Abstract
Networks are increasingly used to study the impact of drugs at the systems level. From the algorithmic standpoint, a drug can ‘attack’ nodes or edges of a protein-protein interaction network. In this work, we propose a new network strategy, “The Interface Attack”, based on protein-protein interfaces. Similar interface architectures can occur between unrelated proteins. Consequently, in principle, a drug that binds to one has a certain probability of binding others. The interface attack strategy simultaneously removes from the network all interactions that consist of similar interface motifs. This strategy is inspired by network pharmacology and allows inferring potential off-targets. We introduce a network model which we call “Protein Interface and Interaction Network (P2IN)”, which is the integration of protein-protein interface structures and protein interaction networks. This interface-based network organization clarifies which protein pairs have structurally similar interfaces, and which proteins may compete to bind the same surface region. We built the P2IN of p53 signaling network and performed network robustness analysis. We show that (1) ‘hitting’ frequent interfaces (a set of edges distributed around the network) might be as destructive as eleminating high degree proteins (hub nodes); (2) frequent interfaces are not always topologically critical elements in the network; and (3) interface attack may reveal functional changes in the system better than attack of single proteins. In the off-target detection case study, we found that drugs blocking the interface between CDK6 and CDKN2D may also affect the interaction between CDK4 and CDKN2D.
INTRODUCTION
Currently, the “one drug one target” approach is often considered an exception rather than the rule 1. With a few exceptions, there is no one-to-one mapping between a gene (and the protein it encodes) and a disease state, and disease processes often involve crosstalk between proteins in different pathways. This is mainly because a protein can have many functions, and many proteins can have a similar function. Drugs are often multi-targeted and a recent study suggested that the average number of target proteins per drug is 6.3 2. Off-targets are the proteins that a drug can bind other than its primary target. Cohen et al.3 note that cancer drug therapy might change the behavior of nearly 1000 different proteins, suggesting that the disturbance of a signaling network through a single protein may affect other proteins. Recently, system-wide approaches are increasingly being considered 4.
Polypharmacology is a new concept in drug discovery that seeks drugs acting on multiple targets. One of the aims of polypharmacology is to discover multi-target drugs that will disturb disease-associated networks 1, similar to aspirin, Metformin, nonsteroidal anti-inflammatory drugs (NSAID's), and Gleevec. Combination therapy showed success in diseases such as AIDS, atherosclerosis, cancer and depression. Although attacking more than one ‘strategic’ point of the system might be a useful approach, multi-target drugs also have increased toxicity. Putative off-targets have been found through different computational methods, such as docking 5,6, pharmacophore mapping 7, ligand structure similarity 8,9, side effect similarity 10,11, ligand binding site similarity 12-17, text mining 18 and integrated methods 19-21. Proteins with similar binding sites often recognize similar ligands 21,22. Earlier works suggested that even weak binding to multiple targets may have profound effects on the biological system23-25. Eventually, all structure-based drug discovery strategies including those targeting protein-protein interactions (PPIs) 26 should consider protein flexibility on the atomistic level 27,28.
The interface attack strategy proposed in this work focuses on protein-protein interface motifs. Currently protein–protein interfaces are increasingly becoming targets in drug discovery 29,30, and it was suggested that the high flexibility of monomers may lead to overlooking small highly populated pockets that may occur when in the complex form 30. Finding small-molecule drugs that hit protein–protein interactions is still highly challenging 31-35. Although generally interfaces of PPIs (~1500 - 3000 Å2) are larger than protein-small molecule interactions (~300 - 1000 Å2), an optimized small molecule may bind with an affinity comparable to that of the native partner protein or peptide 32.
Our interface attack is inspired by interface motifs and by multi-target drugs. Since drugs may disrupt protein interactions which have structurally similar interfaces, we aim to develop a strategy which may take a first step toward prediction of the outcome of disabling a set of structurally similar interactions in protein-protein interaction networks (PINs). Our study is the first to target interfaces in a network attack. A few successful PPI drugs on the market 34 such as tirofiban targeting the integrins (cardiovascular conditions) 36; and maraviroc targeting CCR5–gp120 interactions (HIV) 37, and several new drugs entering Phase II clinical trials 38, suggest that protein interfaces can be druggable.
Biological systems are robust to damage of their components. In the case of the protein-protein interaction network, the topology of the network appears responsible for its robustness 39. The scale-free network characteristics make it resilient to accidental failures: even if 80% of randomly selected proteins fail, the remaining 20% still continue to stay as a compact cluster 40. However, protein networks are vulnerable to systematic attacks on highly connected proteins. Experimental studies indicate that these proteins are also essential41. Integrating structural knowledge can help the abstract protein interaction networks: knowledge of protein interfaces helps not only in understanding the relations among proteins and thus function, but also in identifying drug targets, in inhibitor design and off-targets prediction.
Beyond the introduction of strategy of interface attack, this article presents a new network model, which we name “Protein Interface and Interaction Network (P2IN)”. Similar network models were used by our group previously to analyze interface properties of cancer-related proteins 42 and topological properties of hubs 43. This new model introduces structural information into PINs. All network nodes have 3D interface structures. This representation illustrates which proteins may compete for the same binding site on a protein, and all protein pairs with structurally similar interface topologies.
Here our goal is not to design drugs; instead, we aim to introduce an interface motif-based first step in a systems strategy that seeks to identify potential side effects. Eventually, to more reliably predict drug binding, comprehensive long time scales simulations are essential. Introducing atomistic flexibility into systems-level studies is a challenging aim, beyond current computational feasibility. An alternative could be selection of few likely targets obtained by a strategy such as the one proposed here, and subjecting them to such detailed analysis.
This paper is organized as follows: the Concepts section provides an overview of the notion of similar structural motifs in interfaces, the new network model (P2IN) and the interface attack strategy. The Results and Discussions section consists of four case studies: the first is the P2IN model applied to the p53 signaling pathway, the second relates to off-target prediction and the third case study focuses on comparison of node and interface attacks from a biological standpoint. These case studies are followed by the robustness measures used in analyzing the response of the p53 P2IN to attack strategies. Lastly, the case study on comparison of network attack strategies is presented.
