Abstract
Purpose
Tissue Phase Velocity Mapping (TPVM) is capable of reproducibly measuring regional myocardial velocities. However acquisition durations of navigator gated techniques are long and unpredictable while current breath-hold techniques have low temporal resolution. This study presents a spiral TPVM technique which acquires high resolution data within a clinically acceptable breath-hold duration.
Methods
Ten healthy volunteers are scanned using a spiral sequence with temporal resolution of 24ms and spatial resolution of 1.7x1.7mm. Retrospective cardiac gating is used to acquire data over the entire cardiac cycle. The acquisition is accelerated by factors of 2 and 3 by use of non-Cartesian SENSE implemented on the Gadgetron GPU system resulting in breath-holds of 17 and 13 heartbeats respectively. Systolic, early diastolic and atrial systolic global and regional peak and time-to-peak longitudinal, circumferential and radial velocities are determined.
Results
Global and regional peak and time-to-peak velocities agree well with those previously reported. The two acceleration factors show no significant differences for any quantitative parameter and the results also closely match previously acquired higher spatial resolution navigator-gated data in the same subjects.
Conclusion
By using spiral trajectories and non-Cartesian SENSE high resolution TPVM data can be acquired within a clinically acceptable breath-hold.
Introduction
Healthy functioning of the left ventricle involves complex motion and in order for the heart to pump effectively all areas of the myocardium must work efficiently. In many pathologies, early regional myocardial dysfunction may be compensated for by altered motion in other areas so that ventricular function is preserved. This can make the dysfunction hard to detect using the global parameters which are commonly used in clinical practice, until the disease has progressed to a stage where compensation is no longer possible (1). By measuring regional myocardial mechanics the health of the myocardium can be more directly assessed and disease can therefore potentially be identified earlier.
Within MR, there are several techniques which are capable of measuring regional myocardial mechanics (2). Of these, tissue phase velocity mapping (TPVM) offers potentially high spatial and temporal resolution and has the advantage of already being widely used clinically for blood velocity measurements. Peak and time-to-peak (TTP) velocity measurements made with TPVM have been shown to correlate well with measurements made with tissue Doppler imaging (TDI) (3) which can be limited by the absence of acoustic windows. Navigator gated sequences have allowed the acquisition of high spatial and high temporal resolution data but with long scan times. An acquisition with spatial and temporal resolutions of 2.6x1.3mm and 13.8ms (with view sharing), for example, requires a nominal acquisition time (assuming 100% respiratory efficiency) of 128 heartbeats per slice (4). Spiral k-space coverage has recently been used to greatly reduce durations, resulting in spatial and temporal resolutions of 1.4x1.4mm and 21ms in a nominal acquisition duration of just 53 heartbeats per slice (5). However, poor respiratory efficiency can cause the actual acquisition durations of navigator gated scans to be long and unpredictable in patients with irregular breathing patterns.
Breath-hold sequences generally have lower resolution than navigator gated sequences due to the restriction of having to be completed within an acceptable breath-hold time. This time is typically 20–30 seconds for healthy volunteers (6) but is lower for patients, particularly for the repeated breath-holds that are necessary for a full MR examination. Previous breath-hold TPVM sequences have been used to measure regional myocardial velocities in healthy volunteers but have had low temporal resolution (eg 2.7x1.3mm and 37–87ms using view sharing acquired in a 17–29 heartbeat breath-hold (7)). They have also been implemented with prospective cardiac gating which does not allow the entire cardiac cycle to be analysed.
Optimised GRAPPA (8) and k-t BLAST (9) have previously been applied to Cartesian TPVM to speed up acquisitions, however the inefficient k-space trajectories used in these studies meant that the resulting accelerated scan times were still too long for a breath-hold acquisition. While parallel imaging acceleration techniques such as SENSE (10) or GRAPPA (11) were originally developed for Cartesian imaging they have also been extended to allow use in combination with other, more efficient k-space trajectories such as spirals (12,13).
The aim of this work is to develop a high temporal and spatial resolution spiral TPVM sequence capable of measuring regional myocardial velocities within a breath-hold. The use of retrospective cardiac gating allows analysis of the entire cardiac cycle while an implementation of non-Cartesian SENSE on a Graphics Processing Unit (GPU) allows acquisition and reconstruction within a clinically acceptable time. Basal, mid and apical slices in ten healthy volunteers are scanned with acceleration factors of 2 and 3 (breath-hold durations of 17 and 13 heartbeats respectively) and quantitative regional and global parameters of motion are compared. The results are also compared to those obtained with a high resolution, navigator-gated sequence with known inter-study reproducibility previously acquired in the same volunteers (5).
Methods
A retrospectively gated phase velocity mapping sequence with three orthogonal directions of velocity encoding was developed based on a previous navigator-gated sequence (5) (see Figure 1). K-space was covered in 8 spiral interleaves, each of 14ms duration, leading to an acquired spatial resolution of 1.7x1.7mm and a temporal resolution of 24ms with a slice thickness of 8mm (the slice thickness was chosen as a playoff between signal to noise ratio and through plane resolution). Zero-filling was used to reconstruct at a pixel size of 0.85x0.85mm. Bipolar velocity encoding gradients prior to the spiral readouts resulted in velocity sensitivities of 30cm/s through-plane and 20cm/s in-plane. A black blood pulse of duration 6ms was output on every third sequence repeat to ensure good blood-myocardium contrast and to reduce any artefacts due to beat-to-beat variations in blood flow (14). A 1-1 binomial water excitation pulse (flip angle = 15 degrees) was implemented to reduce off-resonance blurring of fat.
Figure 1.
