Abstract
Krüppel-like factor 8 (KLF8) is a transcriptional factor critical for metastatic progression of breast cancer. Epithelial stromal interaction 1 (EPSTI1), a recently identified stromal fibroblast-induced gene in non-invasive breast cancer cells is highly overexpressed in invasive breast carcinomas. The function and regulation of EPSTI1, however, remain largely unknown. In this paper, we report a novel KLF8 to EPSTI1 signaling pathway in breast cancer. Using various expression analyses, we revealed a high co-overexpression of KLF8 and EPSTI1 in invasive human breast cancer cells and patient tumors. Ectopic overexpression of KLF8 in the non-invasive, MCF-10A cells induced the EPSTI1 expression, whereas KLF8 knockdown from the invasive, MDA-MB-231 cells decreased the EPSTI1 expression. Promoter activation and binding analyses indicated that KLF8 promoted the EPSTI1 expression by directly acting on the EPSTI1 gene promoter. EPSTI1 knockdown dramatically reduced the KLF8-promoted MCF-10A cell invasion and ectopic expression of EPSTI1 in the non-invasive, MCF-7 cells is sufficient to induce the cell invasion. Experiments using nude mice demonstrated that the ectopic EPSTI1 granted the MCF-7 cells capability of both invasive growth in the breasts and metastasis to the lungs. Using co-immunoprecipitation coupled with mass spectrometry, we discovered that EPSTI1 interacts with the valosin containing protein (VCP), resulting in the degradation of IκBα and subsequent activation of NF-κB in the nucleus. These findings suggest a novel KLF8 to EPSTI1 to VCP to NF-κB signaling mechanism potentially critical for breast cancer invasion and metastasis.
Keywords: KLF8, EPSTI1, VCP, NF-κB, invasion and metastasis, breast cancer
Introduction
Metastasis remains the main cause of death of breast patients. Better understanding of the underlying molecular and signaling mechanisms is critical for developing more effective therapeutic strategies to improve patient survival.
Krüppel-like factor 8 (KLF8) is a dual transcription factor (1-11) that is highly overexpressed in invasive human cancers and plays an important role in various types of human cancer including breast cancer by promoting the cell cycle progression (6, 11-14), transformation (3, 14), epithelial to mesenchymal transition (EMT) (1, 9, 15, 16) and DNA damage response (2). Although substantial progress has recently been made in understanding the regulation of its expression, post-translational modification and nuclear localization (1, 2, 5, 6, 10, 17), the molecular and signaling mechanisms by which KLF8 promotes human breast cancer metastasis remain largely uninvestigated.
Epithelial stromal interaction 1 (EPSTI1) was initially identified as an induced gene in the human breast cancer cell line MCF-7 by co-incubation of the cells with a telomerase (hTERT) – activated human breast fibroblast cell line in a three-dimensional culture (18, 19). Despite the subsequent studies showing that EPSTI1 is highly overexpressed in breast patient tumor tissues (19, 20) and suggesting that EPSTI1 promotes EMT, tumor progenitor cell properties and anchorage-independent growth in vitro (19, 20), molecular mechanisms that are responsible for the aberrant high expression of EPSTI1 in the breast cancer cells and whether and how EPSTI1 plays a role in vivo for breast cancer progression have not been reported to date.
In this study, we provide strong evidence showing that EPSTI1 is a novel target of transcriptional activation by KLF8 and plays a critical role in the progression of invasive growth and metastasis of breast cancer through VCP-dependent activation of NF-κB.
Results
KLF8 and EPSTI1 are highly co-overexpressed in human metastatic breast cancer cell lines and patient tumors
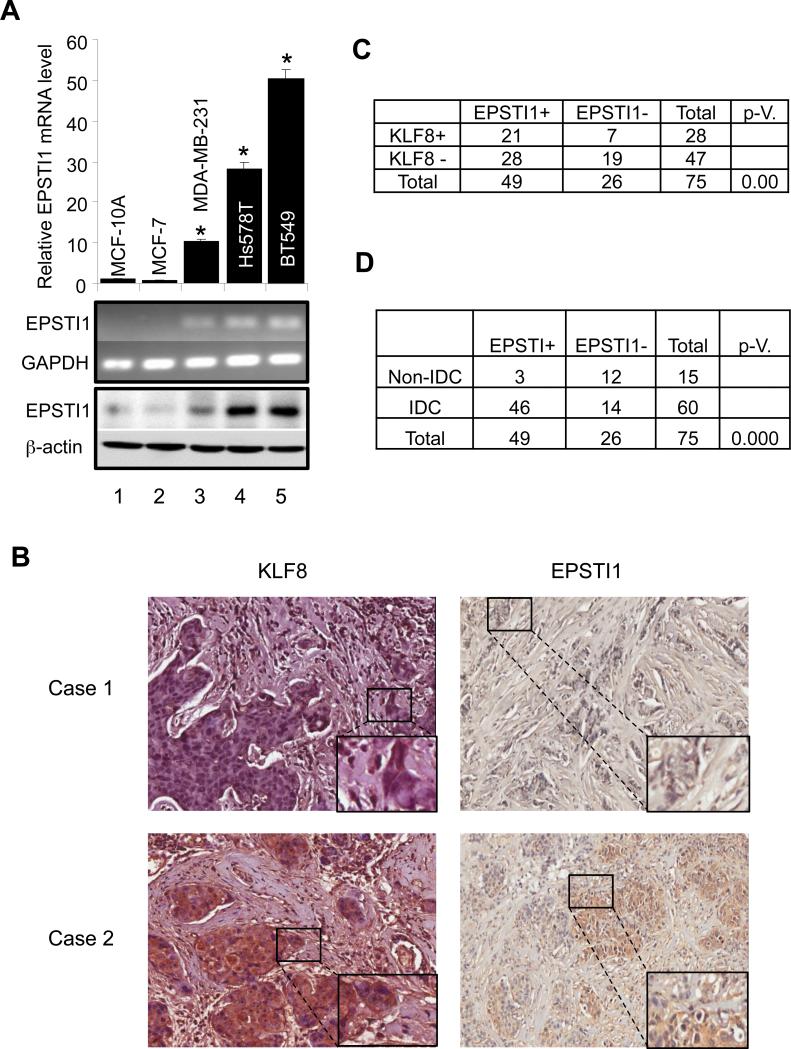
Our cDNA microarray analysis has demonstrated that EPSTI1 is one of the highly upregulated genes in the 10A-iK8 cells (8) when KLF8 expression is induced as compared to the uninduced cells (submitted elsewhere). In addition, we have also recently shown a co-overexpression of KLF8 and EPSTI1 in the MDA-MB-231 cells (21). Since both KLF8 and EPSITI1 have been shown to be aberrantly high in breast cancer cells or tissues (8, 9, 14, 19, 20), we asked if EPSTI1 is upregulated in breast cancer cell lines and tumor specimens known to express high levels of KLF8. We first examined the expression of EPSTI1 in a panel of human breast cancer cell lines in which the KLF8 expression was determined in our previous work (8, 9, 14). The results clearly indicated that there is a high expression of EPSTI1 at both the mRNA and protein levels in the invasive cell lines including the MDA-MB-231, Hs578T and BT549 cells as compared to the barely detectable expression in the non-invasive cancer cell line MCF-7 and the immortalized non-tumorigenic human mammary epithelial cell line MCF-10A (Figure 1A).
Figure 1.
Correlated upregulation of KLF8 and EPSTI1 expression in human metastatic breast cancer cell lines and patient tumors. (a) Aberrant high expression of EPSTI1 in invasive breast cancer cells is well correlated with KLF8 expression. Total RNA and protein lysate were prepared from sub-confluent cells for qRT-PCR (Top panel, *P < 0.01 compared to column 1), semi-quantitative RT-PCR (middle panel) or western blotting (bottom panel) analysis of EPSTI1 expression. (b-e) Positive correlation of protein expression between EPSTI1 and KLF8 in metastatic breast cancer patient tumors. IHC staining of KLF8 or EPSTI1 (brown) in the human breast cancer tissue array containing specimens in duplicate from 75 patient tumors or normal tissues was performed. Images representing a sample negative for both KLF8 and EPSTI1 (case 1) or positive for both KLF8 and EPSTI1 (case 2) were shown in (b). Correlation of EPSTI1 and KLF8 expression was shown in (c). Positive correlation of EPSTI1 expression with the invasive potential was outlined in (d). Statistical significance was determined by χ2-test. (See Supplemental Table 1 for details.)