CONCEPTS
The novelty of our strategy comes from the concept of similar interfaces. Similar interfaces can occur between unrelated proteins, and this is a common occurrence44,45. Consequently, a drug that binds to one has a certain probability of binding others. Our network attack strategy is inspired by multi-target drugs, and makes use of structurally similar interfaces. In this section we give a brief overview of this similar-interfaces-different-global-structures paradigm. We describe how we find similar interfaces on a network of protein interactions; introduce the structural protein-protein interaction network model; explain what a network attack is; and define interface attack, which is the focus of this study.
PRISM May Discover Similar interfaces
The 3D structures of the protein-protein complexes and their interfaces are obtained through the application of the Protein Interactions by Structural Matching (PRISM) method. PRISM aims to predict protein-protein interactions and protein interfaces by identifying structural similarities between protein surfaces and known interfaces which are derived from the Protein Databank (PDB) 46. The algorithm has four steps. First, the surfaces of all target proteins are extracted. Second, using the MultiProt engine47, the surfaces of the target proteins are structurally aligned with known interfaces (templates) obtained from the PDB. In this step PRISM checks whether any surface region of the monomers is structurally similar to one of the complementary chains of the template interfaces, disregarding the order of the residues in the protein chain. Third, it places the two chains that are structurally similar to the template interface onto the template complex. This leads to a putative complex. The fourth step involves flexible refinement of the putative complexes by FiberDock 48,49. This resolves steric clashes and ranks the predicted protein complexes by their energies. Combining geometric complementarity with docking tools makes the prediction more physical. The prediction performance of PRISM algorithm was recently analyzed on standard docking benchmarks, and found to be comparable to other rigid docking strategies, however considerably more efficient (see Tuncbag et al.50).
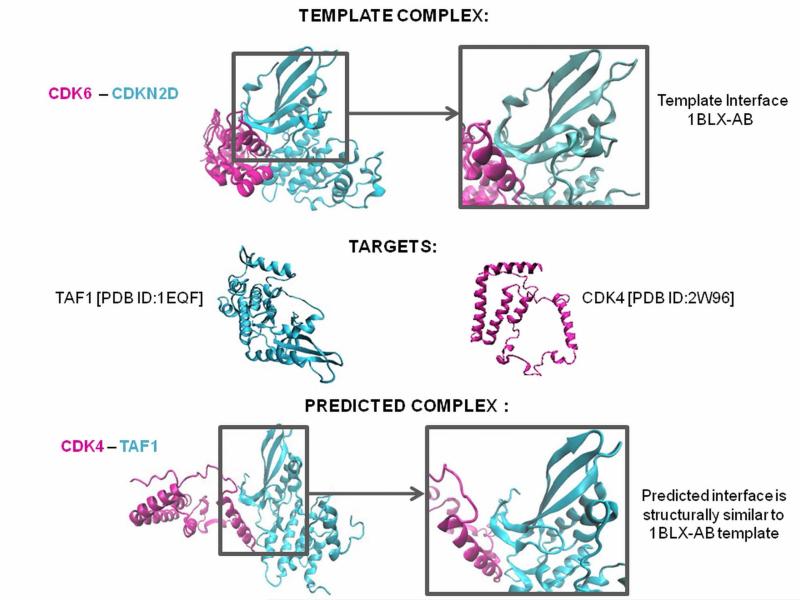
PRISM finds the similarity scores between the surface of each target in our datasets and each side of a PDB template (a template has two sides, i.e. the two complementary surfaces in the complex, in cyan and magenta, Figure 1, top line). From this output, it predicts the interface (Figure 1, bottom line). For instance, take the target protein pair in Figure 1, “TAF1” and “CDK4”, and template interface “1BLXAB”; if “TAF1” has a region on its surface which is similar to the binding site on one chain of “1BLXAB” and “CDK4” on the second chain, then they are predicted to interact similar to the interface “1BLX-AB”. This means that the binding sites of proteins “TAF1” and “CDK4” are similar to those of the protein chains of interface “1BLX-AB”.
Figure 1. Interface Structure Prediction for Interacting Target Proteins.
Interface information is obtained from the “Protein Interactions by Structural Matching” (PRISM) server. PRISM searches for spatial motif similarity on target proteins’ surfaces using geometric complementarity and considers evolutionary conservation of hot spots based on a non-redundant protein-protein interfaces template dataset derived from the PDB. Its prediction principle is to compare both sides of a template interface with surface regions of any given two monomers, and if they are similar these two proteins are predicted to interact with each other via this interface region. In the above example the CDK6 [PDB:1BLX-A] and CDKN2D [PDB:1BLX-B] complex is derived from PDB and the target proteins CDK4 and TAF1 are found to be interacting via an interface structurally similar to 1BLX-AB interface. CDK 4 and TAF1 are predicted to be interacting via 1BLX-AB interface.
A New Structural PPI Network Model: Protein Interface and Interaction Network (P2IN)
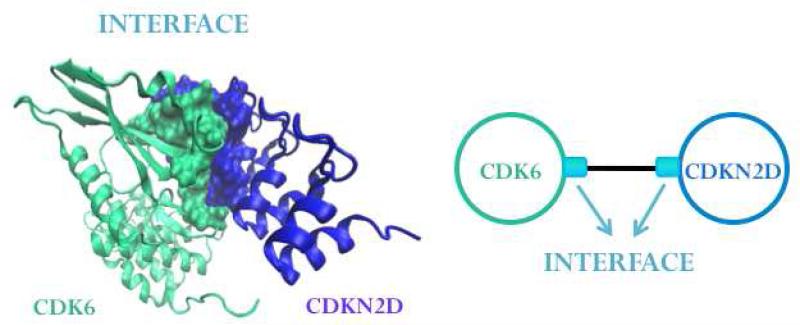
Protein interaction networks (PINs) give binary information relating to whether two proteins communicate. Being enriched with structural information, P2IN is a more physical and realistic version of PIN. Unlike the PINs whose nodes are proteins and the interactions are the connecting edges, P2IN have interface information linked to its edges and each protein in the network has a 3D structure. Interactions between the proteins are represented by edges going through the interfaces of the two chains (Figure 2). Similar interfaces may exist between different protein pairs and the same protein pair may interact through different interfaces44,51,52.