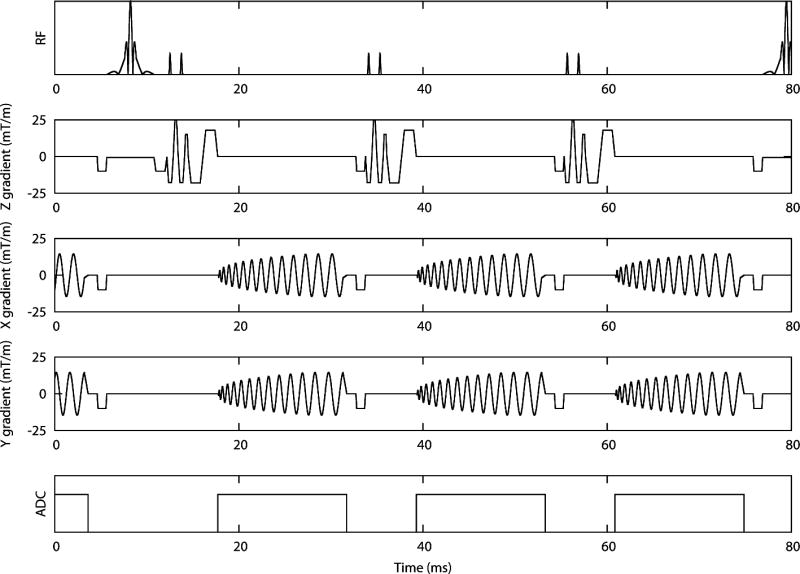
Sequence diagram for R2 and R3 with longitudinal velocity encoding. The different directions of encoding are acquired in separate heartbeats. The black blood pulse is played out on every third sequence repeat.
Reference and velocity encoded data were acquired in consecutive heartbeats leading to a scan time of 33 heartbeats for fully acquired data (including a single dummy cycle at the beginning of the acquisition, as explained below). In order to acquire the data in an acceptable breath-hold duration only a subset of the spirals was acquired. Full field of view images were then reconstructed using a non-Cartesian SENSE algorithm implemented on the Gadgetron GPU system (15). Data were acquired with an acceleration factor of 2 by acquiring 4 out of the 8 spiral interleaves (referred to as R2) and with an acceleration factor of 2.67 by acquiring 3 out of the 8 spiral interleaves (referred to as R3) leading to acquisition durations of 17 and 13 heartbeats per slice respectively. By using the same full FOV trajectories for R2 and R3 (rather than designing separate 4 and 3 interleave trajectories with reduced FOV), the effect of accelerating the acquisition is isolated. The dummy cycle was used to acquire low temporal resolution coil sensitivity maps by playing out all 8 spirals consecutively. These maps were then used in the GPU SENSE reconstruction (16).
The retrospective binning of data is carried out in the Siemens image calculation environment (ICE) and interpolates linearly between the acquired raw data. The data is binned so that 50 phases which are equally spaced through the cardiac cycle can be reconstructed. The binned data is then passed to the Gadgetron which runs the non-Cartesian SENSE algorithm to reconstruct the full images. The reconstructed images are then passed back to the Siemens system for display and analysis.
Ten healthy volunteers (mean age 32 years, range 25–57) were scanned on a clinical scanner (3 T, MAGNETOM Skyra, Siemens AG Healthcare Sector, Germany). The study was approved by an institutional review committee (East London REC 3). All subjects gave written informed consent.
6 elements of an anterior cardiac coil were used for all acquisitions. Second-order shimming and frequency adjustment based on the signal from a user-defined adjustment box situated over the whole heart was performed to reduce off-resonance artefacts. Phase velocity maps were acquired in the basal, mid and apical short axis planes (defined as 25%, 50% and 75% of the long axis length of the LV) with R2 and R3 sequence variations (SENSE acceleration factors of 2 and 2.67 respectively). Heart rates were noted for each acquisition. . In one volunteer a full stack of nine short-axis slices were also acquired as preliminary data to show a more comprehensive analysis of regional variations of motion through the left ventricle. All volunteers had previously been scanned with a high resolution, navigator-gated version of the sequence (referred to as HighRes) having spatial and temporal resolutions of 1.4 × 1.4mm and 21ms and a nominal acquisition duration 53 heartbeats per slice assuming 100% respiratory efficiency (5). The HighRes scans were part of a previous study performed several months before the R2 and R3 acquisitions were carried out. The scan parameters for R2/R3 and HighRes can be seen in Table 1.
Table 1.
Scan parameters for R2, R3 and HighRes
| Parameter | R2 | R3 | HighRes |
|---|---|---|---|
| Black blood | Every third repeat | Every third repeat | Every third repeat |
| Flip angle | 15° | 15° | 15° |
| FOV | 360mm | 360mm | 360mm |
| Velocity encoding | 30cm/s through plane 20cm/s in plane |
30cm/s through plane 20cm/s in plane |
30cm/s through plane 20cm/s in plane |
| Spiral interleaves | 8 | 8 | 13 |
| Acquired spirals | 1,3,5,7 | 1,4,7 | All |
| Acquired spatial resolution | 1.7 × 1.7mm | 1.7 × 1.7mm | 1.4 × 1.4mm |
| Acquired temporal resolution | 24ms | 24ms | 21ms |
| Reconstructed phases | 50 | 50 | 60 |
| Respiratory compensation | 17 heartbeat breath-hold | 13 heartbeat breath-hold | Navigator gated, 53 heartbeat nominal duration |
Background phase errors due to uncorrected eddy current effects or field inhomogeneities were determined for each acquired dataset by scanning a large homogeneous stationary phantom (31cm×22cm×20cm, gelatine with 5mM concentration of Gadolinium to shorten the T1) using the same sequence parameters and the same slice positions as in the volunteer study (17). A simulated ECG with the same heart-rate as the subject was used to trigger the acquisitions. The velocity maps from these phantom acquisitions were smoothed using a median filtering technique to reduce noise and then subtracted from the volunteer data on a pixel-by-pixel basis (17).
Post-processing
All images were analysed using custom software programmed using MATLAB (The Mathworks Inc, Natick MA, USA). The phantom velocity maps were subtracted from the volunteer maps to perform background correction. The velocities were then transformed into a cylindrical co-ordinate system composed of radial (motion towards the centre of the slice defined as positive), circumferential (clockwise motion as viewed from the apex defined as positive) and longitudinal (motion towards the apex defined as positive) components. Endo- and epicardial borders were manually segmented in each frame using a spline drawing algorithm (18).
