the most highly expressed transcription factor in both T84 and HPAFII cell lines (Fig.3B top and bottom). Therefore, STAT5 was selected for investigation of MUC13 regulation in subsequent studies. The predicted promoter of MUC13 contains the STAT5 binding site (GTAGTTCTGAGAATCC) (Fig. 3C). To determine if STAT5 binds to the MUC13 promoter, we performed ChIP assays in T84 and HPAFII cells. Compared to negative control non-immune IgG antibody, ChIP analysis in T84 cells revealed approximately 2 fold (15.3% to 30.0%) enrichment of MUC13 with the STAT5 antibody (Fig. 3D, left). The same studies in HPAFII cells revealed approximately 2.4 fold (8.6% to 20.4%) enrichment of MUC13 with STAT5 antibody compared to negative control IgG antibody (Fig. 3D, right). Enrichment of the MUC13 gene was further confirmed by Q-PCR in T84 and HPAFII cells (Fig. 3E). This data suggest that STAT5 binds to the predicted promoter of MUC13 and may regulate its expression in these cancer cells.
Fig. 3. IL6 induces MUC13 expression via binding of transcription factor STAT5 to the promoter of MUC13.

(A) MUC13 expression levels vary among cell lines: RNA was isolated from colon cancer (SW48, SW480, SW620, T84), ovarian cancer (SKOV-3, CaOV-3) and pancreatic cancer (HPAFII, MiaPaca) cell lines and analyzed by Q-RT-PCR for MUC13 mRNA. β2-microglobulin was used as the housekeeping gene control. For quantitative analysis, the relative MUC13 RNA expression is normalized to SW48 cell line. (B) Expression profile of transcription factors predicted to bind to the MUC13 promoter: RNA isolated from T84 (top) and HPAFII (bottom) cell lines was analyzed by Q-RT-PCR to determine the expression level of the panel of potential transcription factors identified by in silico analysis. Relative mRNA expression of transcription factors were calculated using ΔΔCt method. (C) Schematic representation of STAT5 DNA binding sequence in the predicted promoter of MUC13: Location of the STAT5 binding sequence is boxed and the STAT5 binding sequence is in red and underlined, illustrating the STAT5 binding site is in a 1 kbp upstream region from the translational start site (TSS). (D) STAT5 binds to the predicted promoter of MUC13: ChIP analysis was performed as described in experimental procedures and MUC13 DNA that was pulled down by anti- STAT5 was amplified using semi quantitative PCR in T84 (D, top left) and HPAFII (D, top right) cells. Densitometry analysis of PCR product is shown in T84 (D, bottom left) and HPAFII (D, bottom right). (E) The MUC13 amplification was confirmed by real time PCR in T84 (right) and HPAFII (left).
IL6 enhances MUC13 expression via JAK2/STAT5
The cytokine IL6 is over-expressed in colorectal cancer [43-45] and is a well known activator of the JAK-STAT signaling pathway. In order to determine the effects of IL6 and STAT5 activation on MUC13 regulation, we treated a colon cancer cell line, HT29 cells, with variable doses of IL6 (25-300 ng/ml). We chose to use HT29 cells for this experiment because they have a moderate level of endogenous MUC13 expression and they express JAK2 [7, 46]. After 48 hrs of IL6 treatment, Q-RT-PCR analysis revealed increased MUC13 expression in a dose dependent manner compared to the vehicle (DMSO) control (Fig. 4A). Western blot and confocal microscopy analyses also revealed increased MUC13 expression following IL6 treatment (Fig. 4B, top). As expected, Western blot analysis also showed that IL6 treatment increased the expression of P-JAK2 (Fig. 4C) and P-STAT5 (Fig. 4D), while total STAT5 levels remained unchanged. In addition, increased nuclear localization of P-STAT5 was observed when cells were treated with IL6 compared to DMSO control (supplementary Fig. 1). To further confirm the involvement of JAK2/STAT5, we suppressed JAK2 and STAT5 using JAK2 (AG490) and STAT5 inhibitors. Western blot analyses indicate that JAK2 and STAT5 inhibitors substantially decreased MUC13 expression, in the presence of IL6, in a dose dependent manner (Fig. 4E and F). Altogether, this data suggest that the over-expression of IL6 might be involved in MUC13 over-expression in cancer cells via activation of the JAK2/STAT5 signaling pathway.
Fig. 4. IL6 treatment enhances MUC13 expression via the JAK2/STAT5 pathway.