Figure 2. The P2IN Representation.
Interactions between proteins are represented by the edges going through the interfaces whose two chains represent the binding site regions of the proteins.
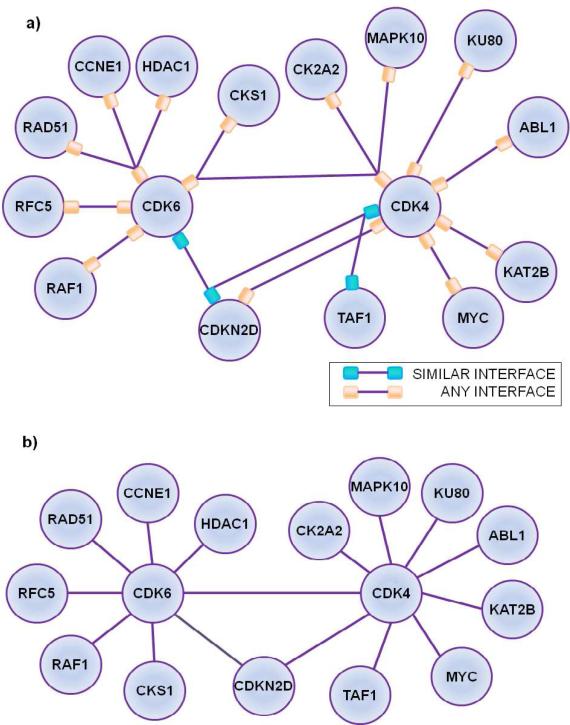
P2IN is capable of providing structural details that a PIN is not able to describe. Some of these details are exemplified in Figure 3: different protein pairs interacting via the same interface (CDK6 – CDKN2D and CDK4 – TAF1 interact via same interface); a protein pair interacting using different interfaces (CDKN2D and CDK4) and multiple proteins competing to bind the same region on a protein (RAD51, CCNE1 and HDAC1 going for the same binding site on CDK6). This additional knowledge may allow identification of interactions which cannot take place simultaneously. Partners of a protein interacting with the same binding site cannot coexist. In addition, since ligands tend to bind proteins that have similar binding sites53-55, locating protein pairs that interact via similar interfaces may help to predict additional, off-targets of these drugs. Thus, P2IN might be one step closer to mimicking systems-wise drugs effects 56.
Figure 3. Protein – Protein Interactions and Interface Networks (P2IN) versus Protein-Protein Interaction Network (PIN).
(a) A subset of PRISM predictions represented with P2IN and (b) its PIN counterpart. In P2IN the same interface may exist between different protein pairs (CDK6 – CDKN2D; CDK4 – TAF1 interact via same interface) and the same protein pair may interact using different interfaces (CDKN2D and CDK4). Moreover many proteins may compete to bind the same binding on a protein (RAD51, CCNE1 and HDAC1 bind the same site on CDK6). PIN's are not capable of depicting such structural information of protein interactions.
Network Attacks
An attack on a network is executed in order to disrupt the information flow locally or globally, to disable a pathway or to destroy the network as a whole. An attack implies deletion or attenuation of an edge or a node of the network25.
i. Node Attack
A node attack on the network removes edges focused at a single node. We studied two different node attacks: complete knockout (Figure 4a) and partial knockout (Figure 4b). Complete knockout refers to removing a node with all of its edges; partial attack involves removing randomly selected half of the edges of a node. Complete node attacks are commonly used attack strategies. The targets of these attacks vary according to the network topology. Complex networks were believed to be randomly linked 57 until Barabasi et al. discovered a common topology58. This discovery introduced scale-free networks into network theory. While in a random network nodes have roughly the same number of edges, in a scale-free network there are many nodes with a small number of edges and a few nodes (hubs) with a large number of connections. Random node attacks may be destructive to networks that are randomly linked, whereas scale-free networks are highly robust under these attacks. Scale-free networks are defenseless upon few vital node removals 39. Accordingly, targeting hub nodes is a preferred approach in network attacks59-62. Detaching those nodes, which have many neighbors, will disrupt the information flow.
Figure 4. Network Attacks.
(a) Complete knockout (b) and partial knockout target a hub node. (c) Distributed attack.
Partial knockout was performed by Agoston et al.25 on E. coli and S. cerevisiae networks. They removed randomly half of the edges of target node or attenuated all the edges of the node. This study suggested that partial weakening of a small number of nodes (3- 5) might have a stronger effect than completely removing a selected node; in both cases the most damaging nodes were selected. Zhang et al. 63 questioned whether this result is a general concept for complex networks and retested all attack strategies on the Barabasi-Albert (BA) scale-free network 58 and the Erdös-Renyi (ER) random network 57. They confirmed that multi target partial attacks may disturb complex networks more than single target complete attacks and ER random networks are more resistant to multi target partial attacks than the BA networks.
ii. Edge Attack
An edge attack removes one or multiple edges from the network, where the edges do not have to be incident to a node. Depending on the network topology, attacking a high betweenness edge may damage the system more than attacking a hub node with many edges. Thus, deleting a number of edges scattered in different regions of the network might be a more efficient attack strategy than targeting a node25. This attack is a ’distributed attack’ (Figure 4c).
Interface Attack
Here we propose an attackstrategy which is based on the expectation that PPI-targeting drugs may disrupt a number of protein-protein interactions which have structurally similar interfaces. Interface attack is the graphical representation of this strategy and removes interactions with similar interfaces from the network (Figure 5).