Global peak and time to peak (TTP) velocities were calculated for the main peaks in the cardiac cycle in all three directions. In the longitudinal and radial directions there are three main peaks; the systolic peak (referred to as SL and SR for longitudinal and radial directions respectively), the early diastolic peak (DL and DR) and the atrial systolic peak (ASL and ASR). In the circumferential direction there are two systolic peaks (C1 and C2) and one main diastolic peak (C3). In order to reduce the effect of different heart-rates on TTP values the length of systole was measured and systole and diastole were separately normalized, as described previously (5). TTP values are therefore quoted in units of ‘normalised ms’.
Regional analysis of velocities was also performed by dividing the myocardium in each slice circumferentially into 24 regions. By plotting circumferential position along the y-axis against time on the x-axis colour coded regional velocity-time plots can be produced (5). Healthy motion plots were made by averaging the velocities over all volunteers; in order to stop small features of motion being blurred by averaging the cardiac cycle was split into systole, early diastole, diastasis and atrial systole and each section was normalised and averaged separately. Transmural velocities were also assessed and compared by splitting the myocardium into endocardial, mid-myocardial and epicardial layers, and averaging velocities in each layer.
The quantitative parameters obtained with R2, R3 and HighRes were compared with statistical tests carried out using SPSS (SPSS 19, SPSS, Chicago, Ill). After checking for normality using the Shapiro-Wilk test, repeated measures analysis of variance (with Greenhouse-Geisser correction in the case of non-sphericity) and paired t-testing (with Bonferroni correction for multiple testing) was performed to test for significant differences in each parameter.
Results
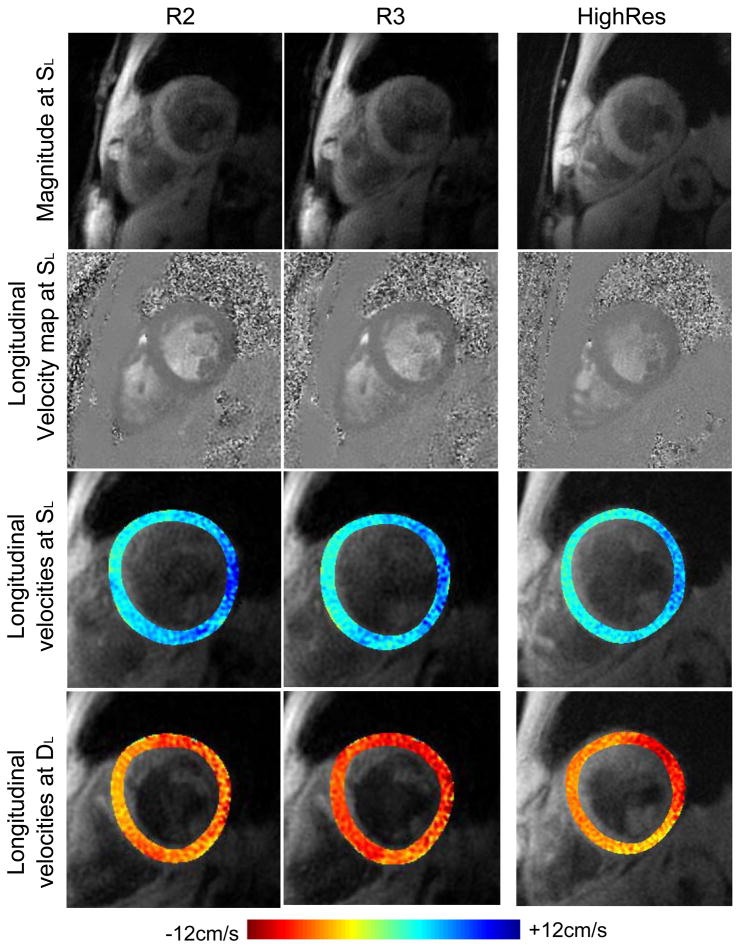
All scans were performed successfully with 3 slices acquired with both R2 and R3 in all ten volunteers. Example images and global velocity-time curves can be seen in Figures 2 and 3 respectively. Also shown in Figure 2 are colour coded overlays of the longitudinal velocities at the time of SL (third row) and DL (fourth row). Equivalent data previously acquired with the navigator-gated HighRes sequence are included for comparison and show similar image quality and features of motion. The suppression of blood and fat signals is good (while in this example blood suppression may appear to be better in R2 and R3 than HighRes this was not found to be the case in general). Average global velocities over the whole cycle were close to zero (R3 values, for example, were 0.22±0.49cm/s for longitudinal, 0.05±0.27cm/s for radial and −0.02±0.24 cm/s for circumferential velocities in the mid slice) indicating that velocity offsets caused by phase errors are effectively removed by the phantom subtraction. Mean RR duration for R2 acquisitions was 947±13ms, for R3 was 953±13ms and for previously acquired HighRes was 966±57ms.
Figure 2.
Top two rows: Example systolic magnitude images (taken at the time of the peak SL) and longitudinal velocity maps for R2 and R3 acquisitions plus HighRes for comparison. Blood suppression is good and off resonance artefacts have been minimised for both. The two acquisitions are of similar quality. Bottom two rows: Longitudinal velocities at the time of SL and DL are shown colour coded and overlaid on the magnitude images. Consistent regional variation is seen between the acquisitions.