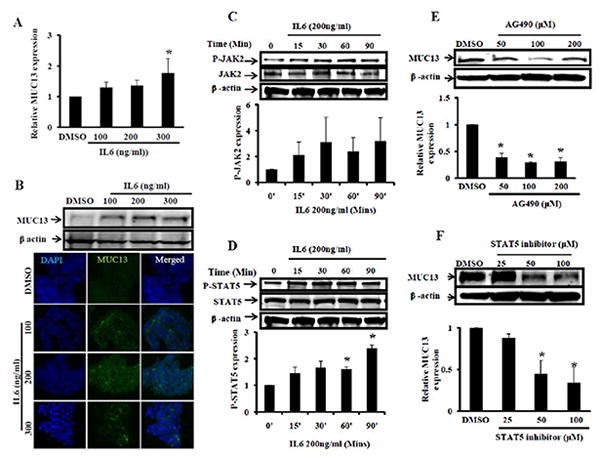
(A and B) IL6 increases MUC13 expression HT-29 cells were serum starved and treated with IL6 (100-300 ng/ml) and vehicle control (DMSO) for 48-72 hrs. A) At 48 hrs RNA was isolated and analyzed by Q-RT-PCR for MUC13 expression. B) At 72 hrs cell lysates were collected and analyzed by Western blot (top) or cells were fixed and processed for immunofluorescence analysis (bottom). (C and D) IL6 treatment increases P-JAK2 and P-STAT5 expression: Cells were treated with IL6 (200 ng/ml) or DMSO for 15-90 mins after an 8 hr serum starvation. SDS lysates were collected and subjected for Western blot analysis to detect total JAK2, P-JAK2 (4C), total STAT5 and P-STAT5 (4D) expression. Quantification of P-JAK2 and P-STAT5 expression is shown below corresponding Western blots. (E and F) Treatment with JAK2 and STAT5 inhibitors attenuated MUC13 expression: Cells were serum starved for 8 hrs and then treated with JAK2 (AG490) and STAT5 inhibitors. Cells were further treated with JAK2 and STAT5 inhibitors in the presence of IL6 for 48-72 hrs. Expression of MUC13 was detected by Western blot analysis following treatment with JAK2 inhibitor (4E) and STAT5 inhibitor (4F). Quantitative analysis is shown below corresponding Western blots. All experiments were repeated at least two times and representative blot is shown. Bar indicates the mean, error bar indicates the SEM, * P<0.05.
MUC13 is associated with P-STAT5 in colon cancer
We and others have previously reported that MUC13 is over-expressed in colon cancer compared to very faint expression in adjacent normal tissue [10, 13]. In addition, we have also shown that MUC13 localization (membrane vs. cytoplasm vs. nucleus) is altered in colon cancer metastasis [7]. Given our current findings showing that STAT5 binds to the MUC13 promoter and that activation of P-STAT5 increases MUC13 expression, we investigated the association of MUC13 and P-STAT5 in colon cancer tissue from patients with non-metastatic and metastatic disease. IHC analysis showed that both MUC13 and P-STAT5 were significantly up-regulated in non-metastatic colon cancer compared to adjacent normal tissue (Fig. 5A and B). Additionally, metastatic colon cancer and liver metastasis showed significantly higher MUC13 (cytoplasmic, and nuclear) and P-STAT5 expression compared to adjacent normal colon tissue. All together this data suggest that there may be an association between MUC13 and P-STAT5 in colon cancer.
Fig. 5. P-STAT5 expression is associated with MUC13 expression in colon cancer tissues.
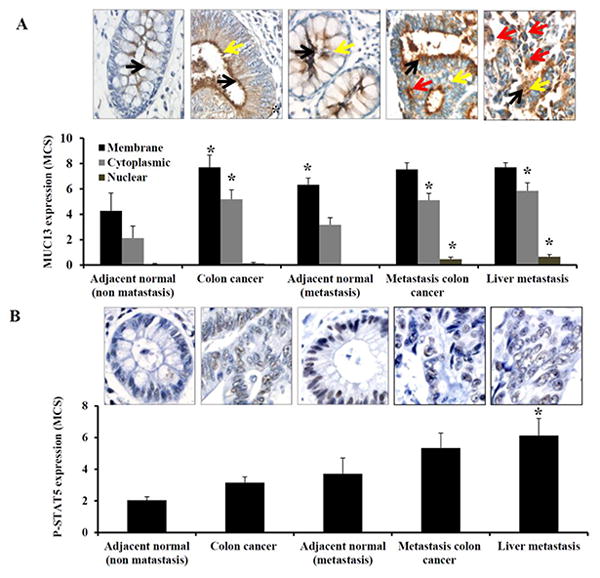
Tissue microarrays containing tissues from patients with non-metastatic disease (adjacent normal and colon cancer), and from patients with metastatic disease (adjacent normal, metastatic colon cancer, and liver metastasis tissues) were processed for immunostaining using MUC13 and P-STAT5 antibodies. Representative images of (A) MUC13 and (B) P-STAT5 immunostaining are shown and quantification was done as described in experimental methods. Black, yellow and red arrow indicates membranous, cytoplasmic and nuclear MUC13 expression respectively. Bar indicates the mean, error bar indicates the SEM, * P<0.05.