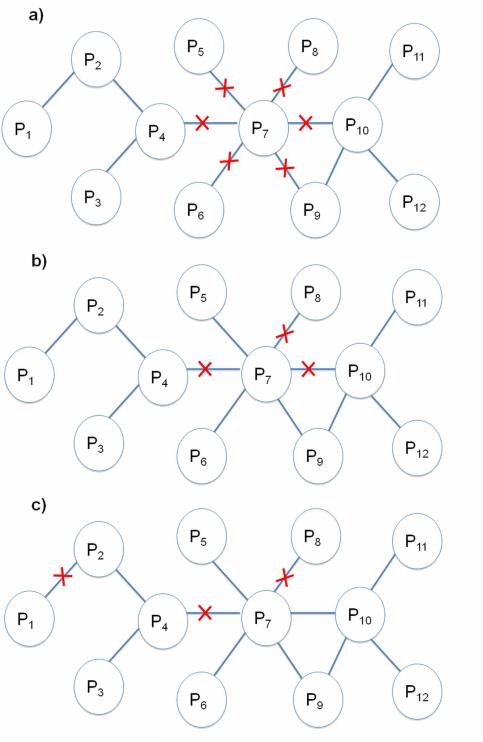
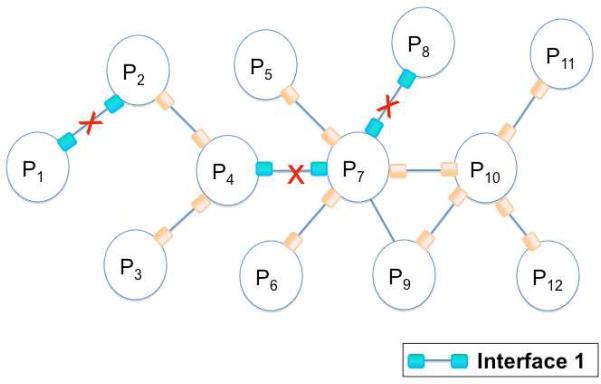
Figure 5. Interface Attack.
Interface attack hits the set of edges, which interact via structurally similar interfaces (marked with red crosses). When the interaction between P1 and P2 is targeted, the interactions between P4 and P7; P7 and P8 are also hit, since they all interact through interface 1.
Interface attack is a kind of distributed attack, since it targets one or more interactions between protein pairs. However, instead of selecting random edges or the ones which lead to the most damage, structurally similar interfaces are targeted. Interface attack is a knowledge-based distributed attack.
RESULTS AND DISCUSSION
As a first case study, we built the p53 protein interface and interaction network (P2IN), showing how knowledge of similar interfaces may help to detect drug off-targets. A second case study compares the consequences of the interface attack and the complete node attack. To obtain the hypothetical biological impact of interface attack, we built a new network in which the interactions and nodes without structural knowledge were also present. Lastly the network attack scenarios are practiced on P53 P2IN and changes in the network robustness are analysed.
P53 P2IN
The p53 tumor suppressor is a center of a protein interaction network. Under cellular stress, it is a key factor in the decision between cell cycle progression or apoptosis 61. Stress signals may be due to failures in DNA replication, chromosome segregation and cell division 64. Malfunction of p53 causes uncontrolled growth 65. p53 is inactivated in more than 50% of human cancers 66,67. We constructed the p53 signaling P2IN using the PRISM 68,69 predictions for this signaling pathway. Our network has 251 interactions among 81 proteins (please refer to Table SI1 for the list of PRISM interaction predictions for p53 network). 46 different types of interface structures are observed for these interactions. 26 out of the 251 are present in Kohn's molecular interaction map (MIM) 70; 59 are in PPI databases such as HPRD 71, Mint 72, IntAct 73, Reactome 74, BioGrid 75, Pathway Commons 76 and NCI-Nature PID 77. 66 interaction predictions are directly experimentally validated and there is evidence in the STRING 78 database for 90 of the interactions predicted by PRISM. Overall, 104 interactions out of 251 are validated experimentally or via STRING.
Knowledge of Similar Interfaces May Help Off-Target Detection
This section describes a case study for off-target prediction application on the interfaces of p53 P2IN. CDK6 is a regulator of cell cycle progression and affects the activity of tumor suppressor protein RB which inhibits it and keeps the cell growing in G1 phase. Inactivation through phosphorylation by CDK leads to cell cycle progression. Some CDK6 inhibitors that block the G1/S transition of cell are listed in Table 1. The drugs in this table have 3D structures in complex with CDK6 46.
Table 1.
List of CDK6 Inhibitors.
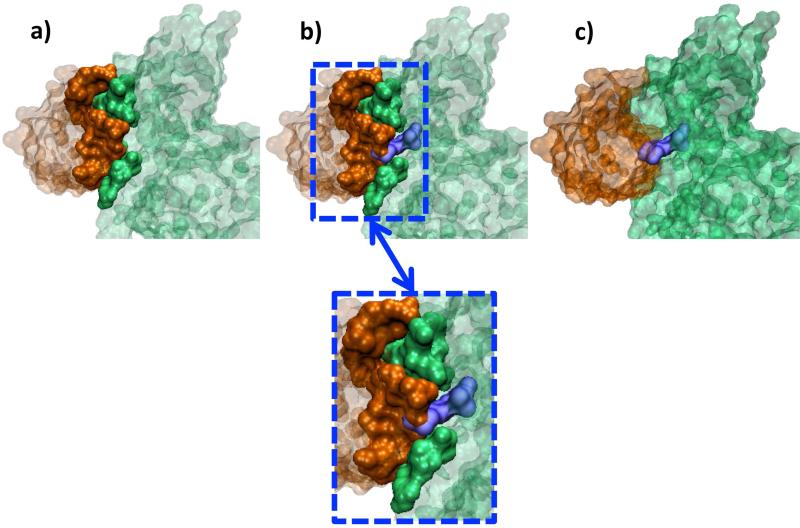
CDKN2D is a cyclin dependent kinase inhibitor, which forms a stable complex with CDK6 (Figure 6a). The drugs listed in the table (Aminopurvalanol, PD-0332991, CHEBI: 792519, CHEBI: 792520 and Fisetin) seem to interfere with CDK6 and CDKN2D interface, when the CDK6–CDKN2D complex is superimposed on CDK6 and drug complexes present in PDB (Figure 6b - 6c, Figure SI1). The crystal structure of CDK6 and CDKN2D interface is available (PDB ID: 1BLX, chains: A, B86. 1BLX is a complex between human CDK6 and mouse CDKN2D. The same complex is also available for human CDK6 and human CDKN2D (PDB ID: 1BI8, chains: A, B) 87. We considered the mouse and human CDKN2D as homologs, with 87% sequence similarity and 0.41 RMSD and used the 1BLX complex in this study since it has a better X-ray resolution). PRISM predicts an interaction between CDK4 and CDKN2D, with a structurally similar interface to the CDK6-CDKN2D interface. The interaction of CDK4 and CDKN2D is detected by in vitro and in vivo assays88, but the 3D structure of their complex is unavailable. The interface attack by the five drugs blocking the interaction of CDK6-CDKN2D may disturb the CDK4-CDKN2D interaction.