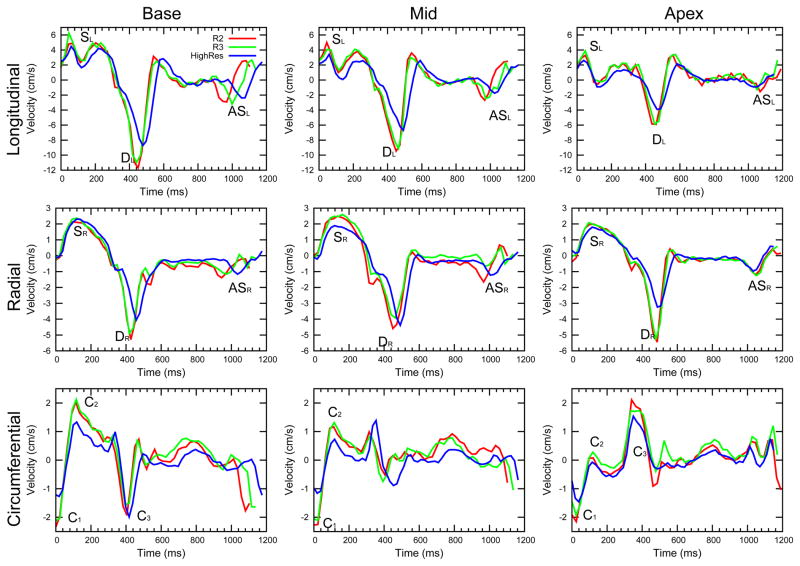
Figure 3.
Example longitudinal, radial and circumferential curves for R2 and R3 acquisitions for a single volunteer. Basal, mid and apical curves are shown, and in all plots the main peaks are labelled. The velocity-time curves from the two accelerated acquisitions show highly similar features. Also shown are the same plots from the previously acquired HighRes acquisitions in the same volunteer for comparison.
Global peak and TTP velocities
The main peaks in all three directions are labelled in Figure 3, and the mean (+/− standard deviation (SD)) peak velocity values can be found in Table 2, with TTP values in Table 3. The peak and TTP mean and SD are shown graphically in Figure 4. For comparison, Tables 2 and 3 and Figure 4 also include equivalent results from previously acquired acquisitions with the HighRes sequence in the same subject group. Paired analysis of R2 and R3 data show no significant differences for any of the peak or TTP values measured. Although some differences between HighRes and the other sequences can be seen in the example shown in Figure 3, average results for R2 and R3 are in good agreement with the previously acquired HighRes data apart from peak values for ASL and C1 which show small but statistically significant differences (see discussion). The remaining results presented in this section are derived from the R3 version of the sequence, since this is the most clinically applicable version (shortest breath-hold) and shows no significant differences from the longer breath-hold duration version, R2.
Table 2.
Average ± SD peak velocity values for R2 and R3 in the basal, mid and apical slices showing no statistically significant differences between the two acquisitions. HighRes values are also shown together with any statistically significant p values for comparisons with R2 and R3. A p value of < 0.05 is considered significant.
| Peak | R2 | R3 | R2vsR3 | High Res | HighResvsR2 | HighResvsR3 |
|---|---|---|---|---|---|---|
| BASE | ||||||
| SL (cm/s) | 8.00±1.91 | 7.63±1.25 | NS | 6.94±1.87 | NS | NS |
| DL(cm/s) | − 9.97±2.49 | − 10.11±2.27 | NS | − 9.45±1.97 | NS | NS |
| ASL(cm/s) | − 3.86±0.99 | −4.10±1.08 | NS | − 2.62±0.99 | 0.047 | 0.021 |
| SR(cm/s) | 2.54±0.32 | 2.34±0.24 | NS | 2.35± 0.28 | NS | NS |
| DR(cm/s) | − 4.01±1.44 | −4.69±0.84 | NS | −4.01 ±0.65 | NS | NS |
| ASR(cm/s) | − 1.41±0.68 | −1.64±0.46 | NS | −1.41 ±0.41 | NS | NS |
| C1(cm/s) | − 3.57±1.46 | −3.50±1.46 | NS | − 2.88±1.35 | 0.033 | 0.034 |
| C2(cm/s) | 2.74±1.13 | 2.48±1.10 | NS | 2.33±0.91 | NS | NS |
| C3(cm/s) | − 0.93±1.20 | −0.73±1.43 | NS | − 1.58±0.52 | NS | NS |
| MID | ||||||
| SL (cm/s) | 6.79±1.35 | 6.53±1.61 | NS | 5.87±1.89 | NS | NS |
| DL(cm/s) | − 7.24±1.66 | −7.46±1.55 | NS | − 6.50±1.85 | NS | NS |
| ASL(cm/s) | − 3.05±1.08 | −2.83±1.30 | NS | − 1.66±0.67 | 0.024 | NS |
| SR(cm/s) | 2.62±0.31 | 2.48±0.70 | NS | 2.38±0.38 | NS | NS |
| DR(cm/s) | − 4.55±0.72 | −4.52±0.84 | NS | − 3.61±0.77 | NS | NS |
| ASR(cm/s) | − 1.94±0.66 | −1.91±0.84 | NS | − 1.59±0.43 | NS | NS |
| C1(cm/s) | − 4.14±1.48 | −4.07±1.27 | NS | − 3.52±1.50 | 0.002 | 0.006 |
| C2(cm/s) | 1.59±0.87 | 1.48±1.07 | NS | 1.40±0.83 | NS | NS |
| APEX | ||||||
| SL (cm/s) | 5.31±1.06 | 5.34±1.06 | NS | 4.83±1.70 | NS | NS |
| DL(cm/s) | − 4.38±1.46 | −4.50±1.27 | NS | − 4.27±1.67 | NS | NS |
| ASL(cm/s) | − 1.63±0.78 | −1.83±0.72 | NS | − 0.81±0.29 * | 0.002 | 0.006 |
| SR(cm/s) | 2.24±0.19 | 2.24±0.24 | NS | 2.14±0.27 | NS | NS |
| DR(cm/s) | − 4.58±0.88 | −4.70±0.70 | NS | − 3.99±0.94 | NS | NS |
| ASR(cm/s) | − 1.64±0.68 | −1.45±1.27 | NS | − 1.57±0.45 | NS | NS |
| C1(cm/s) | − 4.01±1.09 | −4.09±1.10 | NS | − 3.50±1.23 | 0.027 | 0.027 |
| C2(cm/s) | 0.31±0.68 | 0.39±0.68 | NS | 0.39±0.69 | NS | NS |
| C3(cm/s) | 2.12±0.42 | 2.09±0.56 | NS | 2.09±0.72 | NS | NS |
Table 3.