We have recently performed tissue array analysis of KLF8 protein expression in patient tumors and correlated it with the invasive potential of the tumors (8). To test if expression of EPSTI1 protein is also correlated with that of KLF8 protein and/or the tumor invasive potential in the same set of tissue array, we carried out similar IHC staining for EPSTI protein. The result showed that 75% of the KLF8 positive tumors express EPSTI1 (Figure 1B and 1C). Among the KLF8-negative samples, 59% express EPSTI1, suggesting that in the absence of KLF8 other factors could play a role for the upregulation of EPSTI1 (Figure 1C). EPSTI1 expresses in 77% of the invasive tumors but only 20% of the non-invasive tumors (Figure 1D)..
These results strongly suggested that EPSTI1 expression is positively regulated by KLF8 in the invasive breast cancer tissues.
EPSTI1 is a direct target of transcriptional activation by KLF8
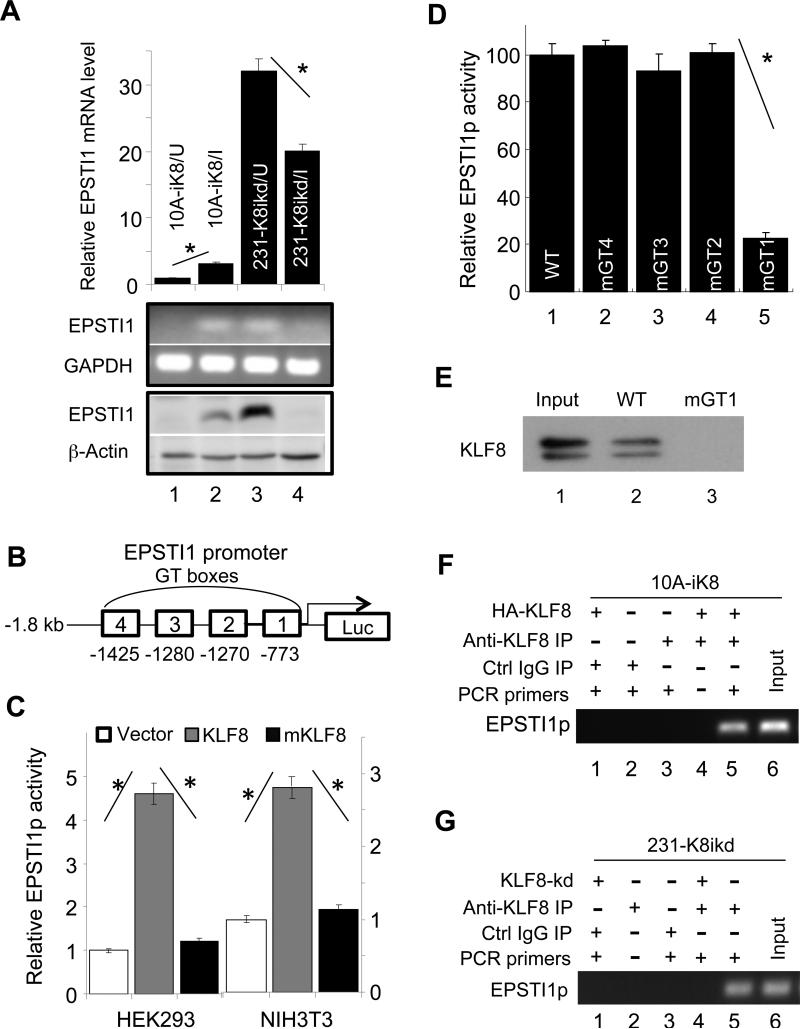
We further validated the microarray results by qRT-PCR and western blotting and verified the high expression of EPSTI1 in the 10A-iK8 cells only when the KLF8 expression was induced (Figure 2A, compare lanes 2 with 1). Conversely, when KLF8 was knocked down in the 231-K8ikd cells (8), EPSTI1 expression was significantly reduced (Figure 2A, compare lanes 4 with 3).
Figure 2.
KLF8 upregulates EPSTI1 expression at the transcriptional level. (a) KLF8 expression is both sufficient and necessary for EPSTI1 expression. The 10A-ik8 and 231-k8ikd cells were grown under induced (I) or uninduced (U) conditions for 48 h. Analyses of EPSTI1 expression were performed similarly as described in Figure 1A. (b and c) KLF8 activates the EPSTI1 gene promoter (EPSTI1p). The human EPSTI1p was cloned into a luciferase reporter plasmid (b) and co-transfected into the indicated cells with wild-type KLF8, it's transactivation-defective mutant (mKLF8) (6) or the control vector plasmid for 16 h. The reporter activity was determined as described in the Material and Methods (c, *P < 0.01). (d) The GT-box 1 (GT1) is required for the activation of EPSTI1p by KLF8. The GT-boxes were individually mutated (CACCC to TCTCA). The wild-type (WT) or mutant (mGT) reporters were used for similar reporter assay. *P < 0.01. (e) KLF8 directly binds EPSTI1p at the GT-box 1 site. HEK293 cell lysate containing HA-KLF8 and oligos spanning the wild-type GT1 (WT) or its mutant (mGT1) were used for BOP assay as described in the Material and Methods. (f and g) KLF8 binds EPSTI1p in vivo. The 10A-iK8 or 231-K8ikd cells were cultured under the uninduced (U) or induced (I) conditions for 72 h. ChIP assay was done as described in the Material and Methods.
To test if EPSTI1 is a KLF8 transcriptional activation target, we first searched the human EPSTI1 gene promoter sequence (EPSTI1p) and found 4 GT-boxes, the potential KLF8 acting sites. We then cloned the EPSTI1p into a luciferase reporter vector (Figure 2B). Co-transfection of the wild-type KLF8 caused more than 3 times increase in the promoter activity in both the HEK293 and NIH3T3 cells. In contrast, the EPSTI1p did not respond to the mutant KLF8 lacking transactivation function (6) (Figure 2C). These results indicated that EPSTI1 is direct transactivating target of KLF8.
To determine if any of the GT-boxes mediates KLF8 activation of the EPSTI1p, we mutated each of the GT-boxes individually and tested their response to KLF8. We found that mutation of the GT-box 1, but none of the other three, prevented the activation of the EPSTI1p by KLF8 (Figure 2D), suggesting that the GT-box 1 is required for KLF8 activation of the EPSTI1p. To determine the possible interaction between KLF8 protein and the GT-box 1 site of the EPSTI1p, we performed biotinylated oligonucleotide precipitation (BOP) as well as chromatin immunoprecipitation (ChIP) assays. The results demonstrated that both ectopic KLF8 in (Figure 2F) and endogenous KLF8 (Figure 2G) can bind the EPSTI1p in vivo at the promoter region containing the GT-box 1 and the interaction was abolished when the GT-box 1 was disrupted (Figure 2E).
Taken together, these results strongly suggested that KLF8 directly activates the EPSTI1p to upregulate the expression of EPSTI1.