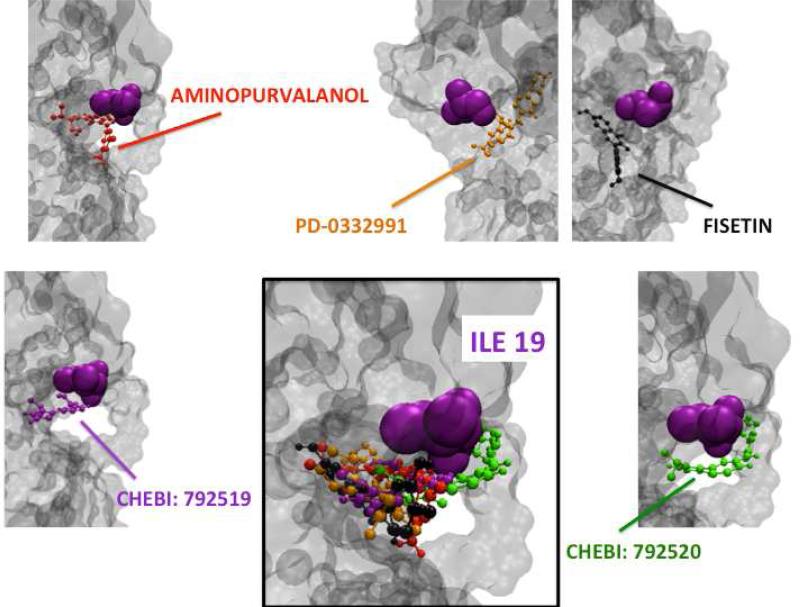
Figure 6. The CDK6 (green) - CDKN2D (orange) Complex and CHEBI: 792520 (purple) Interference.
(a) The interface of CDKN2D - CDK6 is from PDB ID:1BLX. (b,c) In the PDB, CHEBI: 792520 has a 3D structure in complex with CDK6 (PDB ID: 3NUX). When CDK6 proteins of 3NUX and 1BLX are superimposed, CHEBI: 792520 interferes with the CDK6 and CDKN2D interface. These two figures are predicted outcomes; no structural data are available.
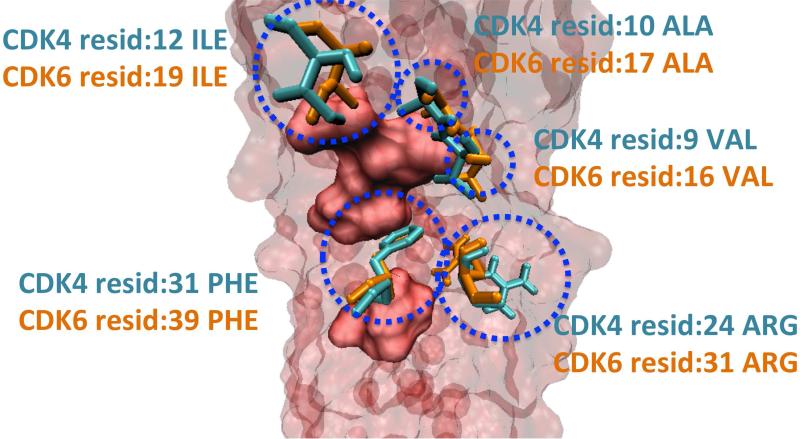
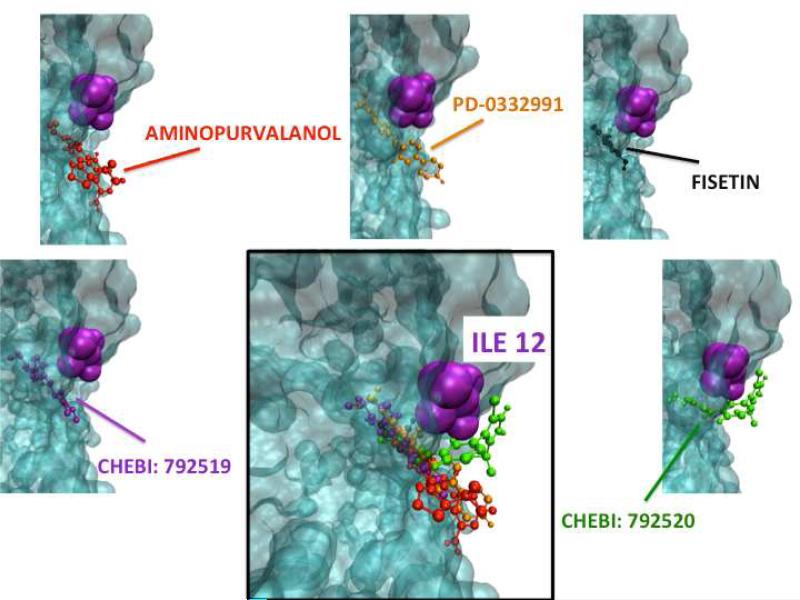
Using the HotPoint server 89, we identified the computational hotspots of CDK4, CDK6 and CDKN2D. When the interfaces with CDKN2D are superimposed by using Multiprot engine 47, CDK4 (obtained from PRISM predictions) and CDK6 (obtained from the PDB) have a number of identical hotspots (Figure 7). CDKN2D interacts with them via the same surface area. Lastly, we found that the hotspot (CDK6 residue Ile19) that is closest to the ligand binding region on CDK6, is also present on the binding region of CDK4 (residue Ile12) (Figures 8-9). These drugs are also close to hotspots Gln98, and Asp97 on CDK4, and Gln103 (hotspot), Asp102 (non-hotspot) on CDK6 (please refer to Figures SI2 - SI3). These residues overlap when CDK4 (PDB ID: 2W96, chain: B) and CDK6 (PDB ID: 1BLX, chain: A) are superimposed with Multiprot engine (RMSD: 1.28 Å). At this point we propose that CDK4 may be an off-target candidate for drugs targeting CDK6. In order to see how alike the binding pockets of CDK4 and CDK6 are, we superimposed the ligand binding sites using VMD90 (Figure SI4). The results revealed that CDK4 has a binding pocket which is similar to that of CDK6, with RMSD 0.87 Å.