Average ± SD TTP velocity values for R2 and R3 in the basal, mid and apical slices showing no statistically significant differences between the two acquisitions. HighRes values are also shown together with any statistically significant p values for comparisons with R2 and R3. A p value of < 0.05 is considered significant.
| Peak | R2 | R3 | R2vsR3 | High Res | HRvsR2 | HRvsR3 |
|---|---|---|---|---|---|---|
| BASE | ||||||
| SL (ms) | 58.4±14.1 | 52.4±14.6 | NS | 60.6±6.7 | NS | NS |
| DL(ms) | 470.9±23.7 | 470.7±29.3 | NS | 467±26 | NS | NS |
| ASL(ms) | 851.4±33.9 | 847.3±41.5 | NS | 864.8±34.5 | 0.02 | 0.02 |
| SR(ms) | 114.8±19.6 | 121.9±26.4 | NS | 120.4±15.8 | NS | NS |
| DR(ms) | 472.0±49.4 | 452.7±28.4 | NS | 456.0±18.2 | NS | NS |
| ASR(ms) | 854.9±50.7 | 839.2±34.5 | NS | 857.0±46.2 | NS | NS |
| C1(ms) | 40.9±19.7 | 35.8±20.1 | NS | 44.5±9.1 | NS | NS |
| C2(ms) | 149.7±20.3 | 146.8±20.4 | NS | 140.7±20.0 | NS | NS |
| C3(ms) | 436.0±55.5 | 435.3±60.0 | NS | 410.5±16.3 | NS | NS |
| MID | ||||||
| SL (ms) | 54.8±8.9 | 51.1±10.6 | NS | 59.9±6.3 | NS | NS |
| DL(ms) | 475.5±27.7 | 473.9±36.8 | NS | 483.3±24.1 | NS | NS |
| ASL(ms) | 853.4±30.0 | 860.4±36.8 | NS | 869.4±30.6 | NS | NS |
| SR(ms) | 116.2±8.9 | 122.0±30.3 | NS | 123.6±16.1 | NS | NS |
| DR(ms) | 481.8±31.2 | 473.7±33.0 | NS | 487.2±20.2 | NS | NS |
| ASR(ms) | 852.6±33.2 | 862.4±38.7 | NS | 875.9±29.9 | NS | NS |
| C1(ms) | 37.2±17.2 | 39.0±16.6 | NS | 48.7±9.8 | 0.02 | NS |
| C2(ms) | 144.8±23.3 | 143.5±25.2 | NS | 147.4±21.4 | NS | NS |
| APEX | ||||||
| SL (ms) | 45.6±9.4 | 46.7±10.3 | NS | 51.5±7.7 | NS | NS |
| DL(ms) | 448.9±30.9 | 453.8±48.0 | NS | 449.5±41.0 | NS | NS |
| ASL(ms) | 853.9±44.5 | 835.9±58.3 | NS | 859.0±30.6 | NS | NS |
| SR(ms) | 114.2±20.3 | 96.6±20.5 | NS | 114.8±22.1 | NS | NS |
| DR(ms) | 496.5±41.4 | 488.2±45.7 | NS | 482.6±30.6 | NS | NS |
| ASR(ms) | 864.8±30.8 | 858.7±30.9 | NS | 880.4±28.6 | NS | 0.01 |
| C1(ms) | 38.8±15.3 | 38.6±12.9 | NS | 48.3±6.3 | NS | NS |
| C2(ms) | 145.5±25.2 | 140.8±17.0 | NS | 141.8±41.6 | NS | NS |
| C3(ms) | 407.1±83.1 | 392.6±63.2 | NS | 359.1±21.5 | NS | NS |
Figure 4.
Average ± SD peak and TTP values for the main longitudinal, radial and circumferential peaks for R2 and R3 acquisitions in the basal, mid and apical slices. Note the different time scale for the circumferential direction. Also shown are the same plots for the previously acquired HighRes acquisitions for comparison.
Global velocity-time curves show the same patterns of motion that have been seen previously (5),(19); longitudinal velocities (see Table 2 and Figure 4) are significantly higher at base than mid levels and are higher at mid than apical levels (for example DL values are −10.11±2.27cm/s at base, −7.46±1.55cm/s at mid and −4.50±1.27cm/s at apex, p<0.01 for both, with significant differences between all levels also being present for SL and ASL, p<0.02 for all). As found previously, radial velocities do not show significant differences between slices, however, SR occurs earlier at apex than base (p=0.011) and DR occurs earlier at base than apex (p=0.011). SR is less sharp than SL (see Figure 3) and occurs later in the cardiac cycle. Circumferential velocities show many small peaks through the cardiac cycle, the timings of which are variable between subjects. However all subjects show the early systolic peaks C1 and C2 indicating a quick reversal of circumferential direction as the ventricle contracts. Basal and apical slices rotate in opposite directions during early diastole, shown by the opposite polarities of C3.
Regional variation
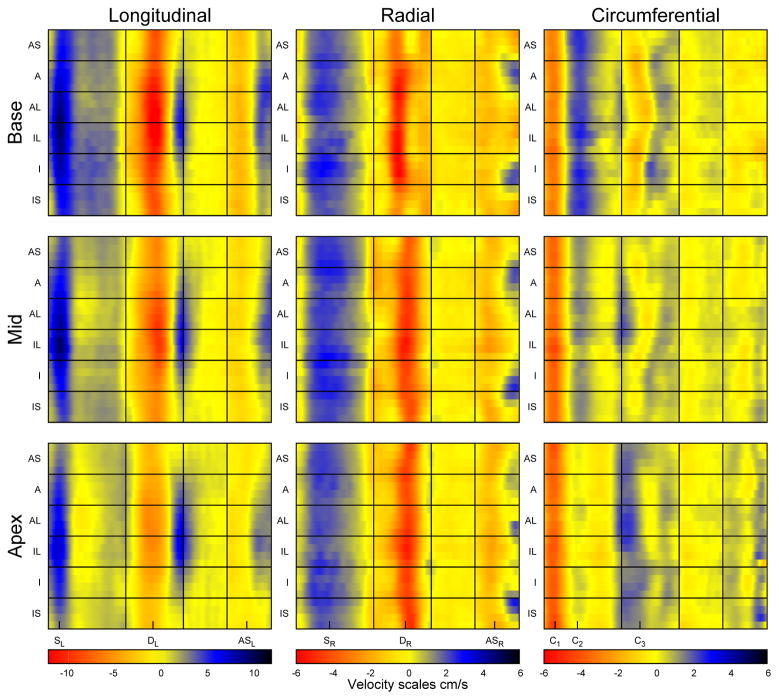
The regional variation of myocardial motion is complicated and best described graphically. Colour plots of the velocities over the cardiac cycle (5) averaged over all volunteers can be seen in Figure 5. Plots for three individual volunteers are also included as Supplemental Figures. Vertical lines indicate the four portions of the cardiac cycle that were used to time-normalise velocities before averaging. They also split the cycle into four sections (systole, early diastole, diastasis and atrial systole) that make reference easier. Each direction of motion has a colour scale which is consistent between slices, although the scale for each direction is different and designed to cover the range of velocities in that direction without masking fine features of motion.