EPSTI1 promotes the cell invasiveness downstream of KLF8
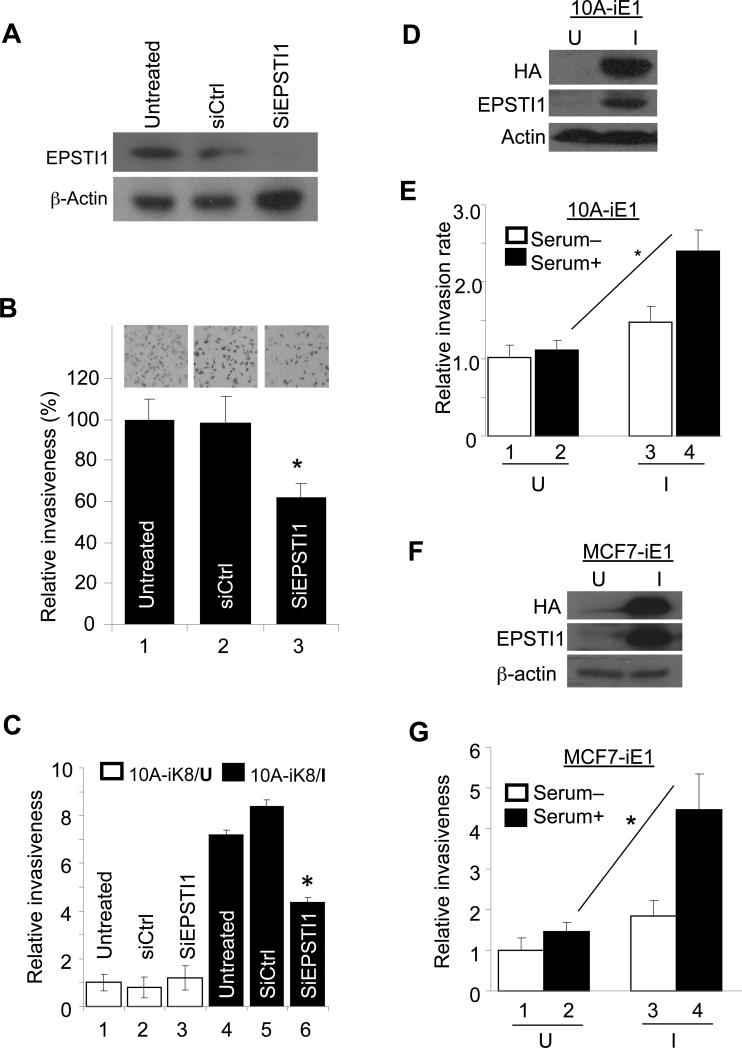
To determine a potential role of EPSTI1 in the cell invasiveness in the context of KLF8 expression, we first knocked down EPSTI1 from the MDA-MB-231 cells known to express aberrant high levels of KLF8 (8, 9, 14) and examined the cell invasiveness using the Matrigel invasion chambers. We found that upon the EPSTI1 knockdown (Figure 3A), the cell invasieveness was drastically reduced as compared to the cells treated with a control siRNA or untreated cells (Figure 3B). To test if EPSTI1 plays this role downstream of KLF8, EPSTI1 was knocked down similarly from the 10A-iK8 cells and the cell response to the induction of KLF8 expression for invasion was examined. We found that the silence of EPSTI1 expression caused approximately 50% reduction in the cell invasiveness regardless of the KLF8 expression (Figure 3C, compare columns 6 with 4 or 5). These results suggested that EPSTI1 plays a critical role downstream of KLF8 in promoting the cell invasion.
Figure 3.
EPSTI1 promotes the cell invasiveness downstream of KLF8. (a and b) EPSTI1 knockdown decreases the invasiveness of MDA-MB-231 cells. The cells were transfected with siRNA for 72 h. A fraction of the cells was prepared for western blotting (a). Another fraction of the cells was used for Matrigel invasion for 16 h (b) as described in the Material and Methods. Data was normalized to the untreated cells. *P < 0.05. (c) EPSTI1 knockdown inhibits KLF8-promoted invasiveness. The 10A-iK8 cells were cultured under uninduced (U) or induced (I) conditions for 48 h and then treated with or without the indicated siRNA for 72 h. Matrigel invasion was done similarly. Data was normalized to the uninduced, untreated cells. (d-g) EPSTI1 overexpression is sufficient to promote invasiveness. The 10A-iE1 and MCF7-iE1 cells were generated as described in the Material and Methods. After grown under U or I conditions for 24 h, the cells were prepared for western blotting (d, f) and invasion assay (e, g). The invasion data were normalized to the U and serum-free conditions. *P < 0.05. All the data are representatives of at least three independent experiments in duplicate.
To test if ectopic overexpression of EPSTI1 alone is sufficient to promote the cell invasion, we generated the MCF-10A and MCF-7 cell lines that express inducible EPSTI1 (10A-iE1 and MCF7-iE1, respectively). The cell invasiveness was then compared between the induced and uninduced cells. The results revealed that after EPSTI1 expression was induced (Figure 3D and 3F) the cell invasiveness was significantly enhanced (Figure 3E and 3G, compare columns 4 with 2). These results indicated that EPSTI1 overexpression alone is adequate to promote cell invasion.
EPSTI1 promotes the lung metastasis of breast cancer
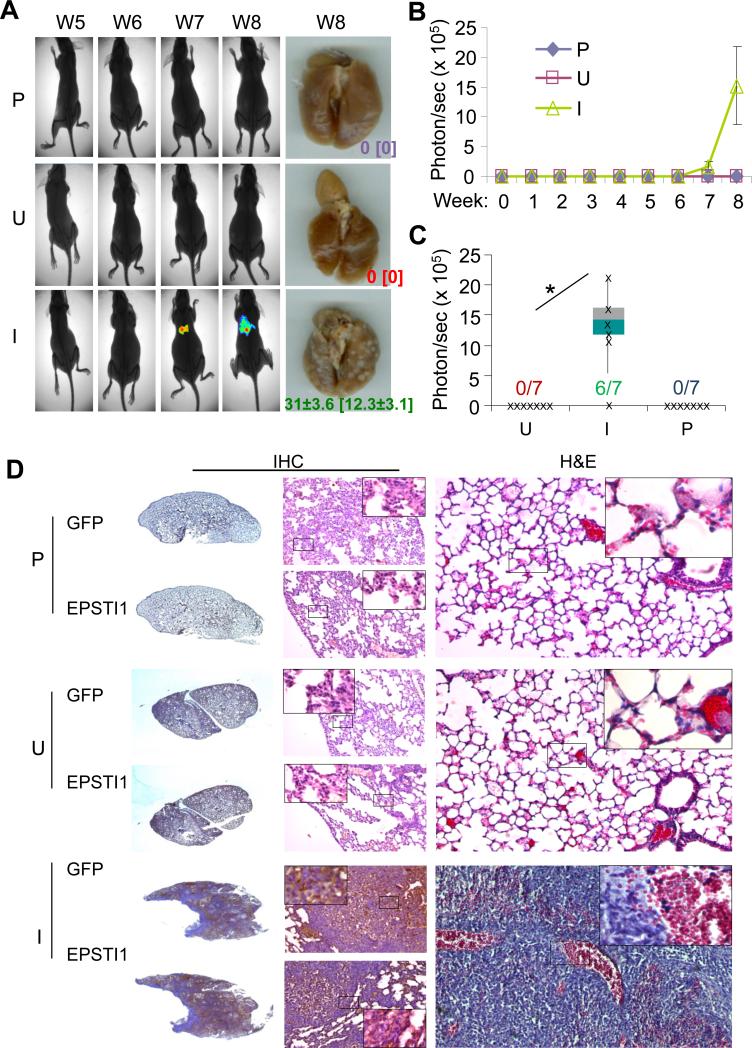
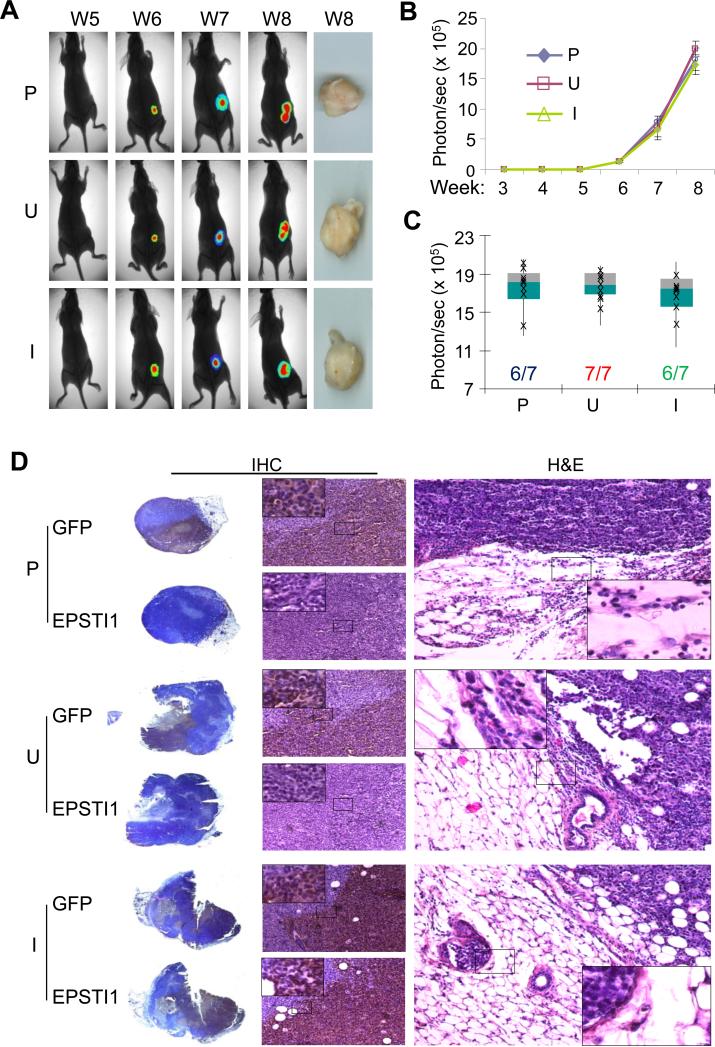
We have recently demonstrated that KLF8 promotes breast cancer lung metastasis (3, 8). To see if EPSTI1 plays a similar role, we injected the MCF7-iE1 cells (uninduced or induced) or the parental cells into the tale vein and followed up the lung metastasis by bioluminescent imaging (BLI) (Figure 4A – FC), stereomicroscopy (Figure 4A), as well as histological analyses (Figure 4D). We found that the lung metastases began to be detectable by BLI at week 6 or week 7 and became significantly more obvious at week 8 even by surgical microscopy (Figure 4A – 4C). Notably, the lung metastases were formed by the MCF7-iE1 cells, at an incidence of 86%, only when EPSTI1 expression was induced (I), whereas the uninduced cells (U) or the parental cells (P) did not form any detectable lung metastasis (Figure 4A - 4C). The human origin of the metastases (indicated by GFP expression) and the effective induction of EPSTI1 expression in the metastases were verified by the H&E and IHC staining (Figure 4D). These results supported the notion that EPSTI1 serves as one of the driving forces for the lung metastasis of breast cancer.