Figure 7. Hotspots of CDK4-CDKN2D and CDK6-CDKN2D Interfaces.
The predicted hotspots of CDK4 (cyan) and CDK6 (orange) proteins are represented with “Licorice” and the hotspots of CDKN2D are drawn as a (red) surface, using VMD 90. The red, transparent body in the background is also CDKN2D protein. CDK4 and CDK6 have a number of identical hotspots, when their interfaces with CDKN2D are superimposed.
Figure 8. CDK4 Docking Simulations.
AutoDock 91 is used to dock the drugs (Aminopurvalanol, PD-0332991, CHEBI: 792519, CHEBI: 792520 and Fisetin) to candidate off target CDK4. The hotspot (CDK6 residue Ile19) that is closest to the ligands’ binding region on CDK6, is also present on the binding region of CDK4 (residue Ile12).
Figure 9. CDK6 Docking Simulations.
AutoDock91 is used to dock the mentioned drugs (Aminopurvalanol, PD-0332991, CHEBI: 792519, CHEBI: 792520 and Fisetin) to primary target CDK6. The hotspot (CDK6 residue Ile19) that is closest to the ligands’ binding region on CDK6, is also present on the binding region of CDK4 (residue 12).
Docking simulations may suggest if a ligand is capable of binding to a protein. AutoDock91 is used to dock these drugs to candidate off-target CDK4 (Figure 8) and primary target CDK6 (Figure 9). As shown in Table 2, the binding free energies between CDK4 and the drugs are promising; they are comparable to the binding energies between CDK6 and its inhibitors. The listed energies are the lowest binding free energies of the most populated clusters. The RMSD values of superimpositions of the best poses of each drug molecule docked to CDK4 compared to CDK6 are also provided in Table 2 (Figure SI5). These findings strengthen our proposition that CDK4 is an off-target for the drugs targeting CDK6.
Lastly, we searched for the inter-relationship between CDK6 inhibitors and CDK4 in the literature. We found that PD-0332991 has been designed to turn off both CDK4 and CDK692. Moreover, SuperTarget states that CDK4 is a target of CHEBI: 792520 93. Accordingly, we are able to verify two of our off-target predictions. To conclude, we may now suggest that CDK6 binding drugs that block the interface between CDK6 and CDKN2D, may also bind to CDK4 and disrupt the interaction between CDK4 and CDKN2D. Therefore, when CDK6-CDKN2D interaction is hit in the interface attack, we may also break the interaction between CDK4 and CDKN2D.
Biological Consequences of Interface Attack versus Complete Node Attack
Networks of protein interactions are vital tools for explaining a series of events in the cell which may be triggered by a drug. A drug which inhibits protein-protein interactions may be represented in the network by removing the respective edges. To foresee the effects of a drug designed to inhibit all the interactions of a single protein, one can simply remove this node from the network and investigate the changes. For making an accurate functional analysis, we need all known protein interactions in the p53 pathway. We constructed a p53 network which, regardless of the structural availability, contains all known protein interactions and proteins. We simulated the changes in the network when subject to node and interface attacks. We partitioned the network using the “Affinity Propagation” algorithm 94. This clustering algorithm determines the representative examples (examplars) of the graph and then partitions the network according to these examplars.
We mapped the experimentally validated PRISM interface predictions of the p53 pathway on Kohn's MIM 70 as the starting point for constructing an experimentally validated network of protein interactions enriched with interfaces. We obtained a p53 PIN with 109 nodes and 227 edges. We expanded this network with the 66 PRISM predicted interfaces that were experimentally validated (26 interactions present in Kohn's MIM, 33 additional interactions from various experimental databases). We gathered a network of 115 nodes and 269 edges. Recall that there were a number of proteins from databases other than Kohn's MIM in our PRISM target. As a result the number of nodes also increased (Figure 10). The clusters generated by the Affinity Propagation algorithm are shown using pie charts (Figure 11 top row). Clusters are named according to the highest degree node of that partition.
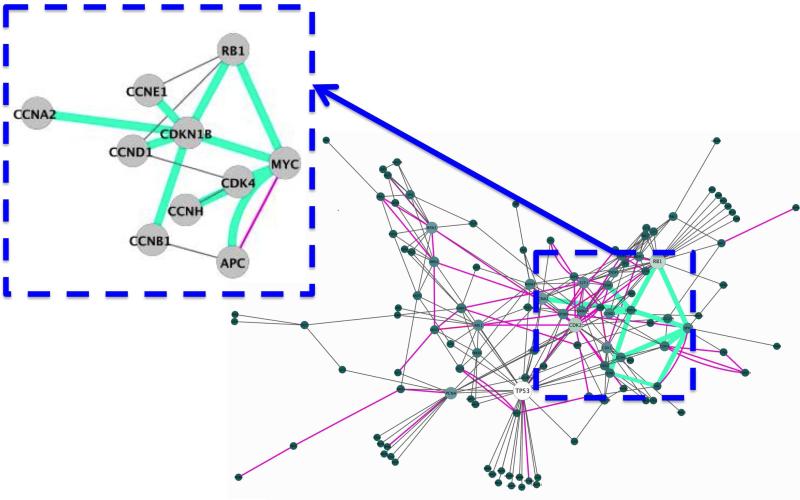
Figure 10. Structurally Enriched MIM Attacked Based on the 1jsuBC Interface.
Experimentally validated edges of p53 P2IN mapped on the Kohn's MIM 70. The edges with interface structures are shown in pink color and the edges with 1jsuBC interface is highlighted in green. In the close-up figure edges with 1jsuBC interfaces are also can be seen in green.