Discussion
Colon cancer is one of the most common malignancies with considerably high mortality. The five year survival rate is extremely low (<10%) in stage IV colon cancer [2, 3], with the majority of deaths occurring from metastases to vital organs, such as liver and lungs [2]. These facts accentuate the need to understand the basic molecular mechanisms regarding colon cancer progression and metastasis to design effective treatments in order to increase the duration of survival and minimize mortality. MUC13 is known to be over-expressed and aberrantly localized in colon cancer [7, 10]; in the present study we sought to investigate the regulation of MUC13 and its role in colon cancer progression and metastasis.
Mucins are implicated in colorectal carcinogenesis and metastasis [47, 48]. MUC1 expression is increased in colon cancer and is correlated with poor prognosis [47]. Bresalier et al. reported that increased mucin production by colon cancer cells increased the ability of colon cancer cells to metastasize to the liver in a mouse model, indicating that mucins may have a role in colorectal cancer metastasis [49]. In order to investigate the functional roles of MUC13 in colon cancer, we took advantage of the paired colon cancer cells lines: SW480 (low MUC13 expressing) and SW620 (high MUC13 expressing) and created stable MUC13 over-expressing and knock-down cells, respectively. Our results show that the over-expression of MUC13 increased in vitro indicators of tumorigenesis and metastasis (cell growth, doubling time, colony formation, cell migration, and cell invasion). These results are in accordance with our previous report where we observed increased cytoplasmic MUC13 expression in colon cancer tissues with liver metastases as compared to colon cancer that had not metastasized [7]. Additionally, a prior study from our laboratory has also shown that MUC13 increases cell motility and invasion in pancreatic cancer cells via modulating adhesion to proteins such as fibronectin, basement membrane complex (BMC), collagen IV, laminin and changing the expression of metastasis associated proteins (S100A4) [8]. The over-expression and aberrant localization of mucins, such as MUC13, may block cell-cell and cell-extracellular matrix adhesion and thereby facilitate cellular migration and invasion of cancer cells [9, 50], suggesting that MUC13 may play a role in colon cancer metastasis.
The development and progression of cancer requires the dysregulation of multiple oncogenic pathways and often the modulation of one protein can initiate a cascade of events with extensive downstream consequences. Although much of the mucin glycoprotein is extra-cellular, the ability of mucins to modulate cell signaling events is now well appreciated [51]. In our current study MUC13 influenced the expression of multiple proteins, including SHH, TERT, BMI-1, GATA1, and Bcl-xl. These proteins are known to be involved in colon cancer progression and metastasis [52-58]. In our previous publications we have shown that MUC13 increases HER2 and P-ERK expression in ovarian and pancreatic cancers, indicating that MUC13 affects the HER2/MAPK signaling pathway in these cancer cell lines [8, 13]. Similarly, our current results show that MUC13 over-expression increases HER2 and P-ERK expression in colon cancer cell lines. Taken together these findings suggest that MUC13 may influence colon cancer tumorigenesis and metastasis via multiple oncogenic proteins.
Given the important functional effects observed upon MUC13 over-expression, we sought to investigate the regulation of MUC13 expression in colon cancer cells. Other mucins, such as MUC1 and MUC4 are regulated by ERK/MAPK, STATs, TGF beta-SMAD and NFκB [59-62]. However, the regulatory mechanisms of MUC13 expression are not yet known. Herein, we identified STAT5 as a potential transcription factor which is highly expressed in MUC13 expressing cells. As shown by ChIP analysis, STAT5 binds to the predicted MUC13 promoter. STAT5 is often activated via JAK family members and JAK-STAT signaling regulates various oncogenes important in promoting colorectal tumor growth [63-65]. IHC analysis of colon cancer tissues support our in vitro findings as we found expression of both MUC13 and P-STAT5 increases in colon cancer and liver metastatic tissues as compared to adjacent normal tissues, suggesting that P-STAT5 may increase MUC13 expression in vivo.
IL6 activates STAT5 in colon epithelial cells and high levels of IL6 are reported in colorectal and gastric cancers [27, 66, 67]. Increased expression of IL6 is associated with enhanced tumorigenesis [68, 69] and IL6 signaling is also correlated with development of colitis induced premalignant lesions in colon epithelial cells in a murine model [45]. In a recent report, using ex vivo cultures of gastrointestinal tissue from Muc13 knock-out mice, Sheng et al. showed that loss of MUC13 caused a decrease in cytokine IL8 secretion in response to tumor necrosis factor-α, indicating a potential role for MUC13 in inflammation induced cancer [70]. These reports, and our current data, support the notion that IL6 and other cytokines may aggravate inflammatory conditions, increasing MUC13 expression, and predisposing colon tissue to malignant transformation.
