Abstract
Current models of T-helper-cell differentiation depict the generation of effector cells from a naïve T cell based on the cytokine environment upon T-cell-receptor engagement. We propose a new model of CD4+ T-cell activation, differentiation, and function whereby the outcome of antigen recognition is dictated by mTOR activity and the subsequent up-regulation of selective metabolic function. Such a model more readily explains the generation of effector and memory cells including the concept of effector and memory Foxp3+ regulatory cells.
Current models of CD4+ T-cell immunity are dominated by the concept that upon antigen recognition naïve T cells are instructed by combinations of cytokines to differentiate into effector cells or Foxp3+ regulatory T cells (Weaver et al. 2006). As defined in vitro, activation in the presence of sets of cytokines results in terminally differentiated effectors characterized by canonical transcription factors. Although these paradigms have served as a meaningful model to dissect the role of cytokines, signaling molecules and transcription factors in influencing the development and function of CD4+ effector cells, recently a much greater complexity with regard to plasticity among the different subsets of cells has become appreciated (O’Shea and Paul 2010). Furthermore, these models poorly account for the concept of CD4+ T-cell memory.
Recently, studies from our laboratory and others have showed a critical role for the mechanistic target of rapamycin (mTOR) in guiding the outcome of TCR recognition (Powell et al. 2012). mTOR is a member of the PI3-kinase family that is conserved from yeast to mammalian cells and functions as an integrator of environmental signals to regulate cellular function (Zoncu et al. 2011). mTOR plays an important role in regulating the expression and function of proteins involved in metabolism. Indeed, it is becoming increasingly clear that metabolic programs play a critical role in regulating activation, differentiation, and function of cells of the immune system (Pearce 2010; Waickman and Powell 2012). Herein, we lay out the rational for a new model of CD4+ T-cell differentiation based on mTOR activation and the subsequent regulation of metabolic programs.
SIGNAL 1 + 2
A hallmark of the adaptive immune response is the exquisite specificity of antigen receptor recognition. This specificity is highlighted by the ability of the T-cell receptor (TCR) to interrogate peptide–major histocompatibility complexes (MHCs) on the surface of a cell and “find” the “correct” interaction. However, as remarkable as this ligand–receptor interaction is, this recognition imparts only partial information concerning the nature of the peptide and the ensuing response. By invoking a second signal, it was possible to devise a model whereby TCR engagement heralds recognition and the outcome of the recognition is determined by the presence or absence of a second ligand (Bretscher and Cohn 1970; Lafferty and Cunningham 1975). That is, TCR engagement (Signal 1 alone) leads to tolerance, and TCR engagement in the setting of the second signal (Costimulation, Signal 2) leads to a full-blown immune response (Schwartz 1992). In such a model, the outcome of recognition is dictated by regulation of the expression of the second signal. The insight into “what controlled the presence of the second signal?” was revealed in part by Janeway’s model of infectious nonself (Janeway 1989). Succinctly summarized, TCR engagement leads to full-blown immune responses when antigen is accompanied by pathogen-associated molecular patterns (PAMPS) that activate APCs and that upregulate costimulatory ligands. Subsequently, the Danger theory sought to expand this view to account for transplantation and tumor rejection (Matzinger 2001).
One of the outcomes of TCR engagement in the absence of costimulation (Signal 1 alone) is T-cell anergy (Jenkins et al. 1990). Anergic T cells fail to respond upon subsequent full stimulation. Initially it was proposed that anergy was the result of TCR engagement in the absence of proliferation (Jenkins 1992). To test this hypothesis, we stimulated T cells in the presence of the mTOR inhibitor rapamycin which at the time was felt to suppress immune responses by inhibiting T-cell proliferation in G1 (Powell et al. 1999). Indeed, Signal 1 + 2 in the presence of rapamycin promoted anergy. However, subsequent experiments employing other inhibitors of proliferation failed to induce anergy (Allen et al. 2004). This led us to conclude that anergy was the result of TCR engagement in the absence of mTOR activation. Inas-much as Signal 2 was not necessarily one distinct ligand–receptor interaction, but rather the net sum of multiple costimulatory and coinhibitory signals, we proposed that mTOR-induced activation was an important component of “Signal 2” (Powell et al. 2012).