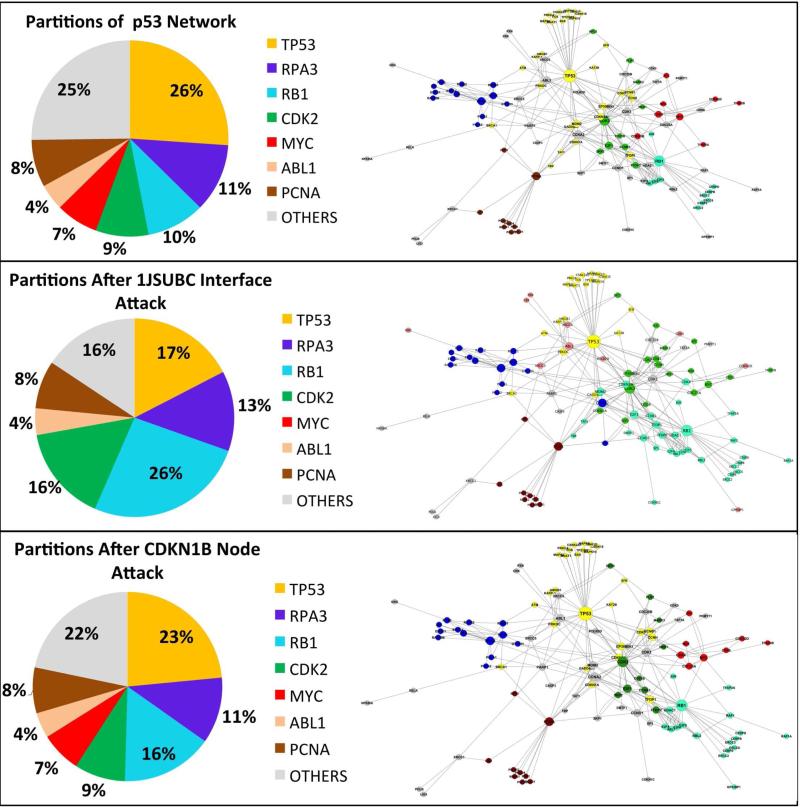
Figure 11. Pie Charts of Clusters Generated with Affinity Propagation Algorithm.
In the pie charts each slice represents a cluster and they are named with the clusters’ hub nodes. Percentages of the slices are the ratio of the node number in the corresponding cluster to the total number of nodes in the network. (Top row) Clusters of the network generated by mapping the experimentally validated PRISM predictions of p53 pathway onto Kohn's MIM. (Middle row) The clusters after 1jsuBC interface attack.
When the 1jsuBC interface (template interface is between the CCNA2 and CDKN1B proteins) is attacked, 11 edges are removed from the network. Six of these are around the CDKN1B node. Therefore, this node is completely removed from the network by the 1jsuBC interface attack, in addition to the removal of 5 edges around other nodes. One can see that this attack causes the cluster, with the RB1 hub node, to get significantly bigger (please refer to the slices of RB1cluster in the top and middle rows of Figure 11). RB1 now has a greater influence on the network. MYC is no more the hub node of a cluster (red slice present in the top row of Figure 11 dissapears in the middle row of Figure 11) and the cluster of CDK2 enlarges from 9% of the nodes of network to 16% (Figure 11 middle row). A complete node attack targeting the CDKN1B protein, means breaking all of this node's interactions detaching it from the network. PRISM predicts that all 6 interactions of CDKN1B have a similar structure to 1jsuBC interface. Thus, to block all of the interactions of CDKN1B, a drug has to attack the 1jsuBC interface, which affects 5 more edges in the network. However, in the case of complete node attack on CDKN1B, only edges of this node are discarded from the network. We do not observe a significant change in the sizes of the clusters following complete node attack (see top and bottom rows of Figure 11).
The changes observed after the interface attack appear reasonable. During the 1jsuBC interface attack, CDKN1B is removed from the network, CDK2 cluster gets bigger and the influence of this protein on other nodes increases. Since CDKN1B has inhibitory activity on some CDK2 complexes95, this change is expected. Once MYC is not a hub in a cluster, the RB1 cluster expands. In the presence of MYC, the RB1 transcription is suppressed and MYC activates a set of miRNAs, which in turn inhibit the translation of RB1 96. Finally, CDKN1B and RB1 are tumor-suppressors. The RB1 cluster gets bigger when CDKN1B loses all of its interactions, possibly suggesting that RB1 may be involved in an alternative pathway. (Bottom Row) The clusters after CDKN1B node attack.
Network Attack Scenarios Applied to P53 P2IN and Changes in the Network Robustness
The robustness of a network relates to its ability to withstand the damage caused by attacks. It can be expressed by topological parameters. The most commonly used robustness parameters are the average inverse geodesic length (AIGL) 59,97 and the giant component size 59 (GCS). To monitor the change in the connectedness of the nodes in the system, we use both.
For the p53 P2IN survivability analysis, several attack types and target selection strategies are used. These attack scenarios refer to partial or complete knockout of hub nodes and deletion of multiple edges that are scattered around the network. At each step a new target is hit and the topological parameters are recalculated until the network is left without interactions.
i. Hub Node Attack
A hub is the highest degree node of the network; it is the node that has the largest number of interactions. This attack type targets the largest degree node of the network. Hitting this element also affects its interacting partners and causes a serious disturbance in the network communication.
ii. Frequent Interface Attack
In P2IN, the number of occurrences of each interface type is known. In this strategy the most frequently observed interface is selected as the target of interface attack.
iii. Maximal Damage Strategy
The maximal damage strategy is a greedy algorithm, which was studied by Agoston et al. 25. It hits the component that will harm the network the most in each attack. This tactic may be used in both node and edge attack types. Removing multiple edges that are selected according to the maximal damage target selection strategy is a kind of a distributed attack. It targets the node or interface that is expected to cause the greatest possible harm.
Frequent Interface Attack is as Harmful as Complete Hub Knockout and it is a More Realistic
Scenario
Breaking an edge can be considered as the graphical representation of a drug blocking the interaction of two proteins. If we were to map node-targeted attacks (complete or partial knockout) to a drug mechanism, it would be a “magic bullet”; even if a drug would specifically bind to one protein, in most of the cases it may not obstruct all of its interactions. It seems that complete knockout is rarely observed in realistic drug action. The common “similar binding sites should recognize similar ligands” strategy22, motivated us to develop the interface attack.