A prognostic indicator itself, P-STAT5 is over-expressed in colon cancer and is associated with poor prognosis [30, 71]. Additionally, Xiong et al. have shown that inhibition of STAT5 reduces tumor cell invasion in colorectal cells. They also concluded that STAT5 formed a complex with p44/42 MAPK in colorectal cancer cells, suggesting crosstalk between MAPK and STAT5 [72]. In accordance with this report, our current findings show that MUC13 over-expression increases P-ERK expression, which is a downstream target of the MAPK signaling cascade, indicating a complex interplay between JAK2/STAT5, MAPK and MUC13. These reports also support our findings regarding regulation of MUC13 via P-STAT5 activation and suggest that targeted inhibition of MUC13 via suppression of STAT5 may be a potential therapeutic modality.
In conclusion, the results of this study suggest MUC13's role in colon tumorigenesis may be through modulation of various oncogenic pathways. Furthermore, for the first time, we demonstrate the regulation of MUC13 via IL6 induced JAK2/STAT5 signaling pathway in colon cancer. Finally, we show an association of MUC13 with P-STAT5 in colon cancer. The potential mechanistic pathways pertaining to the oncogenic roles and regulation of MUC13 are shown in Fig. 6. Future studies are needed to elucidate the molecular details regarding the interactions between MUC13 and oncogenic proteins that were modulated by MUC13 expression in our current study. Our data suggest that MUC13 over-expression in colon cancer could be attenuated by JAK2/STAT5 inhibitors which may increase the efficacy of colon cancer treatment.
Fig. 6. Schematic representation of MUC13 regulation and associated cancer pathways influencing MUC13 induced tumorigenesis.
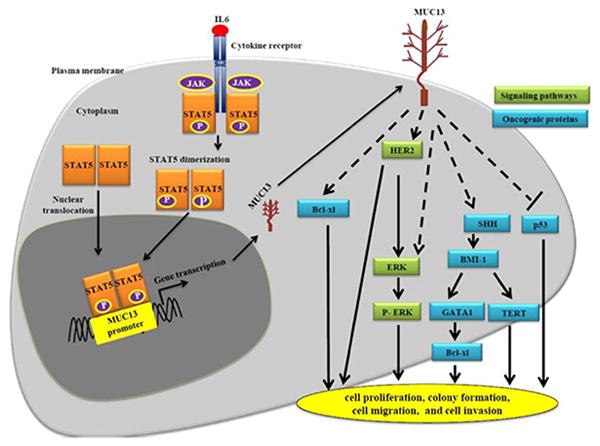
IL6 treatment induces phosphorylation of P-JAK2 and P-STAT5. Once phosphorylated, P-STAT5 translocates from cytoplasm to nucleus where it binds to the predicted promoter of MUC13 and increases MUC13 production at RNA and protein levels. MUC13 modulates the expression of various oncogenic proteins, resulting in increased tumorigenic features such as cell growth, cell doubling time, cell migration and cell invasion.
Supplementary Material
Acknowledgments
The authors thankfully acknowledge Sanford Research/USD Core facilities which are supported by P20 RR17662 and P20RR024219 COBRE grants. This work was supported from grants to SCC NIH RO1 CA142736.
Footnotes
Conflict of interest: Authors declare no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62(1):10–29. doi: 10.3322/caac.20138. Epub 2012/01/13. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–36. doi: 10.3322/caac.20121. Epub 2011/06/21. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. Epub 2010/07/09. [DOI] [PubMed] [Google Scholar]
- 4.Hollingsworth MA, Strawhecker JM, Caffrey TC, Mack DR. Expression of MUC1, MUC2, MUC3 and MUC4 mucin mRNAs in human pancreatic and intestinal tumor cell lines. International journal of cancer Journal international du cancer. 1994;57(2):198–203. doi: 10.1002/ijc.2910570212. Epub 1994/04/15. [DOI] [PubMed] [Google Scholar]
- 5.Ogata S, Uehara H, Chen A, Itzkowitz SH. Mucin gene expression in colonic tissues and cell lines. Cancer research. 1992;52(21):5971–8. Epub 1992/11/01. [PubMed] [Google Scholar]
- 6.Devine PL, Birrell GW, Whitehead RH, Harada H, Xing PX, McKenzie IF. Expression of MUC1 and MUC2 mucins by human tumor cell lines. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 1992;13(5-6):268–77. doi: 10.1159/000217775. Epub 1992/01/01. [DOI] [PubMed] [Google Scholar]
- 7.Gupta BK, Maher DM, Ebeling MC, Sundram V, Koch MD, Lynch DW, et al. Increased expression and aberrant localization of mucin 13 in metastatic colon cancer. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2012;60(11):822–31. doi: 10.1369/0022155412460678. Epub 2012/08/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chauhan SC, Ebeling MC, Maher DM, Koch MD, Watanabe A, Aburatani H, et al. MUC13 mucin augments pancreatic tumorigenesis. Molecular cancer therapeutics. 2012;11(1):24–33. doi: 10.1158/1535-7163.MCT-11-0598. Epub 2011/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher DM, Gupta BK, Nagata S, Jaggi M, Chauhan SC. Mucin 13: structure, function, and potential roles in cancer pathogenesis. Molecular cancer research : MCR. 2011;9(5):531–7. doi: 10.1158/1541-7786.MCR-10-0443. Epub 2011/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh MD, Young JP, Leggett BA, Williams SH, Jass JR, McGuckin MA. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Human pathology. 2007;38(6):883–92. doi: 10.1016/j.humpath.2006.11.020. Epub 2007/03/16. [DOI] [PubMed] [Google Scholar]