INTEGRATING ENVIRONMENTAL CUES TO DICTATE THE OUTCOME OF ANTIGEN RECOGNITION
The two signal model provides a framework for determining immunogenic versus tolerogenic responses to antigen. However, particularly in the last decade, it has become clear that for CD4+ T cells there are numerous fates that can result upon antigen encounter (Weaver et al. 2006). Depending on cues from the immune microenvironment (i.e., cytokines), upon TCR engagement, CD4+ T cells can differentiate into Th1, Th2, and Th17 effector cells or alternatively become inducible T regulatory cells. Within this context, we view Signal 2 as the net sum of cues (cytokines, ligand–receptor interactions, and even, as discussed below, nutrients) that accompany TCR engagement and thus guide the outcome of antigen recognition.
mTOR is an evolutionarily conserved serine threonine kinase that integrates signals from the environment to dictate cellular function (Zoncu et al. 2011). From yeast cells to mammalian cells a diversity of signals, including growth factors, cytokines, amino acids, stress responses, and ligand–receptor interactions, all converge on mTOR leading to its activation or inhibition (Powell et al. 2012). In turn, mTOR signaling has been implicated in a wide variety of cellular function ranging from cell size, proliferation, survival, mobility, protein synthesis, and transcription (Sengupta et al. 2010). In light of the generalized role of mTOR in integrating cues from the environment to dictate cellular function, we hypothesized that mTOR might also play a critical role in integrating cues from the immune microenvironment to guide the outcome of antigen recognition. To this end we selectively deleted mTOR in T cells. In the absence of mTOR signaling, T cells failed to differentiate into Th1, Th2, and Th17 cells under skewing conditions (Delgoffe et al. 2009). Instead, even in the absence of exogenous TFG-β, antigen recognition in the absence of mTOR signaling led to the generation of Foxp3+ regulatory T cells. That is, the default pathway for TCR engagement in the absence of mTOR signaling was to differentiate into a regulatory T cell.
mTOR signals via two distinct signaling complexes, mTOR Complex 1 (mTORC1) and mTORC2, which are distinguished by the scaffolding proteins Raptor and Rictor, respectively, and are involved in phosphorylating different downstream substrates (Zoncu et al. 2011). We wanted to determine if these complexes played differential roles in regulating CD4+ T-cell differentiation. To this end we created T cells that selectively lacked mTORC1 or mTORC2 signaling (Delgoffe et al. 2011). Strikingly, the mTORC1-deficient T cells were unable to differentiate into Th1 and Th17 cells but retained their ability to become Th2 cells. Alternatively, the mTORC2-deficient T cells maintained their ability to differentiate into Th1 and Th17 cells, but failed to differentiate into Th2 cells under appropriate skewing conditions. Thus, mTORC1 and mTORC2 selectively regulate CD4+ T-cell differentiation.