Figure 5.
Colour plots showing regional variation of myocardial velocities averaged over the R3 acquisitions in all volunteers. Note the different scales for each direction of velocity. The main peaks used for characterising global velocities are indicated below the plots. (AS = anteroseptum, A = anterior, AL = anterolateral, IL = inferolateral, I = inferior, IS = inferoseptal).
Many interesting features of motion can be seen in the plots which concisely display the regional motion of the myocardium over the entire cardiac cycle and allow quick interpretation (5). For example, in the longitudinal direction it is immediately obvious that septal velocities are lower than lateral velocities, whereas this is not the case for radial velocities. Interesting regional variation can be seen in the circumferential direction, particularly during early diastole. Peak and TTP velocities for the mid slice transmural regions can be found in Table 4. In the radial direction endocardial velocities are significantly higher than mid-myocardial velocities, which are in turn significantly higher than epicardial velocities for all three peaks (p<0.05). C1 shows no transmural gradient, however C2 peak velocities are higher at epicardium than epicardium. No differences are seen transmurally in the longitudinal direction.
Table 4.
Peak velocity values for the different transmural regions in the mid slice for R3 acquisitions. Also shown are any significant differences between regions.
| Peak velocity(cm/s) | Endocardium | Mid | Epicardium | p(Endo-Mid) | p(Mid-Epi) | p(Endo-Epi) |
|---|---|---|---|---|---|---|
| SR | 3.03±0.32 | 2.50±0.23 | 2.06±0.18 | 0.00 | 0.00 | 0.00 |
| DR | −5.28±0.77 | −4.59±0.70 | −3.89±0.65 | 0.00 | 0.00 | 0.00 |
| ASR | −2.11±0.94 | −1.93±0.85 | −1.73±0.76 | 0.02 | 0.01 | 0.01 |
| SL | 6.40±1.56 | 6.66±1.67 | 6.52±1.61 | 0.02 | NS | NS |
| DL | −7.32±1.60 | −7.56±1.56 | −7.50±1.57 | NS | NS | NS |
| ASL | −2.76±1.21 | −2.88±1.38 | −2.86±1.32 | NS | NS | NS |
| C1 | 3.86±1.11 | 4.16±1.30 | 4.16±1.39 | 0.02 | NS | NS |
| C2 | −1.24±1.08 | −1.47±1.07 | −1.71±1.07 | 0.00 | 0.00 | 0.00 |
Stack of slices
The global velocity curves for the stack of slices acquired in a single volunteer can be seen in Figure 6. The entire acquisition (including reconstruction time) took less than thirty minutes. In the longitudinal curves, SL and DL both decrease in magnitude steadily from base to apex. This gradient is not seen in the radial peaks, however the timing of DR is progressively later when moving from base to apex (as seen earlier in the global TTP values averaged over all volunteers). In the circumferential direction C1 is always negative and approximately the same magnitude at all levels. However, C2 is positive at base but gradually decreases towards the apex. C3 does the opposite, starting negative at base and ending up positive (but with a similar magnitude) at apex.
Figure 6.
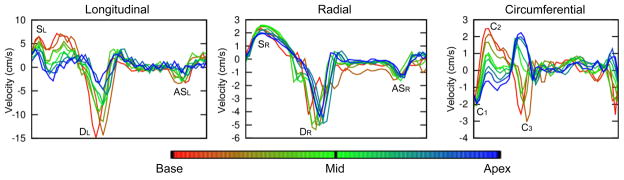
Global longitudinal, radial and circumferential velocity-time curves for a full short axis stack of slices acquired in a single volunteer.
Discussion
The combination of efficient spiral trajectories and non-Cartesian SENSE reconstruction has allowed the acquisition of TPVM data within a clinically acceptable breath-hold duration (13 cardiac cycles) and with higher temporal resolution than has previously been possible. As shown in Figure 2, image quality for R2 and R3 is high, and is similar to HighRes which uses shorter spiral interleaves and has higher spatial resolution, but which takes much longer to acquire, requiring the use of respiratory navigators. The use of retrospective cardiac gating has allowed analysis of the entire cardiac cycle including atrial systole. The loss of SNR which is expected at increasing levels of acceleration can be seen in Figure 2, however, comparisons between R2 and R3 show that, at this level of acceleration, the non-Cartesian SENSE reconstruction is not affecting quantitative measurements. In addition, comparing the results with those obtained from a previously developed, higher resolution navigator-gated sequence with known inter-study reproducibility shows that the resolution is still high enough to make accurate measurements of global and regional peak and TTP velocities.
Acquired spatial and temporal resolution (1.7x1.7mm and 24ms) are better than previously reported Cartesian breath-hold sequences (for example 2.7x1.3mm and 37–87ms resolution acquired in 17–29 heartbeats (7)) allowing the detection of fine features of motion which previously could not be detected within a breath-hold. However the use of long readout times could be limiting the resolution achieved in vivo (see below). Coverage of the entire cardiac cycle has recently been achieved with a self navigated TPVM sequence using golden angle spirals which allows the reconstruction of real time images from data acquired during free breathing (20). Despite the advantages gained by using real time imaging the acquisition and reconstruction times are currently long (about 8.7 minutes and 1 hour per slice respectively) compared with R3 which is acquired in 13 seconds and reconstructed at the time of imaging within one minute and twenty seconds.