- 11.Packer LM, Williams SJ, Callaghan S, Gotley DC, McGuckin MA. Expression of the cell surface mucin gene family in adenocarcinomas. Int J Oncol. 2004;25(4):1119–26. [PubMed] [Google Scholar]
- 12.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001;276(21):18327–36. doi: 10.1074/jbc.M008850200. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, et al. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer research. 2009;69(3):765–74. doi: 10.1158/0008-5472.CAN-08-0587. Epub 2009/01/30. [DOI] [PubMed] [Google Scholar]
- 14.Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96(5):265–73. doi: 10.1111/j.1349-7006.2005.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rainczuk A, Rao J, Gathercole J, Stephens AN. The emerging role of CXC chemokines in epithelial ovarian cancer. Reproduction. 2012;144(3):303–17. doi: 10.1530/REP-12-0153. Epub 2012/07/10. [DOI] [PubMed] [Google Scholar]
- 16.Camporeale A, Poli V. IL-6, IL-17 and STAT3: a holy trinity in auto-immunity? Frontiers in bioscience : a journal and virtual library. 2012;17:2306–26. doi: 10.2741/4054. Epub 2012/06/02. [DOI] [PubMed] [Google Scholar]
- 17.Knupfer H, Schmidt R, Stanitz D, Brauckhoff M, Schonfelder M, Preiss R. CYP2C and IL-6 expression in breast cancer. Breast. 2004;13(1):28–34. doi: 10.1016/j.breast.2003.07.002. Epub 2004/02/05. [DOI] [PubMed] [Google Scholar]
- 18.Lee SO, Lou W, Johnson CS, Trump DL, Gao AC. Interleukin-6 protects LNCaP cells from apoptosis induced by androgen deprivation through the Stat3 pathway. The Prostate. 2004;60(3):178–86. doi: 10.1002/pros.20045. Epub 2004/06/04. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo K, Oka M, Murase K, Soda H, Isomoto H, Takeshima F, et al. Expression of interleukin 6 and its receptor in human gastric and colorectal cancers. The Journal of international medical research. 2003;31(2):69–75. doi: 10.1177/147323000303100202. Epub 2003/05/23. [DOI] [PubMed] [Google Scholar]
- 20.Azevedo A, Cunha V, Teixeira AL, Medeiros R. IL-6/IL-6R as a potential key signaling pathway in prostate cancer development. World journal of clinical oncology. 2011;2(12):384–96. doi: 10.5306/wjco.v2.i12.384. Epub 2011/12/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. The Journal of clinical investigation. 2007;117(12):3660–3. doi: 10.1172/JCI34237. Epub 2007/12/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. The Biochemical journal. 1998;334(Pt 2):297–314. doi: 10.1042/bj3340297. Epub 1998/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen EA, Slewa A, Gudlaugsson E, Jonsdottir K, Skaland I, Soiland H, et al. Biologic profiling of lymph node negative breast cancers by means of microRNA expression. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23(12):1567–76. doi: 10.1038/modpathol.2010.177. Epub 2010/09/08. [DOI] [PubMed] [Google Scholar]
- 24.Koptyra M, Gupta S, Talati P, Nevalainen MT. Signal transducer and activator of transcription 5a/b: biomarker and therapeutic target in prostate and breast cancer. The international journal of biochemistry & cell biology. 2011;43(10):1417–21. doi: 10.1016/j.biocel.2011.06.007. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, Cooper JC, Yu CL. A constitutively active Lck kinase promotes cell proliferation and resistance to apoptosis through signal transducer and activator of transcription 5b activation. Molecular cancer research : MCR. 2006;4(1):39–45. doi: 10.1158/1541-7786.MCR-05-0202. Epub 2006/02/01. [DOI] [PubMed] [Google Scholar]
- 26.Tormo AJ, Letellier MC, Sharma M, Elson G, Crabe S, Gauchat JF. IL-6 activates STAT5 in T cells. Cytokine. 2012;60(2):575–82. doi: 10.1016/j.cyto.2012.07.002. Epub 2012/08/03. [DOI] [PubMed] [Google Scholar]