mTOR LINKS T-CELL ACTIVATION WITH METABOLIC PROGRAMS
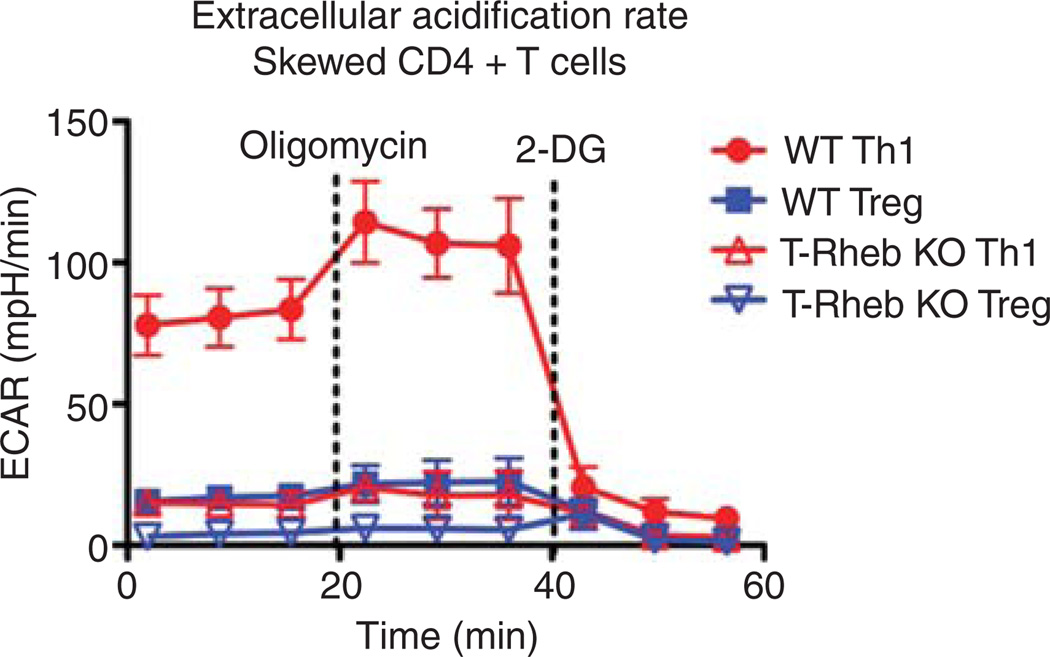
T-cell activation is metabolically demanding. Indeed, as is the case for cancer cells, effector T cells use aerobic glycolysis as a means of generating energy (Fox et al. 2005). It has been proposed that by employing glycolysis for energy, this allows for TCA cycle intermediates to be used as substrates for amino acids, lipids, and nucleotides to support the tremendous proliferation that accompanies activation. mTOR plays an important role in upregulating the enzymes necessary for glycolysis. Th1 cells are highly glycolytic (Fig. 1). Deleting Rheb, a small GTPase necessary for mTORC1 activity, inhibits Th1 generation. Furthermore, Rheb deletion inhibits glycolysis. Alternatively, resting regulatory T cells are not very glycolytic. Likewise, we have showed the deletion of mTOR leads to the generation of regulatory T cells. These data are consistent with a model whereby mTOR regulates T-cell fate in part by controlling the metabolic programs necessary for various effector functions. Along these lines, anergic T cells, which fail to show Th1 effector function upon rechallenge, also fail to upregulate various metabolic pathways upon activation (Zheng et al. 2009b). These observations suggest that one mechanism that maintains T-cell anergy is a block in metabolism.
Figure 1.
T cells lacking Rheb−/− fail to become Th1 cells and are less glycolytic. CD4+ T cells from WT C57Bl/6 and T-Rheb−/− were isolated and stimulated under Th1 or T regulatory skewing conditions for 48 h. The cells were then isolated and the metabolic profile of the cultured cells was determined using a Seahorse XF96 analyzer, measuring extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) following treatment with oligomycin, an ATP synthase inhibitor and 2-deoxyglucose to inhibit glycolysis.
TRACKING CELL FATE BY TRACKING mTOR ACTIVATION
By deleting selective mTOR signaling in T cells, we could interrogate the role of mTOR in promoting T-cell effector fate. However, we were also interested in tracking the fate of wild-type T cells based upon mTOR activation. To this end, we took advantage of the fact that mTOR controls cell size. We observed that when naïve T cells are activated with peptide, we could identify cells that had robust mTOR activation from those that had low levels of mTOR signaling by gating on activated large and small cells, respectively (manuscript in preparation). Indeed, the large cells show markedly increased phosphorylation of S6, S6 K, and 4E-BP1 as determined by phospho-flow staining.