R2 and R3 show no differences in measured peak or TTP values indicating that the level of acceleration is not affecting the accuracy of velocity measurements. This agrees with previous studies using Cartesian imaging and other acceleration schemes with similar acceleration factors (8,9) which have found that accelerated TPVM acquisitions give results that are comparable to those found from unaccelerated acquisitions.
While no significant differences were found between R2 and R3 for any peak or TTP values, the R2 and R3 scans show a tendency towards measuring higher peak velocity values than HighRes which is unexpected since the resolution is lower. This is possibly due to the fact that the HighRes acquisitions are much longer and so will be more affected by natural heart rate variation. Small but statistically significant differences between R2/R3 and HighRes were found for C1 and for ASL in all three slices. C1 occurs at the very beginning of the cardiac cycle, which is where the crossed-pairs navigator is being acquired in the HighRes sequence. The navigator sequence has been specifically adapted to make it as short as possible (just 9ms plus 10ms feedback time), however this interruption in the data acquisition may affect the accuracy of the retrospective reconstruction interpolation at this time and could explain the small differences seen. The effect could be compounded by the fact that a different temporal resolution was used for R2/R3 than was used for HighRes (acquired temporal resolution was 24ms for R2/R3 and 21ms for HighRes, 50 phases were reconstructed for R2/R3 and 60 for HighRes). The small but significant difference seen in ASL values may be due to the fact that the longitudinal peak in atrial systole is an inconsistent feature of motion between volunteers (as shown by the high standard deviation relative to the mean peak size, see Table 2) and is also less reproducible within volunteers as shown by the previous HighRes study (5). Differences may also be caused by differences in slice position. While R2 and R3 scans were performed within the same study (and are consequently well co-registered), the HighRes acquisitions were performed on a different day and some differences in positioning would be expected. However, the fact that the majority of the measured parameters show excellent agreement with different sequences and on different days indicates that TPVM has very good interstudy reproducibility.
Peak and TTP velocity values from the R3 sequence show the same patterns as have previously been shown with TPVM sequences; a significant gradient in peak longitudinal velocities is seen from base to apex, but not in radial velocities, while the transmural gradient is seen in radial but not longitudinal velocities. Normalising the TTP to take into account variations in the length of systole and diastole allows easier comparison between subjects (5). The colour plots provide a way to quickly interpret the motion patterns of the myocardium and the application to patient data would potentially allow regional wall motion abnormalities to be easily seen.
Spiral images require more complicated reconstruction than Cartesian trajectories and this often does not allow images to be reconstructed online at the scanner and seen immediately. However the use of the Gadgetron GPU system has meant that reconstruction time is not a limiting factor. The 50 phases are reconstructed in approximately one minute and twenty seconds on current hardware (HP Z800 Workstation with 4GB Asus GeForce GTX 670 graphics card) which does not delay the scan process significantly. This also allowed the acquisition of a full stack of short axis slices showing features of regional variation that were previously only seen in full 3D acquisitions (21). The reconstruction time could be further reduced by the addition of further inexpensive GPUs.
The spiral interleaves have a 14ms duration which could lead to off-resonance blurring in vivo thereby degrading the resolution of the images to below the 1.7x1.7 mm specified by the spiral trajectory design. Future work will include further optimising the spiral trajectories to reduce this effect (by reducing the spiral durations). A design that uses shorter spiral interleaves to acquire lower nominal spatial resolution could achieve comparable image quality due to the reduced level of off resonance blurring, while further increasing temporal resolution or reducing scan time.
The level of acceleration could be further increased by exploiting temporal correlations in the data using a TSENSE reconstruction (22) (having more up to date coil sensitivity maps could also improve the images). However the need to switch spiral interleaves within each heartbeat could potentially cause additional phase errors. This will be investigated as future work, as higher acceleration factors could also allow the use of much shorter spirals, decreasing the sensitivity of the sequence to off resonance artefacts which currently could be limiting the resolution as discussed above.
Currently a large slice thickness (8mm) is used. As the technique moves towards coverage of the entire ventricle (as shown in the preliminary stack-of-slices data) it may be necessary to use a thinner slice to reduce partial volume effects, particularly near the apex.
A limitation of this study in terms of clinical applicability is that a separate phantom acquisition is used to remove background phase errors. However the use of a correction based on static tissue has previously been used with TPVM (9) and would remove this inconvenience, although the accuracy of the correction is likely to be lower than with a phantom subtraction (23). The time consuming manual segmentation of the images is another limitation to the technique. Future work will implement semi-automatic segmentation algorithms to improve this.
The accelerated sequence that has been produced allows the acquisition of high spatial and temporal resolution data with three directions of velocity encoding in a single, clinically acceptable breath-hold. This has been achieved without compromising the accuracy of measurements when compared with high resolution, navigator gated acquisitions, as shown by quantitative measurements of peak and TTP velocities. The GPU reconstruction allows the images to be seen online without extending the scan time and hence the technique is now suitable for inclusion within a clinical workflow.
Supplementary Material
Supplemental Figure 1. Longitudinal colour plots for three of the volunteers. The same velocity scale is used for all volunteers and for all slices.
Supplemental Figure 2. Radial colour plots for three of the volunteers. The same velocity scale is used for all volunteers and for all slices.
Supplemental Figure 3. Circumferential colour plots for three of the volunteers. The same velocity scale is used for all volunteers and for all slices.