- 27.Pratt SL, Ogle CK, Mao JX, Zhao W, Lovell G, Horseman ND. Interleukin-6 signal transduction in human intestinal epithelial cells. Shock. 2000;13(6):435–40. doi: 10.1097/00024382-200006000-00003. Epub 2000/06/10. [DOI] [PubMed] [Google Scholar]
- 28.Piekorz RP, Nemetz C, Hocke GM. Members of the family of IL-6-type cytokines activate Stat5a in various cell types. Biochemical and biophysical research communications. 1997;236(2):438–43. doi: 10.1006/bbrc.1997.6976. Epub 1997/07/18. [DOI] [PubMed] [Google Scholar]
- 29.Du W, Wang YC, Hong J, Su WY, Lin YW, Lu R, et al. STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. Journal of cellular physiology. 2012;227(6):2421–9. doi: 10.1002/jcp.22977. Epub 2011/08/10. [DOI] [PubMed] [Google Scholar]
- 30.Mao YL, Li ZW, Lou CJ, Pang D, Zhang YQ. Phospho-STAT5 expression is associated with poor prognosis of human colonic adenocarcinoma. Pathology oncology research : POR. 2011;17(2):333–9. doi: 10.1007/s12253-010-9321-3. Epub 2011/01/15. [DOI] [PubMed] [Google Scholar]
- 31.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer research. 1976;36(12):4562–9. Epub 1976/12/11. [PubMed] [Google Scholar]
- 32.Penna C, Nordlinger B. Colorectal metastasis (liver and lung) The Surgical clinics of North America. 2002;82(5):1075–90. x–xi. doi: 10.1016/s0039-6109(02)00051-8. Epub 2003/01/01. [DOI] [PubMed] [Google Scholar]
- 33.Jiang L, Li J, Song L. Bmi-1, stem cells and cancer. Acta biochimica et biophysica Sinica. 2009;41(7):527–34. doi: 10.1093/abbs/gmp040. Epub 2009/07/07. [DOI] [PubMed] [Google Scholar]
- 34.Yu YL, Chiang YJ, Chen YC, Papetti M, Juo CG, Skoultchi AI, et al. MAPK-mediated phosphorylation of GATA-1 promotes Bcl-XL expression and cell survival. The Journal of biological chemistry. 2005;280(33):29533–42. doi: 10.1074/jbc.M506514200. Epub 2005/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo YY, Chang ZF. GATA-1 and Gfi-1B interplay to regulate Bcl-xL transcription. Molecular and cellular biology. 2007;27(12):4261–72. doi: 10.1128/MCB.02212-06. Epub 2007/04/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopp R, Rothbauer E, Ruge M, Arnholdt H, Spranger J, Muders M, et al. Clinical implications of the EGF receptor/ligand system for tumor progression and survival in gastrointestinal carcinomas: evidence for new therapeutic options. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2003;162:115–32. doi: 10.1007/978-3-642-59349-9_10. Epub 2003/06/07. [DOI] [PubMed] [Google Scholar]
- 37.Tong WM, Ellinger A, Sheinin Y, Cross HS. Epidermal growth factor receptor expression in primary cultured human colorectal carcinoma cells. British journal of cancer. 1998;77(11):1792–8. doi: 10.1038/bjc.1998.298. Epub 1998/07/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi Y, Widjono YW, Ohta K, Hanioka K, Obayashi C, Itoh K, et al. Expression of EGF, EGF-receptor, p53, v-erb B and ras p21 in colorectal neoplasms by immunostaining paraffin-embedded tissues. Pathology international. 1994;44(2):124–30. doi: 10.1111/j.1440-1827.1994.tb01696.x. Epub 1994/02/01. [DOI] [PubMed] [Google Scholar]
- 39.Menard S, Casalini P, Campiglio M, Pupa S, Agresti R, Tagliabue E. HER2 overexpression in various tumor types, focussing on its relationship to the development of invasive breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2001;12(Suppl 1):S15–9. doi: 10.1093/annonc/12.suppl_1.s15. Epub 2001/08/28. [DOI] [PubMed] [Google Scholar]
- 40.Lu Y, Jingyan G, Baorong S, Peng J, Xu Y, Cai S. Expression of EGFR, Her2 predict lymph node metastasis (LNM)-associated metastasis in colorectal cancer. Cancer biomarkers : section A of Disease markers. 2012;11(5):219–26. doi: 10.3233/CBM-2012-00282. Epub 2012/12/12. [DOI] [PubMed] [Google Scholar]
- 41.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome research. 2002;12(6):996–1006. doi: 10.1101/gr.229102. Epub 2002/06/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabiosciences. DECODE (Decipherment of DNA Elements) 2010 http://www.sabiosciences.com/chipqpcrsearch.php?species_id=0&factor=Over+200+TF&gene=MUC13&nfactor=n&ninfo=n&ngene=n&B2=Search.