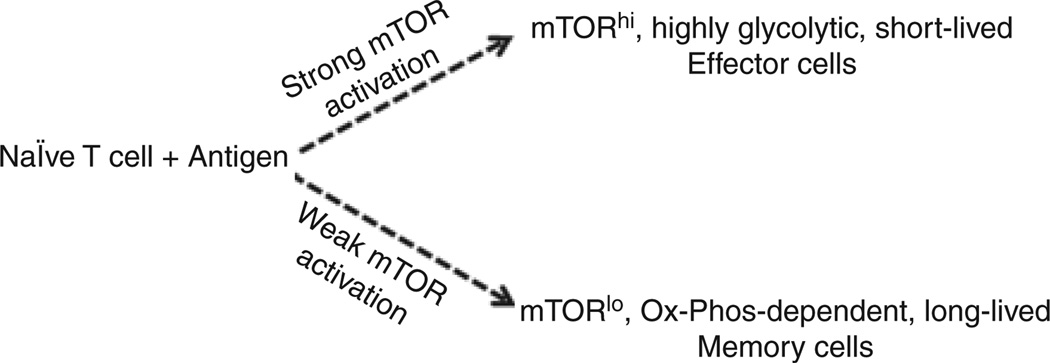
Using this technique, a series of experiments was performed that tracked the fate of the mTORhi and mTORClo cells upon antigen recognition. We observed that the mTORhi cells were metabolically more active, avidly taking up glucose and show increased glycolysis upon activation. Such cells, however, were relatively short-lived. Phenotypically, the mTORhi cells express high levels of CXCR3 on their surface. Furthermore, the mTORhi cells proliferate to a greater extent and express much higher levels of effector cytokines. In contrast, the mTORlo cells were much more long-lived. Metabolically, the mTORlo cells show decreased glucose uptake and glycolysis. However, such cells show increased mitochondria with time and a higher spare respiratory capacity, both of which are associated with long-lived cells (Choi et al. 2009). Although the mTORlo cells did not produce robust levels of cytokines initially, they show increased recall responses days later. Thus, within a clonal T-cell response, some T cells will engage antigen in the setting of robust mTOR activation and are fated to become short-lived effector cells, and some T cells will engage antigen in the setting of minimal mTOR activity and are fated to become long-lived memory cells (Fig. 2).
Figure 2.
mTOR activation and metabolic programs regulate effector and memory T-cell generation. We propose that upon antigen recognition, the degree of mTOR activation and the subsequent activation of glycolytic versus oxidative phosphorylation metabolic programs dictates effector or memory fates. TCR engagement in the setting of robust mTOR activation leads to the generation of short-lived, highly glycolytic effector cells. Alternatively, TCR engagement in the setting of minimal mTOR activation promotes the generation of long-lived, oxidative phosphorylation-dependent memory cells. Importantly, our model also applies to the generation of “effector” and “memory” Foxp3+ regulatory T cells.
As mentioned above, deletion of mTOR in T cells leads to the generation of Foxp3+ regulatory T cells (Delgoffe et al. 2009). Along these lines, it has been shown that T cells activated in the presence of rapamycin promotes the generation of regulatory T cells (Battaglia et al. 2005). Thus, we examined the mTORlo cells for Foxp3+ T cells. Indeed, in addition to the CD4+Foxp3− cells that were fated to become effector memory cells, we also observed an increased population of CD4+ Foxp3+ T cells. That is, upon activation with antigen (in the absence of the addition of any exogenous TGF-β), a population of regulatory T cells emerges from cells that have recognized antigen but show decreased mTOR activation. Previously, it has been showed that in the absence of exogenous TGF-β, stimulation with low doses of peptide can lead to the generation of increased Foxp3+ T cells (Gottschalk et al. 2010). Indeed, we were able to recapitulate such findings and were further able to show that stimulation with low doses of peptide is associated with decreased mTOR activity.
On the other hand, a common technique to generate Foxp3+ T cells in vitro is to stimulate T cells with high doses of peptide (or antiCD3) in the presence of exogenous TGF-β (Chen et al. 2003). Under these conditions, the T cells display high levels of mTOR signaling despite becoming regulatory T cells. As such, we investigated the phenotype of mTORhi (generated with antiCD3 and TGF-β) and mTORlo (generated in the presence of rapamycin) regulatory T cells. As was the case for the mTORhi effector cells, the mTORhi regulatory T cells show increased glycolysis upon activation. Alternatively, the mTORlo Foxp3+ regulatory T cells show decreased glycolysis but increased spare respiratory capacity. Thus, the mTORhi regulatory T cells metabolically resembled effector cells, whereas the mTORlo regulatory T cells metabolically resemble memory cells. Thus, our model (Fig. 2) concerning the ability of mTOR to regulate the effector versus memory fate of a naïve T cell also applies to cells destined to become regulatory T cells. Importantly, the critical decision upon antigen recognition is not necessarily the definition of the type of effector cell (Th1, Th2, Th17, or T regulatory) but rather whether the cell is destined to be a short-lived effector cell or a long-lived memory cell.