Figure 4.
EPSTI1 promotes the lung metastasis of breast cancer. The MCF7-iE1 cells or the parental cells (P) were injected into the tail vein. The mice injected with MCF7-iEPSTI1 cells were fed with the Dox Diet (I, EPSTI1 induction) or Control Diet (U, no EPSTI1 induction). The mice injected with the parental cells were feed with normal food. The lung metastasis was followed up for 8 weeks by BLI or stereomicroscopy at week 8 as described in the Materials and Methods. (a) Representative images Representative images with average number and standard deviation of metastatic tumor nodules per mouse visible on the surface of the lungs or stained positive for GFP in the tissue section shown in the brackets. (b) Quantitative results of the volumes of the metastasis. (c) Metastatic incidence at week 8. *P < 0.05. (d) H&E and IHC staining with GFP or EPSTI1 antibody confirming the human origin of the lung metastases and the effective induction of EPSTI1 expression in the tumors.
EPSTI1 promotes tumor invasion in the breast
To determine how EPSTI1 expression affects the tumor progression in the breast, we injected the MCF7-iE1 cells or the parental cells orthotopically into the mammary fat pad and examined the tumor growth and invasion (Figure 5). We found that the tumors became detectable between week 3 and week 4 and grew at the same rate and incidence across the groups regardless of EPSTI1 expression (Figure 5A – 5C). Interestingly, however, the histological analyses indicated that the tumors formed by the MCF7-iE1 cells with the induction of EPSTI1 (I) have invaded the surrounding adipose tissue of the host mice, which did not occur to the tumors formed by the uninduced MCF7-iE1 cells (U) or the parental cells (P) (Figure 5D). These results suggested that EPSTI1 plays a role primarily in promoting the tumor invasion rather than proliferation.
Figure 5.
EPSTI1 promotes tumor invasion at the orthotopic site. The MCF7-iE1 or parental (P) cells were injected into the mammary fat pad. The mice were fed as described in Figure 4. BLI analysis and stereomicroscopy of the tumors (a-c) and H&E or IHC analyses (d) were performed similarly as described in Figure 4.
EPSTI1 interaction with the N-terminal half of VCP is critical for the cell invasion
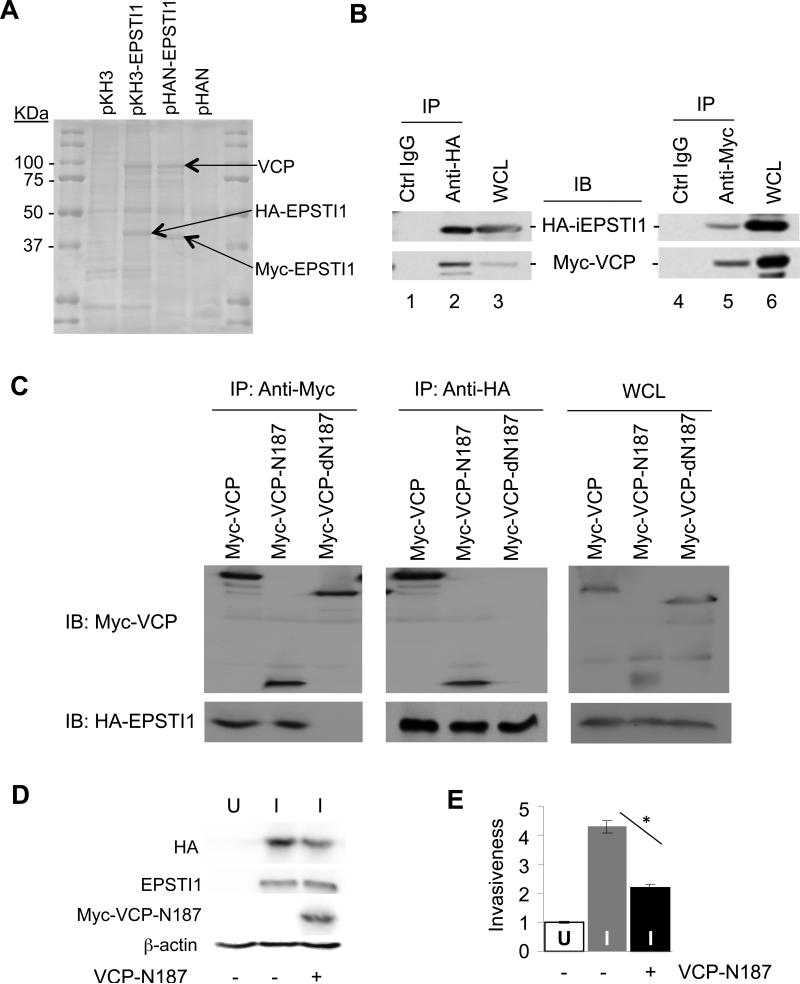
Molecular mechanisms behind EPSTI1 have not been studied to date. To begin with, we performed CoIP coupled with mass spectrometry to identify potential EPSTI1 interacting proteins. One of the strongly interacting candidate proteins identified was VCP (Figure 6A and 6B), a primary function of which is to target proteins for degradation (22). Truncation mutagenesis of VCP indicated that the amino terminal 187 residual stretch (VCP-N187) contains an EPSTI1 binding domain or motif (Figure 6C). To determine the relevance of this VCP region to the function of EPSTI1, the VCP-N187 peptide was co-expressed with EPSTI1 in the MCF7-iE1 cells (Figure 6D). We found that the expression of this peptide inhibited the cell invasiveness induced by EPSTI1 (Figure 6E). This result suggested that the EPSTI1-VCP interaction is critical for the cell invasion and the VCP-N187 peptide reduced the cell invasiveness probably by competing with the endogenous VCP for EPSTI1 interaction.
Figure 6.
EPSTI1 interaction with the 1-187 aa region of VCP (VCP-N187) is critical for the cell invasion. (a) Mass spectrometry identified VCP as an EPSTI1 interaction protein. Plasmid encoding HA- or Myc-tagged EPSTI1 (pKH3-EPSTI1 or pHAN-EPSTI1) or the control vector was transfected into HEK293ft cells for 24 h, followed by anti-HA or anti-Myc coIP and subsequent mass spectrometry as described in the Materials and Methods. (b) CoIP verification of interaction between EPSTI1 and VCP. The HEK293ft cells were cotransfected with pKH3-EPSTI1 and pHAN-VCP for 24 h and prepared for CoIP and western blotting. WCL, whole cell lysate. (c) VCP-N187 is where EPSTI1 binds. The full-length VCP, its N-terminal 187 residual fragment (N187) or its C-terminal half (dN187) was co-transfected and Co-IP were followed similarly as described in b. (d and e) VCP-N187 is crucial for EPSTI1 to promote the cell invasion. The MCF7-iE1 cells were grown under U or I conditions for 24 h. The I-cells were then transfected with the VCP-N 187 peptide or the control vector for another 24 h prior to western blotting (d) or Matrigel invasion assay (e, *P < 0.05).