- 43.Li YY, Hsieh LL, Tang RP, Liao SK, Yeh KY. Interleukin-6 (IL-6) released by macrophages induces IL-6 secretion in the human colon cancer HT-29 cell line. Human immunology. 2009;70(3):151–8. doi: 10.1016/j.humimm.2009.01.004. Epub 2009/03/11. [DOI] [PubMed] [Google Scholar]
- 44.Waldner MJ, Foersch S, Neurath MF. Interleukin-6--a key regulator of colorectal cancer development. International journal of biological sciences. 2012;8(9):1248–53. doi: 10.7150/ijbs.4614. Epub 2012/11/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto S, Hara T, Mitsuyama K, Yamamoto M, Tsuruta O, Sata M, et al. Essential roles of IL-6 trans-signaling in colonic epithelial cells, induced by the IL-6/soluble-IL-6 receptor derived from lamina propria macrophages, on the development of colitis-associated premalignant cancer in a murine model. J Immunol. 2010;184(3):1543–51. doi: 10.4049/jimmunol.0801217. Epub 2010/01/01. [DOI] [PubMed] [Google Scholar]
- 46.Thoennissen NH, Iwanski GB, Doan NB, Okamoto R, Lin P, Abbassi S, et al. Cucurbitacin B induces apoptosis by inhibition of the JAK/STAT pathway and potentiates antiproliferative effects of gemcitabine on pancreatic cancer cells. Cancer research. 2009;69(14):5876–84. doi: 10.1158/0008-5472.CAN-09-0536. Epub 2009/07/17. [DOI] [PubMed] [Google Scholar]
- 47.Byrd JC, Bresalier RS. Mucins and mucin binding proteins in colorectal cancer. Cancer metastasis reviews. 2004;23(1-2):77–99. doi: 10.1023/a:1025815113599. Epub 2004/03/06. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz B, Bresalier RS, Kim YS. The role of mucin in colon-cancer metastasis. International journal of cancer Journal international du cancer. 1992;52(1):60–5. doi: 10.1002/ijc.2910520113. Epub 1992/08/19. [DOI] [PubMed] [Google Scholar]
- 49.Bresalier RS, Niv Y, Byrd JC, Duh QY, Toribara NW, Rockwell RW, et al. Mucin production by human colonic carcinoma cells correlates with their metastatic potential in animal models of colon cancer metastasis. The Journal of clinical investigation. 1991;87(3):1037–45. doi: 10.1172/JCI115063. Epub 1991/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4(1):45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 51.Carraway KL, Ramsauer VP, Haq B, Carothers Carraway CA. Cell signaling through membrane mucins. BioEssays : news and reviews in molecular, cellular and developmental biology. 2003;25(1):66–71. doi: 10.1002/bies.10201. Epub 2003/01/01. [DOI] [PubMed] [Google Scholar]
- 52.Oniscu A, James RM, Morris RG, Bader S, Malcomson RD, Harrison DJ. Expression of Sonic hedgehog pathway genes is altered in colonic neoplasia. The Journal of pathology. 2004;203(4):909–17. doi: 10.1002/path.1591. Epub 2004/07/20. [DOI] [PubMed] [Google Scholar]
- 53.Yoshikawa K, Shimada M, Miyamoto H, Higashijima J, Miyatani T, Nishioka M, et al. Sonic hedgehog relates to colorectal carcinogenesis. Journal of gastroenterology. 2009;44(11):1113–7. doi: 10.1007/s00535-009-0110-2. Epub 2009/08/08. [DOI] [PubMed] [Google Scholar]
- 54.Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK, Choe IS, et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer letters. 2004;203(2):217–24. doi: 10.1016/j.canlet.2003.07.009. Epub 2004/01/21. [DOI] [PubMed] [Google Scholar]
- 55.Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ, Li SX, et al. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. Journal of cancer research and clinical oncology. 2010;136(7):997–1006. doi: 10.1007/s00432-009-0745-7. Epub 2009/12/22. [DOI] [PubMed] [Google Scholar]
- 56.Naito Y, Takagi T, Handa O, Ishikawa T, Matsumoto N, Yoshida N, et al. Telomerase activity and expression of telomerase RNA component and catalytic subunits in precancerous and cancerous colorectal lesions. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2001;22(6):374–82. doi: 10.1159/000050640. Epub 2002/01/12. [DOI] [PubMed] [Google Scholar]
- 57.Ayanbule F, Belaguli NS, Berger DH. GATA factors in gastrointestinal malignancy. World journal of surgery. 2011;35(8):1757–65. doi: 10.1007/s00268-010-0950-1. Epub 2011/01/07. [DOI] [PubMed] [Google Scholar]