A NEW MODEL OF T-CELL DIFFERENTIATION BASED ON mTOR ACTIVATION AND METABOLISM
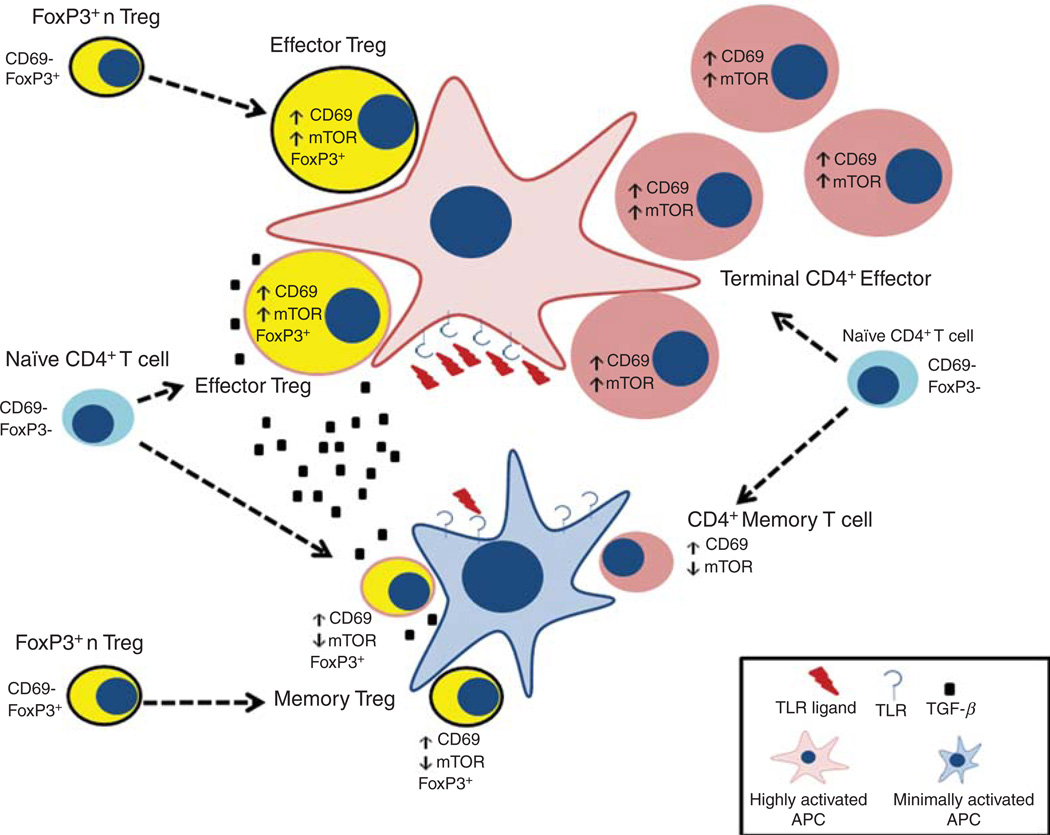
Based on these findings, we propose a modified view of CD4+ T-cell differentiation based on mTOR activation and metabolism (Fig. 3). We propose that the degree of mTOR activation and subsequent metabolic programming plays a critical role in dictating the fate of a naïve T cell upon antigen recognition. T cells that recognize antigen in the setting of robust mTOR activation will become effector cells. Such cells will be relatively short-lived, highly glycolytic, express effector chemokine receptors and acutely produce robust amounts of effector cytokine. Clearly, a key component of this model lies in what controls mTOR activation. To this end, we posit that at the height of an infection, there will be copious amounts of antigen + PAMPS, cytokines, growth factors, and costimulatory molecules, all of which will promote mTOR signaling. This will favor the generation of effector cells (not exclusively though, because even under these conditions some T cells might still recognize antigen in the setting of low mTOR activity). Alternatively, as the pathogen is cleared and there are fewer “danger signals,” our model suggests that TCR engagement will occur under conditions of decreased mTOR activation. This will favor the generation of long-lived memory cells that metabolically use oxidative phosphorylation and fatty acid oxidation and possess increased spare respiratory capacity. In addition, this process is undoubtedly modulated by the availability of nutrients. To this end, it has been shown that regulatory T cells can regulate immune responses in part by decreasing the availability of branched chain amino acids that are critical for mTOR activation (Cobbold et al. 2009). Furthermore, for CD8+ T cells, our laboratory has been able to show that proliferation can lead to the generation of one daughter cell with high mTOR signaling and another with low mTOR activity (in prep.). If such is also the case for CD4+ T cells, then this asymmetric division of mTOR activity might account in part for the simultaneous generation of cells destined to become effector cells and cells destined to become memory cells.