References
- 1.Karamitsos TD, Francis JM, Myerson S, Selvanayagam JB, Neubauer S. The role of cardiovascular magnetic resonance imaging in heart failure. J Am Coll Cardiol. 2009;54(15):1407–1424. doi: 10.1016/j.jacc.2009.04.094. [DOI] [PubMed] [Google Scholar]
- 2.Simpson RM, Keegan J, Firmin DN. MR assessment of regional myocardial mechanics. J Magn Reson Imaging. 2013;37(3):576–599. doi: 10.1002/jmri.23756. [DOI] [PubMed] [Google Scholar]
- 3.Delfino JG, Bhasin M, Cole R, et al. Comparison of myocardial velocities obtained with magnetic resonance phase velocity mapping and tissue Doppler imaging in normal subjects and patients with left ventricular dyssynchrony. J Magn Reson Imaging. 2006;24(2):304–311. doi: 10.1002/jmri.20641. [DOI] [PubMed] [Google Scholar]
- 4.Jung B, Zaitsev M, Hennig J, Markl M. Navigator gated high temporal resolution tissue phase mapping of myocardial motion. Magn Reson Med. 2006;55(4):937–942. doi: 10.1002/mrm.20808. [DOI] [PubMed] [Google Scholar]
- 5.Simpson R, Keegan J, Firmin D. Efficient and reproducible high resolution spiral myocardial phase velocity mapping of the entire cardiac cycle. J Cardiovasc Magn Reson. 2013;15(1):34. doi: 10.1186/1532-429X-15-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott AD, Keegan J, Firmin DN. Motion in cardiovascular MR imaging. Radiology. 2009;250(2):331–351. doi: 10.1148/radiol.2502071998. [DOI] [PubMed] [Google Scholar]
- 7.Petersen SE, Jung BA, Wiesmann F, et al. Myocardial tissue phase mapping with cine phase-contrast mr imaging: regional wall motion analysis in healthy volunteers. Radiology. 2006;238(3):816–826. doi: 10.1148/radiol.2383041992. [DOI] [PubMed] [Google Scholar]
- 8.Peng HH, Bauer S, Huang TY, et al. Optimized parallel imaging for dynamic PC-MRI with multidirectional velocity encoding. Magn Reson Med. 2010;64(2):472–480. doi: 10.1002/mrm.22432. [DOI] [PubMed] [Google Scholar]
- 9.Lutz A, Bornstedt A, Manzke R, Etyngier P, Nienhaus GU, Rasche V. Acceleration of tissue phase mapping by k-t BLAST: a detailed analysis of the influence of k-t-BLAST for the quantification of myocardial motion at 3T. J Cardiovasc Magn Reson. 2011;13:5. doi: 10.1186/1532-429X-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 11.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 12.Pruessmann KP, Weiger M, Bornert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Magn Reson Med. 2001;46(4):638–651. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 13.Heidemann RM, Griswold MA, Seiberlich N, et al. Direct parallel image reconstructions for spiral trajectories using GRAPPA. Magn Reson Med. 2006;56(2):317–326. doi: 10.1002/mrm.20951. [DOI] [PubMed] [Google Scholar]
- 14.Drangova M, Zhu Y, Pelc NJ. Effect of artifacts due to flowing blood on the reproducibility of phase-contrast measurements of myocardial motion. J Magn Reson Imaging. 1997;7(4):664–668. doi: 10.1002/jmri.1880070409. [DOI] [PubMed] [Google Scholar]
- 15.Hansen MS, Sorensen TS. Gadgetron: An open source framework for medical image reconstruction. Magn Reson Med. 2012;68:741–750. doi: 10.1002/mrm.24389. [DOI] [PubMed] [Google Scholar]
- 16.Sorensen TS, Atkinson D, Schaeffter T, Hansen MS. Real-time reconstruction of sensitivity encoded radial magnetic resonance imaging using a graphics processing unit. IEEE Trans Med Imaging. 2009;28(12):1974–1985. doi: 10.1109/TMI.2009.2027118. [DOI] [PubMed] [Google Scholar]
- 17.Caprihan A, Altobelli SA, Benitez-Read E. Flow-Velocity Imaging from Linear Regression of Phase Images with Techniques for Reducing Eddy-Current Effects. J Magn Reson. 1990;90:19. [Google Scholar]
- 18.Lee ETY. Choosing nodes in parametric curve interpolation. Computer-Aided Design. 1989;21(6):7. [Google Scholar]
- 19.Jung B, Föll D, Böttler P, Petersen S, Hennig J, Markl M. Detailed analysis of myocardial motion in volunteers and patients using high-temporal-resolution MR tissue phase mapping. J Magn Reson Imaging. 2006;24(5):1033–1039. doi: 10.1002/jmri.20703. [DOI] [PubMed] [Google Scholar]
- 20.Steeden JA, Knight DS, Bali S, Atkinson D, Taylor AM, Muthurangu V. Self-navigated tissue phase mapping using a golden-angle spiral acquisition-proof of concept in patients with pulmonary hypertension. Magn Reson Med. 2013 doi: 10.1002/mrm.24646. [DOI] [PubMed] [Google Scholar]
- 21.Lutz A, Paul J, Bornstedt A, et al. Volumetric motion quantification by 3D tissue phase mapped CMR. J Cardiovasc Magn Reson. 2012;14:74. doi: 10.1186/1532-429X-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellman P, Epstein FH, McVeigh ER. Adaptive sensitivity encoding incorporating temporal filtering (TSENSE) Magn Reson Med. 2001;45(5):846–52. doi: 10.1002/mrm.1113. [DOI] [PubMed] [Google Scholar]
- 23.Gatehouse PD, Rolf MP, Graves MJ, et al. Flow measurement by cardiovascular magnetic resonance: a multi-centre multi-vendor study of background phase offset errors that can compromise the accuracy of derived regurgitant or shunt flow measurements. J Cardiovasc Magn Reson. 2010;12:5. doi: 10.1186/1532-429X-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Longitudinal colour plots for three of the volunteers. The same velocity scale is used for all volunteers and for all slices.
Supplemental Figure 2. Radial colour plots for three of the volunteers. The same velocity scale is used for all volunteers and for all slices.
Supplemental Figure 3. Circumferential colour plots for three of the volunteers. The same velocity scale is used for all volunteers and for all slices.