- 58.Zhang YL, Pang LQ, Wu Y, Wang XY, Wang CQ, Fan Y. Significance of Bcl-xL in human colon carcinoma. World journal of gastroenterology : WJG. 2008;14(19):3069–73. doi: 10.3748/wjg.14.3069. Epub 2008/05/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Theodoropoulos G, Carraway KL. Molecular signaling in the regulation of mucins. Journal of cellular biochemistry. 2007;102(5):1103–16. doi: 10.1002/jcb.21539. Epub 2007/10/25. [DOI] [PubMed] [Google Scholar]
- 60.Thompson EJ, Shanmugam K, Hattrup CL, Kotlarczyk KL, Gutierrez A, Bradley JM, et al. Tyrosines in the MUC1 cytoplasmic tail modulate transcription via the extracellular signal-regulated kinase 1/2 and nuclear factor-kappaB pathways. Molecular cancer research : MCR. 2006;4(7):489–97. doi: 10.1158/1541-7786.MCR-06-0038. Epub 2006/07/20. [DOI] [PubMed] [Google Scholar]
- 61.Kondo S, Yoshizaki T, Wakisaka N, Horikawa T, Murono S, Jang KL, et al. MUC1 induced by Epstein-Barr virus latent membrane protein 1 causes dissociation of the cell-matrix interaction and cellular invasiveness via STAT signaling. Journal of virology. 2007;81(4):1554–62. doi: 10.1128/JVI.02222-06. Epub 2006/12/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mejias-Luque R, Peiro S, Vincent A, Van Seuningen I, de Bolos C. IL-6 induces MUC4 expression through gp130/STAT3 pathway in gastric cancer cell lines. Biochimica et biophysica acta. 2008;1783(10):1728–36. doi: 10.1016/j.bbamcr.2008.05.020. Epub 2008/06/25. [DOI] [PubMed] [Google Scholar]
- 63.Slattery ML, Lundgreen A, Kadlubar SA, Bondurant KL, Wolff RK. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Molecular carcinogenesis. 2013;52(2):155–66. doi: 10.1002/mc.21841. Epub 2011/11/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spano JP, Milano G, Rixe C, Fagard R. JAK/STAT signalling pathway in colorectal cancer: a new biological target with therapeutic implications. Eur J Cancer. 2006;42(16):2668–70. doi: 10.1016/j.ejca.2006.07.006. Epub 2006/09/12. [DOI] [PubMed] [Google Scholar]
- 65.Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7(6):545–55. doi: 10.1593/neo.04571. Epub 2005/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komoda H, Tanaka Y, Honda M, Matsuo Y, Hazama K, Takao T. Interleukin-6 levels in colorectal cancer tissues. World journal of surgery. 1998;22(8):895–8. doi: 10.1007/s002689900489. Epub 1998/07/23. [DOI] [PubMed] [Google Scholar]
- 67.Lin MT, Lin BR, Chang CC, Chu CY, Su HJ, Chen ST, et al. IL-6 induces AGS gastric cancer cell invasion via activation of the c-Src/RhoA/ROCK signaling pathway. International journal of cancer Journal international du cancer. 2007;120(12):2600–8. doi: 10.1002/ijc.22599. Epub 2007/02/17. [DOI] [PubMed] [Google Scholar]
- 68.Jovanovic M, Vicovac L. Interleukin-6 stimulates cell migration, invasion and integrin expression in HTR-8/SVneo cell line. Placenta. 2009;30(4):320–8. doi: 10.1016/j.placenta.2009.01.013. Epub 2009/03/03. [DOI] [PubMed] [Google Scholar]
- 69.Becker C, Fantini MC, Wirtz S, Nikolaev A, Lehr HA, Galle PR, et al. IL-6 signaling promotes tumor growth in colorectal cancer. Cell Cycle. 2005;4(2):217–20. Epub 2005/01/19. [PubMed] [Google Scholar]
- 70.Sheng YH, Triyana S, Wang R, Das I, Gerloff K, Florin TH, et al. MUC1 and MUC13 differentially regulate epithelial inflammation in response to inflammatory and infectious stimuli. Mucosal immunology. 2012 doi: 10.1038/mi.2012.98. Epub 2012/11/15. [DOI] [PubMed] [Google Scholar]
- 71.Mao Y, Li Z, Lou C, Zhang Y. Expression of phosphorylated Stat5 predicts expression of cyclin D1 and correlates with poor prognosis of colonic adenocarcinoma. International journal of colorectal disease. 2011;26(1):29–35. doi: 10.1007/s00384-010-1090-7. Epub 2010/11/17. [DOI] [PubMed] [Google Scholar]
- 72.Xiong H, Su WY, Liang QC, Zhang ZG, Chen HM, Du W, et al. Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Laboratory investigation; a journal of technical methods and pathology. 2009;89(6):717–25. doi: 10.1038/labinvest.2009.11. Epub 2009/03/18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