Figure 3.
A modified model of CD4+ T-cell differentiation based on mTOR activation and metabolism. Within the inflammatory milieu, we propose that naïve T cells stimulated by antigen and robust mTOR activation become effector CD4+ T cells (right side, top half of the figure). In a Th1 milieu they become Th1 effector cells, in a Th2 milieu they become Th2 effector cells, etc. The presence of high number of pathogens and hence PAMPS would favor the generation of the effector cells. However, as the pathogen is cleared we envision that TCR engagement in the setting of decreased mTOR activation might be favored, thus generating more memory cells (right side, bottom half of the figure). Likewise, we propose that Foxp3+ regulatory T cells arriving at the site of inflammation are activated to become “effector” regulatory T cells or “memory” regulatory T cells depending on the degree of mTOR activation they receive (left side of figure, top and bottom, respectively). Finally, in the presence of increased TGF-β, activation of naïve T cells in the setting of strong mTOR activation results in the generation of inducible effector T regulatory cells, whereas weak mTOR activation in the setting of little inflammatory cytokines results in the generation of inducible memory regulatory T cells (left side, middle of the figure).
We believe that our model has some interesting implications for the generation of Foxp3+ regulatory T cells. Based on our data we propose that Foxp3+ CD4+ T cells that traffick to activated lymph nodes and become robustly stimulated (mTORhi) will become short-lived “effector” regulatory T cells. Such cells would then home to the tissues and control immune responses. Alternatively, these “effector” T regulatory cells might also be generated from naïve T cells when they are robustly stimulated in the presence of increased TGF-β. It has been proposed that Th1-type T regulatory cells control Th1 responses and Th2-like regulatory cells control Th2 responses and so on (Koch et al. 2009; Zheng et al. 2009a). Our model readily takes into account such properties of regulatory T cells. Likewise, our model proposes the generation of memory regulatory T cells. That is, when naïve T cells are stimulated by antigen in the setting of “very low” mTOR activation or TGF-β, such cells would become long-lived Foxp3+ T cells. Likewise, the fate of activation of “natural” T regulatory cells would also be influenced by the degree of mTOR activation. TCR engagement of natural T regulatory cells in the setting of high mTOR activation will lead to effector T regulatory cells. Alternatively, natural T regulatory cells that recognize antigen in the context of low mTOR signaling will lead to the generation of an increased subset of memory T regulatory cells.
CONCLUSIONS
In our modified model of CD4+ T-cell differentiation we posit a central role for mTOR as an integrator of environmental cues guiding the outcome of antigen recognition. Our model to some extent deemphasizes cytokines as the central determinants of cell fate, instead relegating them to contributing signals from the immune microenvironment. Notably, mTOR plays a role in integrating these signals as well (Delgoffe et al. 2011). In our model, antigen recognition results in either the generation of short-lived effector cells or long-lived memory cells. The type of effector cell is then determined by the cytokine milieu that in turn is influenced by the local tissue inflammatory response. Likewise, our model emphasizes the role of metabolic programs in regulating T-cell fate. In this regard, our paradigm is consistent with the emerging role of nutrient availability as a means of regulating immune responses. Also, we propose that inhibitors of metabolism might prove to be potent pharmacologic regulators of immune responses. For example, inhibitors of glycolysis might prove to be potent immunosuppressants, both inhibiting effector function and promoting the generation of regulatory T cells. Alternatively, under the proper stimulatory circumstances, inhibitors of glycolysis might prove to enhance the generation of memory cells in response to vaccines. Along these lines, treatment with low dose rapamycin is being explored as a means of enhancing vaccine responses (Turner et al. 2011). These latter two observations emphasize the fact that from a metabolic perspective, regulatory T cells (memory, regulatory T cells in our model) and memory CD8+ T cells actually are quite similar.