EPSTI1 interaction with VCP is critical for the activation of NF-κB
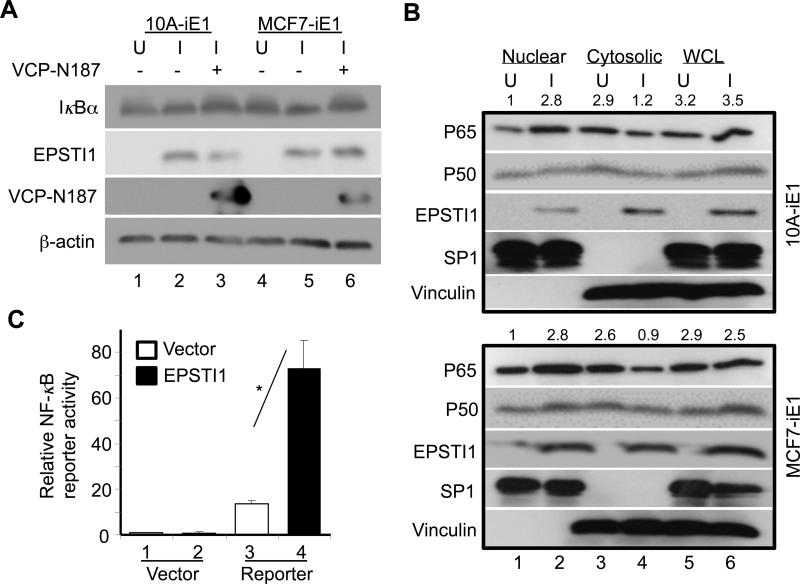
To further understand the signaling mechanisms downstream of the EPSTI1-VCP interaction, we tested if the expression of VCP target proteins is altered by the overexpression of EPSTI1. One of the affected proteins was IκBα whose protein levels were significantly decreased in both the 10A-iE1 and MCF7-iE1 cells upon induction of the EPSTI1 expression (Figure 7A). And this change was rescued by overexpression of the VCP-N187 peptide (Figure 7A, compare lanes 3 or 6 with 2 or 5) (see Figure S2A for quantitative data).
Figure 7.
EPSTI1 interaction with VCP is critical for the activation of NF-κB. (a) EPSTI1 expression decreases the expression of IκBα depending upon its binding to CVP. The 10A-iE1 or MCF7-iE1 cells were cultured under U or I conditions for 24 h. The I-cells were then transfected with the VCP-N187 peptide (lanes 3 or 6) or the control vector (lanes 2 or 5) for another 24 h prior western blotting. (see Figure S2A for quantitative data). (b) EPSTI1 expression promotes the nuclear translocation of NF-κB. The 10A-iE1 or MCF7-iE1 cells were cultured under U or I conditions for 24 h. Then the nuclear and cytoplasmic fractions were isolated for western blotting. SP1 and vinculin were used as a marker for nucleus and cytoplasm, respectively. (see Figure S2B and S2C for quantitative data). (c) EPSTI1 expression increases the transcriptional activity of NF-κB. MCF7 cells were co-transfected with the NF-κB reporter or control vector with EPSTI1 or control vector for 24 h before luciferase activity was determined. Data are representatives of at least three independent experiments in duplicate. *P < 0.05.
We then examined the effect of EPSTI1 expression on the subcellular expression of NF-κB subunits p65 and p50. We found that EPSTI1 expression caused a dramatic increase in the levels of both subunits in the nucleus (Figure 7B, compare lanes 2 with 1) and decrease in the cytoplasm (Figure 7B, compare lanes 4 with 3) (see Figure S2B and S2C for quantitative data).
We then determined the transactivation activity of NF-κB by co-expressing EPSTI1 with an NF-κB-responsive promoter reporter, and found that EPSTI1 expression caused a 3 – 4 times increase in the reporter activity (Figure 7C).
Taken together, these results suggest that EPSTI1 plays a critical role in the degradation of IκBα and subsequent activation of NF-κB.
Discussion
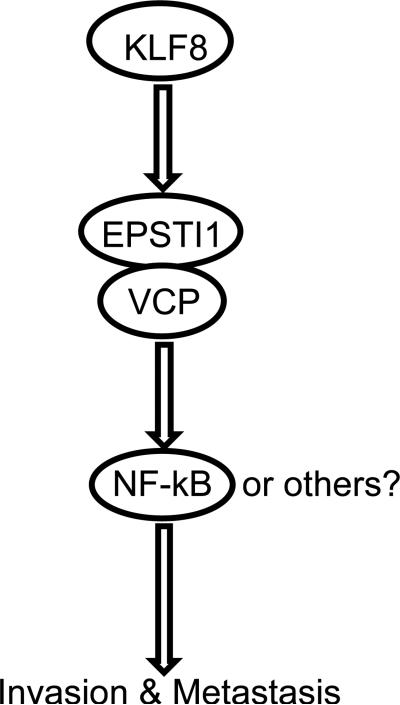
This study identified EPSTI1 as a novel transcriptional target of KLF8 and a critical signaling effecor downstream of KLF8 for the invasive growth and lung metastasis of human breast cancer (Figure 8). In this signaling model of cancer progression, the aberrant high expression of KLF8 in the cancer cells ensures the high levels of EPSTI1 expression through activation of transcription of EPSTI1 by KLF8. EPSTI1 then recruits VCP by an unknown mechanism to IkBα to subsequently induce the degradation of IkBα, resulting in the nuclear translocation of NF-kB to activate certain target genes associated with tumor invasion and metastasis.
Figure 8.
A Model of mechanism of action. KLF8 upregulates EPSTI1 at the transcriptional level. EPSTI1 interacts with VCP to induce the degradation of IkBα, causing the nuclear translocation of NF-κB to activate certain target genes associated with tumor invasion and metastasis.
We have recently demonstrated that the aberrant co-overexpression of KLF8 and EPSTI1 plays a critical part for the invasive growth and drug resistance of breast tumor (21). Our cDNA microarray analysis (submitted to elsewhere) identified EPSTI1 as one of the most highly upregulated genes by KLF8 in the 10A-iK8 cells and this result was verified at both mRNA and protein levels by various means (Figure 1 and 2). In addition, we have correlated the co-overexpression of KLF8 and EPSTI1 with aggressive potential of the patient tumors (Figure 1 and Table S1). Furthermore, we demonstrate that the KLF8-binding consensus site at the EPSTI1p is highly conserved during evolution (Figure S1). Lastly, KLF8 and EPSTI1 share some common targeted cellular functions such as EMT. These lines of evidence, taken together with the earlier reports (18-20), strongly suggests that EPSTI1 is likely a true target of transcriptional activation by KLF8 and the KLF8 to EPSTI1 signaling pathway potentially associated with the tumor microenvironment is critical for breast cancer progression.
EPSTI1 was initially identified due to the induction of its expression in breast cancer cells by co-cultured stromal fibroblasts (19), suggesting that an extracellular factor(s) responsible for the induction of EPSTI1 may be released by the stromal fibroblasts. Our results suggest that this extracellular factor(s) could induce EPSTI1 via KLF8-EPSTI1. Interferon-α (IFN-α) has been associated with the upregulation of EPSTI1 in breast and ovarian cancer tumors (20, 23, 24). TGF-β and Wnt play a role for the induction of KLF8 expression in cancers (1, 9, 11, 14, 15, 25). These data indicate that these stromal factors may act upstream of KLF8-EPSTI1 pathway in breast cancer. Epithelial cells are converted to stromal fibroblast-like cells during EMT. The fact that both KLF8 and EPSTI1 can induce EMT (9, 20) raises a possibility that the KLF8-EPSTI1 signaling induces EMT, causing release of the stromal factor(s) that in turn stimulates the expression of KLF8 and EPSTI1 via an autocrine loop. Experiments are in progress to test these interesting possibilities.
Our recent studies have demonstrated that KLF8 not only promotes invasive tumor growth and metastasis but also plays a role in the growth of tumor sizes of breast cancer (8) (submitted to elsewhere). In contrast, EPSTI1 appears to promote tumor invasion and metastasis only (Figure 5 and 6) and, unlike KLF8, does not appear to play a role in the regulation of cell proliferation (data not shown). These results suggest that EPSTI1 contributes primarily to the invasion/metastasis-promoting function of KLF8.
Our results show that VCP-mediated activation of NF-κB plays an important role downstream of KLF8-EPSTI1 signaling (Figures 6-8). Consistently, the VCP-NF-κB signaling plays a similar role also in the metastasis of osteosarcoma (26). VCP can potentially target many proteins for degradation (22). Similarly, NF-κB targets a wide array of genes associated with a broad range of cellular functions contributing to tumor progression (27). How EPSTI1 interaction regulates VCP-dependent degradation of proteins and how this regulation affects the protein targeting profiles of VCP? In addition to VCP, what other potential EPSTI1 interacting proteins also contribute to the tumor invasion and metastasis and by what distinct mechanisms? What subgroup of NF-κB target genes are involved downstream of EPSTI1 for the invasive and metastatic progression of breast cancer? Although beyond the scope of this report, it is obvious that these interesting questions need to be answered by future investigations.
In summary, this work provides a novel insight into the mechanisms of metastatic progression of breast cancer and opens a new avenue in the research of both KLF8 and EPSTI1. The KLF8 to EPSTI1 to VCP signaling identified here could generally apply to other types of cancer associated with aberrant overexpression of KLF8 and even non-cancerous diseases or disorders associated with expression of the EPSTI1-VCP complex. More investigations are worth of pursuing to understand the details in the signaling pathway as a potentially therapeutic target.
Materials and Methods
Reagents and cell culture
Antibodies
Antibodies used for western blotting were HA-probe (F-7) mouse monoclonal Ab (sc-7392), HA-probe (Y-11), rabbit polyclonal Ab (sc-805) , c-Myc (9E10) mouse monoclonal Ab (sc-40), c-Myc (C-19) rabbit polyclonal Ab (sc-788), β-actin (C4) mouse monoclonal Ab (sc-47778), IκBα (FL) rabbit polyclonal Ab (sc-847), NF-κB p65 (C-20) rabbit polyclonal Ab (sc-372), Sp1 (H-225) rabbit polyclonal Ab (sc-14027), and vinculin (7F9) mouse monoclonal Ab (sc-73614) (Santa Cruz Biotechnology, Inc., Dallas, Texas, USA), EPSTI1 rabbit polyclonal Ab (HPA017362) (Sigma-Aldrich Co., St. Louis, MO, USA), and HRP-conjugated donkey anti-mouse IgG (715-035-150) or anti-rabbit IgG (711-035-152) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA). Antibodies used for CoIP were Anti-HA mouse monoclonal (IP0010) and Anti-c-Myc rabbit polyclonal Ab (IP0020) Immunoprecipitation Kit (Sigma-Aldrich Co., St. Louis, MO, USA). Antibodies used for IHC or tissue array were EPSTI1 rabbit polyclonal Ab (HPA017362) (Sigma-Aldrich Co., St. Louis, MO, USA); GFP mouse monoclonal Ab (sc-101525) and HA-probe (F-7) (Santa Cruz Biotechnology, Inc., Dallas, Texas, USA) and Peroxidase substrate kit (DAB) (SK-4100) (Vector laboratories Inc., Burlingame, CA, USA). Antibodies used for BOP and ChIP assays were HA-probe (F-7) and KLF8 rabbit polyclonal Ab (8477), respectively as previously described (8, 9, 11).
Plasmids
The mammalian expression vectors pKH3 (HA-tagged) and pHAN (Myc-tagged), pKH3-KLF8 and pKH3-mKLF8 were previously described (6). SFG-nTGL encoding the tk-GFP-Luciferase fusion reporter protein was a kind gift from Dr. V. Ponomarev (28). The TetO-FUW-OSKM (29) (# 20321), FUW-M2rtTA (30) (#20342), VCP (wt)-EGFP (31) (# 23971), pLVTHM (32) (#12247) were purchased from Addgene (Cambridge, MA, USA). The ORFEXPRESSTM-shuttle hEPSTI1V2 vector encoding the human EPSTI1 cDNA (GC-T1743) was purchased from GeneCopoeia Inc. (Rockville, MD, US). The CH17-13N15 BAC clone containing the human EPSTI1 gene promoter (EPSTI1p) was purchased from Children's Hospital Oakland Research Institute (CHORI) (Oakland, CA, USA). The NF-κB(2) luciferase reporter vector (LR0052) and the control vector pTL-Luc (LR0000) were purchased from Affymetrix-Panomics (Santa Clara, CA). The pKH3-EPSTI1 and pHAN-EPSTI1 plasmids were generated by PCR using the ORFEXPRESSTM-shuttle hEPSTI1V2 as template and the hEPSTI1-RV-F and hEPSTI1V2-RI-R primer set, cutting the PCR product by EcoRI and EcoRV and ligating it into the pKH3 or pHAN vector between the SmaI and EcoRI sites. TetO-FUW vector was recovered from TetO-FUW-OSKM by EcoRI digestion and self-ligation. TetO-FUW-RFP-EPSTI1 was made by multistep PCR linking mCherry, the T2A and HA-EPSTI1 cDNAs together and ligating it into TetO-FUW. The multistep PCR was done using a mCherry encoding plasmid as template and the Kozak-RFP-F/T2A-RFP-R and Kozak-RFP-F/T2A-R primer sets, the pKH3-EPSTI1 as the template and the T2A-HA-F/pKH3-R and T2A-F/pKH3-R primer sets, and the two PCR product mix as template and the Kozak-RFP-F/pKH3-R primer set. The end PCR product was cut with EcoRI and inserted into the EcoRI site of TetO-FUW. The pGL3b-hEPSTI1p reporter vector was made by PCR using the CH17-13N15 BAC clone as the template and hEPSTI1p–F/ hEPSTI1p–R primer set, cutting the PCR product with HindIII and inserting it into the HindIII site of PGL3b vector. GT-box specific mutants of the promoter were constructed by site-directed mutagenesis PCR (33) using the pGL3b-hEPSTI1p vector as the template and each of the mutant GT-box (mGT) primer sets along with the hEPSTI1p–F/ hEPSTI1p–R primer set. The pHAN-VCP, pHAN-VCP-N187 and pHAN-VCP-dN187 plasmids were constructed by PCR amplifying the human VCP cDNA from the VCP (wt)-EGFP vector using the primer set of hVCP-BHI-F/hVCP-BHI-R, hVCP-BHI-F/hVCP-187-BHI-R, and hVCP-188-BHI-F/hVCP-BHI-R, respectively and cloning into the BamHI site of pHAN. To construct pLVTHM-nGL vector, the tk fragment was removed from the SFG-nTGL by deletion PCR using the dT-F/dT-R. The resulting SFG-nGL was then used as template for PCR using the nGL-F/nGL-R primer set. The PCR product was cut with SpeI and inserted between the PmeI (blunted) and SpeI sites of pLVTHM. Correctness of the constructs was confirmed by insert orientation check with restriction digestion, DNA sequencing, and/or protein expression analysis. See primer sequences in Table S2.
Cell lines
The HEK293 and NIH3T3 (33-35), MCF-10A, MCF-7, MDA-MB-231, Hs578T and BT-549 (9, 14), and the KLF8-expressing Tet-on MCF-10A (10A-iK8) and the KLF8 shRNA-expressing Tet-on MDA-MB-231 (231-K8ikd) (2, 8) cell lines were described previously. These cells were maintained in DMEM/F-12 or DMEM with 10% fetal bovine serum or calf serum. MCF-10A and MCF-7 cell lines expressing the nGL (MCF-10A-nGL and MCF-7-nGL) were generated by infecting the cells with the pLVTHM-nGL lentivirus followed by selection of GFP-expressing cells by fluorescent microscopy. To generate the Tet-on MCF-10A and MCF-7 lines that express inducible EPSTI1 (10A-iE1 and MCF7-iE1), the TetO-FUW-RFP-EPSTI1 and FUW-M2rtTA lentiviruses were generated as previously described (8) and used to co-infect the MCF-10A-nGL or MCF-7-nGL cells. Infected cells were seeded into 96-well plate at a density of 0.7 cells per well. Positive clones were selected by transient induction of RFP expression coupled with fluorescent microscopy. These Tet-on inducible cell lines were maintained under uninduced (U, in the absence of doxycycline) or induced (I, in the presence of doxycycline) conditions depending on the experimental requirement as previously described (2, 8, 9, 11, 33, 35). Doxycycline hydrochloride was purchased from Sigma-Aldrich Corp. (D3072, St. Louis, MO, USA).