Our model takes an unconventional perspective of regulatory T cells. First, we do not view the generation of T-regulatory cells as an alternative outcome of antigen recognition in the presence of TGF-β. Rather, in our model they are potentially generated under many different skewing conditions in part regulated by mTOR activation and metabolism. That is, even under Th1 skewing conditions, TCR engagement in the presence of decreased mTOR activation might result in the generation of a Foxp3+ regulatory cell. Notably, such conditions would also promote the generation of memory T cells. We suspect that the presence of even small amounts of TGF-β might tip a “low mTOR activation encounter” in favor of a T regulatory fate. This hypothesis is based on the observation that although mTOR null T cells become T regs in the absence of exogenous TGF-β, the addition of TGF-β-neutralizing antibodies decreases the generation of T regulatory cells (Delgoffe et al. 2009).
Alternatively robust TCR engagement, resulting in robust mTOR activation can also lead to the generation of a Foxp3+ (effector) regulatory T cells. This population of mTORhi T regs might help to reconcile reports in the literature describing increased mTOR activity in regulatory T cells (Procaccini et al. 2010). We propose that natural T regulatory cells emerging from the thymus have the metabolism of long-lived memory regulatory T cells. This might be the result of the fact that strong stimulation (mTOR activation) in the thymus results in negative selection. Nonetheless, when a natural T regulatory cell encounters antigen in the setting of high mTOR activation we predict it will activate metabolic programs necessary for effector T regulatory cell generation and function.
Although much insight has been gained concerning the development of memory CD8+ T cells, the precise phenotype and function of CD4+ memory cells has been somewhat elusive. We believe that tracking cells based on their metabolic programs (in part regulated by mTOR signaling) will help to dissect the pathways promoting the generation of CD4+ memory cells. Likewise, we believe that this mTOR/metabolic-centric approach might help clarify issues regarding plasticity of CD4+ effector cells.
ACKNOWLEDGMENTS
We thank the other members of the Powell Laboratory and Dr. Drew Pardoll for their advice and suggestions. This work was supported in part by National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH//NIAID) AI77610 and AI091481.
REFERENCES
- Allen A, Zheng Y, Gardner L, Safford M, Horton MR, Powell JD. The novel cyclophilin binding compound, sanglifehrin A, disassociates G1 cell cycle arrest from tolerance induction. J Immunol. 2004;172:4797–4803. doi: 10.4049/jimmunol.172.8.4797. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- Bretscher P, Cohn M. A theory of self–nonself discrimination. Science. 1970;169:1042–1049. doi: 10.1126/science.169.3950.1042. [DOI] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: Spare respiratory capacity and stochastic mitochondrial failure. J Neurochem. 2009;109:1179–1191. doi: 10.1111/j.1471-4159.2009.06055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold SP, Adams E, Farquhar CA, Nolan KF, Howie D, Lui KO, Fairchild PJ, Mellor AL, Ron D, Waldmann H. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: Energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Jenkins MK. The role of cell division in the induction of clonal anergy. Immunol Today. 1992;13:69–73. doi: 10.1016/0167-5699(92)90137-V. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Chen CA, Jung G, Mueller DL, Schwartz RH. Inhibition of antigen-specific proliferation of type 1 murine T cell clones after stimulation with immobilized anti-CD3 monoclonal antibody. J Immunol. 1990;144:16–22. [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty KJ, Cunningham AJ. A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci. 1975;53:27–42. doi: 10.1038/icb.1975.3. [DOI] [PubMed] [Google Scholar]
- Matzinger P. Essay 1: The Danger model in its historical context. Scand J Immunol. 2001;54:4–9. doi: 10.1046/j.1365-3083.2001.00974.x. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol. 2010;22:314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J Immunol. 1999;162:2775–2784. [PubMed] [Google Scholar]
- Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccini C, De Rosa V, Galgani M, Abanni L, Cali G, Porcellini A, Carbone F, Fontana S, Horvath TL, La Cava A, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RH. Costimulation of T lymphocytes: The role of CD28, CTLA-4, and B7/BB1 in interleukin-2 production and immunotherapy. Cell. 1992;71:1065–1068. doi: 10.1016/s0092-8674(05)80055-8. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AP, Shaffer VO, Araki K, Martens C, Turner PL, Gangappa S, Ford ML, Ahmed R, Kirk AD, Larsen CP. Sirolimus enhances the magnitude and quality of viral-specific CD8+ T-cell responses to vaccinia virus vaccination in rhesus macaques. Am J Transplant. 2011;11:613–618. doi: 10.1111/j.1600-6143.2010.03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: An effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature. 2009a;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol. 2009b;183:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]