Quantitative real-time PCR (qRT-PCR), western blotting and co-immunoprecipitation (CoIP)
These assays were done as previously described (9). Cells and antibodies used were described above. Sequences of the PCR primers are listed in Table S2. Sub-cellular fractionation was described previously (13). Specifically, after two washes with 1× PBS and equilibration for 5 min with 0.5 ml of 0.5 mM EDTA in PBS, cells were scraped off and centrifuged (4,500 × g, 2 min, 4°C). The cell pellets were re-suspended in 0.4 ml of low-salt HEPES buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol) for 15 min, lysed in 20 μl of 10% NP-40, and centrifuged (10,000 × g, 30 s, 4°C) to obtain the cytosolic supernatant. The pellet will be re-suspended in 50 μl of high salt buffer (840 mM NaCl, 40mM HEPES [pH 7.8], 2 mM EDTA, 2M EGTA, and 40% glycerol) for 30 min and centrifuged (10,000 × g, 5 min, 4°C) to obtain the nuclear extract.
Affinity precipitation coupled with mass spectrometry, RNA interference, and Matrigel invasion assays
These assays were performed as previously described (8, 9, 17). The pKH3- and pHAN-EPSTI1 plasmids and antibody beads were described above. The human EPSTI1 specific ON-TARGET plus SMARTpool siRNAs (L-015094-01) and control siRNAs (D-001810-0X) were purchased from Thermo Scientific, Dharmacon (West Palm Beach, FL, USA). The siRNAs were delivered into the cells by Oligofectamine-mediated transfection according to manufacturer's instructions (Invitrogen, Grand Island, NY, USA). BD BioCoat™ Matrigel™ Invasion Chambers (354480, BD Biosciences, San Jose, CA, USA), crystal violet (C3886, Sigma-Aldrich Co., St. Louis, MO, USA) and a Zeiss AXIO Observer A1 microscope (Carl Zeiss Inc., Thornwood, NY, USA) were used for the invasion assays.
Promoter reporter assays, chromatin immunoprecipitation (ChIP) and biotinylated oligonucleotide precipitation (BOP)
These assays were performed as previously described (9, 34). The plasmids encoding the wild-type or mutant human hEPSTI1p, the NF-κB(2) luciferase reporter and the control vector, HEK293, NIH3T3, 10A-iK8 and 231-K8ikd cell lines were described above. Sequences of the ChIP primers and BOP oligos for the human EPSTI1p are described in Table S2.
Bioluminescence imaging (BLI) analysis of tumor growth and metastasis
Female ovariectomized athymic nude mice (4-5 weeks old, 7 mice per group, Hsd:Athymic Nude-Foxn1nu, Harlan Laboratories, Indianapolis, IN, USA) were used for all the xenograft studies. The mice were housed and maintained in specific pathogen-free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with current regulations and standards of the United States Department of Agriculture, United States Department of Health and Human Services, and the National Institute of Health. IACUC protocols were approved by the Institutional Animal Care and Use Committee. Human care of the mice was thoroughly considered. A 17β-ESTRADIOL pellet (NE-121, Innovative Research of America, Sarasota, FL, USA) was s.c. implanted. One day later, 106 of the MCF7-iE1 or MCF7-nGL cells were washed and harvested in 0.05 – 0.1 ml PBS and injected into either the mammary fat pad injection or lateral tail vein. The mice were fed with diet (Bio Servs, Frenchtown, NJ, USA) supplemented with doxycycline or Dox Diet (S3888) to induce the EPSTI1 expression in the cells in vivo or with the Control Diet not containing doxycycline (S4207). After injection, tumor growth or lung metastasis was monitored daily or weekly visually and/or by BLI. For BLI, mice were anaesthetized and injected i.p. with 150 mg/kg of D-luciferin (15 mg/ml in PBS) (LUCK-1, Gold Biotechnology, Inc., St. Louis, MO, USA). BLI was then completed between 2 and 5 min using a Kodak In Vivo Imaging System coupled to Molecular Imaging Software (Carestream, Rochester, NY, USA) followed by X-ray imaging. Photon flux (photons/s/cm2 per steradian) was measured with a region of interest drawn around the bioluminescence signal encompassing the thorax or the mammary fat pad. A background value was obtained from a D-luciferin-injected control mouse and subtracted.
Hematoxylin and eosin (H&E) and immunohistochemical (IHC) staining
The mammary tissues and the lungs were surgically collected, fixed in formaldehyde solution (F8775, Sigma-Aldrich Co., St. Louis, MO, USA), embedded in EM-400 Embedding Medium Paraffin (3801320, Leica Microsystems Inc., Buffalo Grove, IL, USA), and sectioned with the Manual Rotary Microtome for Routine Sectioning (Leica RM2235, Leica Microsystems Inc., Buffalo Grove, IL, USA). H&E and IHC staining was performed as previously described (3). Human breast cancer tissue array with invasive data (BR1503, US Biomax, Rockville, MD, USA) was analyzed as previously described (3, 8). Large Volume Myer's Hematoxylin (AMH100420), Eosin Y (17372-87-10) and Xylene (X3P-1GAL) were purchased from Thermo Fisher Scientific (West Palm Beach, FL, USA). The 3,3-diaminobenzidine tetrahydrochloride (868272-85-9) was purchased from Sigma-Aldrich Co., (St. Louis, MO, USA). GFP and EPSTI1 antibodies were described above. The number of tumor nodules on the surface of the whole-mounted lungs or GFP-positive nodules across three independent whole lung sections were counted for each mouse by microscopy. Microscopy was done using Leica DFC295 microscope for sectioned tissues or surgical microscope (LeicaL2) for whole mount tumors or lungs (Leica Microsystems Inc., Buffalo Grove, IL, USA).
Statistical Analysis
A minimum of three observations per group was conducted. Data is presented as mean +/- the standard deviation (SD). Unpaired, paired or single sample Student's t-test with the Bonferroni correction for the multiple comparisons was applied as appropriate. The tissue array data was analyzed by χ2-test. Significance was determined by the alpha level of 0.05.
Supplementary Material
Acknowledgments
We appreciate Dr. Vladimir Ponomarev of Memorial Sloan-Kettering Cancer Center for kindly providing the trimodal imaging reporter vector SFG-nTGL and Dr. Qishan Lin of UAlbany for helping with the mass spectrometry. This work was supported by grants from NCI (CA132977) and Susan G. Komen for Cure breast cancer foundation (KG090444 and KG080616) to J.Z.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lahiri SK, Zhao J. Kruppel-like factor 8 emerges as an important regulator of cancer. American journal of translational research. 2012;4(3):357–63. PubMed PMID: 22937212. Pubmed Central PMCID: 3426389. [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H, Hu L, Li T, Lahiri S, Shen C, Wason MS, et al. A Novel Role of Kruppel-like Factor 8 in DNA Repair in Breast Cancer Cells. The Journal of biological chemistry. 2012 Dec 21;287(52):43720–9. doi: 10.1074/jbc.M112.418053. PubMed PMID: 23105099. Pubmed Central PMCID: 3527957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu H, Wang X, Urvalek AM, Li T, Xie H, Yu L, et al. Transformation of human ovarian surface epithelial cells by Kruppel-like factor 8. Oncogene. 2012 Dec 10; doi: 10.1038/onc.2012.545. PubMed PMID: 23222713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnell O, Romagna A, Jaehnert I, Albrecht V, Eigenbrod S, Juerchott K, et al. Kruppel-like factor 8 (KLF8) is expressed in gliomas of different WHO grades and is essential for tumor cell proliferation. PloS one. 2012;7(1):e30429. doi: 10.1371/journal.pone.0030429. PubMed PMID: 22276196. Pubmed Central PMCID: 3261906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urvalek AM, Lu H, Wang X, Li T, Yu L, Zhu J, et al. Regulation of the oncoprotein KLF8 by a switch between acetylation and sumoylation. American journal of translational research. 2011 Feb;3(2):121–32. PubMed PMID: 21416054. Pubmed Central PMCID: 3056558. [PMC free article] [PubMed] [Google Scholar]
- 6.Urvalek AM, Wang X, Lu H, Zhao J. KLF8 recruits the p300 and PCAF co-activators to its amino terminal activation domain to activate transcription. Cell cycle. 2010 Feb 1;9(3):601–11. doi: 10.4161/cc.9.3.10606. PubMed PMID: 20107328. Pubmed Central PMCID: 2888133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic acids research. 2000 May 1;28(9):1955–62. doi: 10.1093/nar/28.9.1955. PubMed PMID: 10756197. Pubmed Central PMCID: 103308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Lu H, Urvalek AM, Li T, Yu L, Lamar J, et al. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011 Apr 21;30(16):1901–11. doi: 10.1038/onc.2010.563. PubMed PMID: 21151179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, et al. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer research. 2007 Aug 1;67(15):7184–93. doi: 10.1158/0008-5472.CAN-06-4729. PubMed PMID: 17671186. [DOI] [PubMed] [Google Scholar]
- 10.Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, et al. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. The Journal of biological chemistry. 2006 Jun 16;281(24):16664–71. doi: 10.1074/jbc.M513135200. PubMed PMID: 16617055. [DOI] [PubMed] [Google Scholar]
- 11.Zhao J, Bian ZC, Yee K, Chen BP, Chien S, Guan JL. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Molecular cell. 2003 Jun;11(6):1503–15. doi: 10.1016/s1097-2765(03)00179-5. PubMed PMID: 12820964. [DOI] [PubMed] [Google Scholar]
- 12.Mehta TS, Lu H, Wang X, Urvalek AM, Nguyen KH, Monzur F, et al. A unique sequence in the N-terminal regulatory region controls the nuclear localization of KLF8 by cooperating with the C-terminal zinc-fingers. Cell research. 2009 Sep;19(9):1098–109. doi: 10.1038/cr.2009.64. PubMed PMID: 19488069. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Urvalek AM, Liu J, Zhao J. Activation of KLF8 transcription by focal adhesion kinase in human ovarian epithelial and cancer cells. The Journal of biological chemistry. 2008 May 16;283(20):13934–42. doi: 10.1074/jbc.M709300200. PubMed PMID: 18353772. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Oncogene. 2007 Jan 18;26(3):456–61. doi: 10.1038/sj.onc.1209796. PubMed PMID: 16832343. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Liu L, Wang Y, Zhao G, Xie R, Liu C, et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. Journal of cancer research and clinical oncology. 2013 Mar 16; doi: 10.1007/s00432-012-1363-3. PubMed PMID: 23504025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding SZ, Yang YX, Li XL, Michelli-Rivera A, Han SY, Wang L, et al. Epithelialmesenchymal transition during oncogenic transformation induced by hexavalent chromium involves reactive oxygen species-dependent mechanism in lung epithelial cells. Toxicology and applied pharmacology. 2013 May 15;269(1):61–71. doi: 10.1016/j.taap.2013.03.006. PubMed PMID: 23518002. Pubmed Central PMCID: 3664092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Wang X, Li T, Urvalek AM, Yu L, Li J, et al. Identification of poly (ADP-ribose) polymerase-1 (PARP-1) as a novel Kruppel-like factor 8-interacting and -regulating protein. The Journal of biological chemistry. 2011 Jun 10;286(23):20335–44. doi: 10.1074/jbc.M110.215632. PubMed PMID: 21518760. Pubmed Central PMCID: 3121510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudjonsson T, Ronnov-Jessen L, Villadsen R, Bissell MJ, Petersen OW. To create the correct microenvironment: three-dimensional heterotypic collagen assays for human breast epithelial morphogenesis and neoplasia. Methods. 2003 Jul;30(3):247–55. doi: 10.1016/s1046-2023(03)00031-8. PubMed PMID: 12798139. Pubmed Central PMCID: 2933212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen HL, Ronnov-Jessen L, Villadsen R, Petersen OW. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 2002 May;79(5):703–10. doi: 10.1006/geno.2002.6755. PubMed PMID: 11991720. [DOI] [PubMed] [Google Scholar]
- 20.de Neergaard M, Kim J, Villadsen R, Fridriksdottir AJ, Rank F, Timmermans-Wielenga V, et al. Epithelial-stromal interaction 1 (EPSTI1) substitutes for peritumoral fibroblasts in the tumor microenvironment. Am J Pathol. 2010 Mar;176(3):1229–40. doi: 10.2353/ajpath.2010.090648. PubMed PMID: 20133812. Pubmed Central PMCID: 2832145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jun 12;109(24):9623–8. doi: 10.1073/pnas.1121606109. PubMed PMID: 22623533. Epub 2012/05/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nature cell biology. 2012 Feb;14(2):117–23. doi: 10.1038/ncb2407. PubMed PMID: 22298039. [DOI] [PubMed] [Google Scholar]
- 23.Buess M, Rajski M, Vogel-Durrer BM, Herrmann R, Rochlitz C. Tumor-endothelial interaction links the CD44(+)/CD24(−) phenotype with poor prognosis in early-stage breast cancer. Neoplasia. 2009 Oct;11(10):987–1002. doi: 10.1593/neo.09670. PubMed PMID: 19794958. Pubmed Central PMCID: 2745665. Epub 2009/10/02. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moserle L, Indraccolo S, Ghisi M, Frasson C, Fortunato E, Canevari S, et al. The side population of ovarian cancer cells is a primary target of IFN-alpha antitumor effects. Cancer research. 2008 Jul 15;68(14):5658–68. doi: 10.1158/0008-5472.CAN-07-6341. PubMed PMID: 18632618. Epub 2008/07/18. eng. [DOI] [PubMed] [Google Scholar]
- 25.Yang T, Cai2 S, Zhang J, Lu J, Lin C, Zhai J, et al. Krüppel-Like Factor 8 Is a New Wnt/Beta-Catenin Signaling Target Gene and Regulator in Hepatocellular Carcinoma. PloS one. 2012;7(6) doi: 10.1371/journal.pone.0039668. Pubmed Central PMCID: 22761862. Epub 2012 Jun 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asai T, Tomita Y, Nakatsuka S, Hoshida Y, Myoui A, Yoshikawa H, et al. VCP (p97) regulates NFkappaB signaling pathway, which is important for metastasis of osteosarcoma cell line. Japanese journal of cancer research : Gann. 2002 Mar;93(3):296–304. doi: 10.1111/j.1349-7006.2002.tb02172.x. PubMed PMID: 11927012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zubair A, Frieri M. Role of nuclear factor-kB in breast and colorectal cancer. Current allergy and asthma reports. 2013 Feb;13(1):44–9. doi: 10.1007/s11882-012-0300-5. PubMed PMID: 22956391. [DOI] [PubMed] [Google Scholar]
- 28.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. European journal of nuclear medicine and molecular imaging. 2004 May;31(5):740–51. doi: 10.1007/s00259-003-1441-5. PubMed PMID: 15014901. eng. [DOI] [PubMed] [Google Scholar]
- 29.Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America. 2009 Jan 6;106(1):157–62. doi: 10.1073/pnas.0811426106. PubMed PMID: 19109433. Pubmed Central PMCID: 2629226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell stem cell. 2008 Sep 11;3(3):346–53. doi: 10.1016/j.stem.2008.08.014. PubMed PMID: 18786421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010 Feb;6(2):217–27. doi: 10.4161/auto.6.2.11014. PubMed PMID: 20104022. Pubmed Central PMCID: 2929010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. Journal of virology. 2003 Aug;77(16):8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. PubMed PMID: 12885912. Pubmed Central PMCID: 167245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. The Journal of cell biology. 1998 Dec 28;143(7):1997–2008. doi: 10.1083/jcb.143.7.1997. PubMed PMID: 9864370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Pestell R, Guan JL. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Molecular biology of the cell. 2001 Dec;12(12):4066–77. doi: 10.1091/mbc.12.12.4066. PubMed PMID: 11739801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Zheng C, Guan J. Pyk2 and FAK differentially regulate progression of the cell cycle. Journal of cell science. 2000 Sep;113(Pt 17):3063–72. doi: 10.1242/jcs.113.17.3063. PubMed PMID: 10934044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.