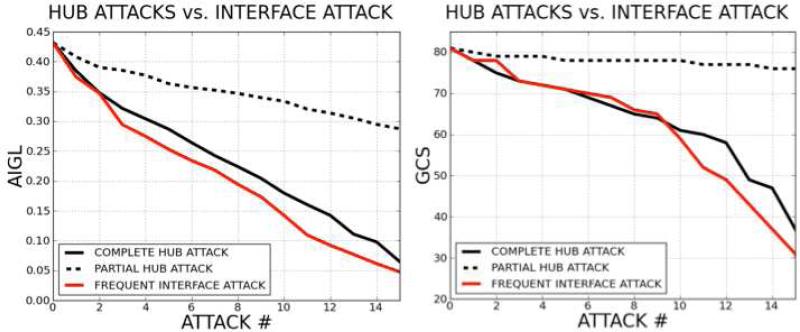
Complete/partial hub node attacks and interface attacks based on their frequencies of occurrence are performed on the p53 P2IN. In Figure 12 the change in the network robustness is plotted according to AIGL and GCS. The x-axis stands for the number of attacks, while the y-axis is the AIGL or GCS values during the attacks. A drop in AIGL or GCS of the network correlates with the damage caused to the system. The plots show that attacking the most frequent interface in the p53 signaling network is at least as harmful as complete removal of the hub nodes from the network. Thus, rather than targeting a well connected protein, which is more likely to be essential 60, we may target edges that have similar interface structures.
Figure 12. Hub Node Attack versus Most Frequent Interface Attack.
The figure plots of the damage to the network following 15 successive complete hub node attacks, partial hub node attacks and frequent interface attacks (for AIGL (on the left) and GCS (on the right) topological parameters). The results suggest that the most frequent interface attack and complete hub knockout lead to roughly the same damage, while the effect of the partial hub knockout is to a lesser extent.
The most frequent interface (PDB ID: 1JSU, chains: B, C) is observed 46 times in the p53 network. 21 of these predictions are validated experimentally or present in the STRING database. If there were a drug designed to disturb one of these 46 interactions, not just that particular edge, all 46 interactions could be hit. Hence, building the interface and interaction network of a biological system may be helpful for drug development.
Interface Attack is not as Harmful as Distributed Attack when Maximal Damage Strategy is Applied
Agoston et al. 25 showed that rather than removing a node completely from the network, one could inflict similar damage by removing a number of edges distributed around the network. They chose the most destructive edges.
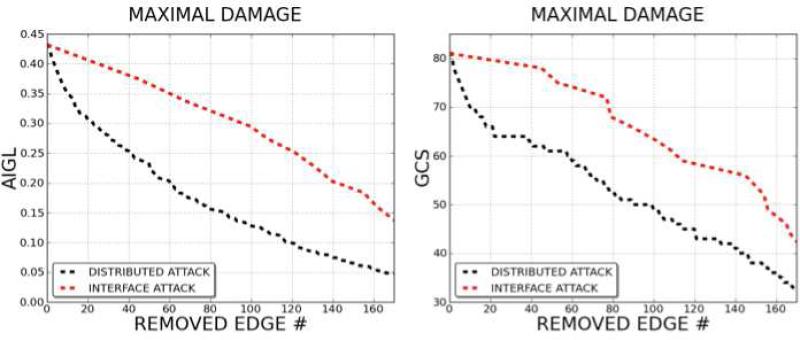
Interface attack is a kind of distributed attack, but it chooses the target edge set based on interface similarity. We performed distributed attacks and interface attacks on the p53 P2IN. In this experiment we followed a maximal damage target selection strategy, by selecting the most damaging edges (distributed attack) or the most damaging interface in successive attacks. The comparison of the damage caused by distributed attack and interface attack is plotted in Figure 13. The x-axis is the number of edges removed during attacks and the y-axis the change in the network GCS and AIGL. It is clear that distributed attack harms the network more than interface attack. However, comparison of interface attack and distributed attack is not straightforward, since distributed attack selects edges one by one, while interface attack chooses between sets of edges. This is why distributed attack is so harmful and is nearly the optimal attack strategy for collapsing the network. However, interface attack seems to be physically more suitable for simulating the impact of multi-target drugs on the network, since the interactions affected by multi-target drugs are not always the most harmful.
Figure 13. Maximal Successive Damage Strategy on Distributed and Interface Attack.
Damage in the network (both according to AIGL (on the left) and GCS (on the right) topological parameters) is monitored, under successive attacks. Distributed attack and interface attack are executed using the maximal damage strategy. The number of edges removed from the network in each attack is parallel to the harm attacks cause on the network. It is obvious that distributed attack is the most harmful strategy.
Frequent Interfaces are not Observed on Topologically Critical Interactions
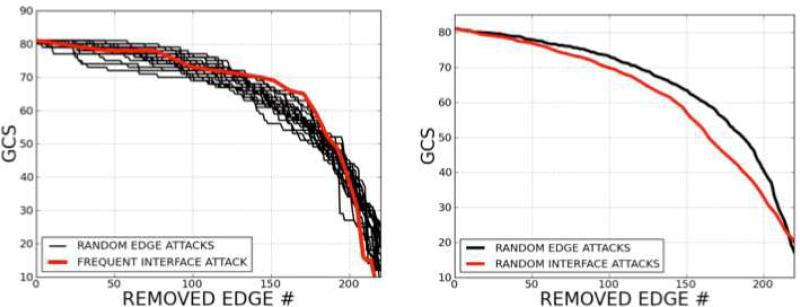
P53 P2IN is a small sub-network, and it does not have a scale-free architecture. When random edge attacks are compared with frequent interface attacks (Figure 14) according to the change in giant component sizes, the most frequent interface attack is less harmful to the p53 P2IN than random edge attacks.
Figure 14. Random Edge Attacks versus Interface Attacks.
The most frequent interface attack is relatively less harmful to the p53 P2IN than random edge attacks (on the left). However, the average of random interface attacks harms the network more than the average of random edge attacks (on the right). Consequently, random interface attacks give more harm to the network than frequent interface attacks.
However, when the attacks are performed on randomly selected interfaces, we observe that on average they harm the network more than random edge attacks. Consequently, random interface attacks are more harmful to the network than frequent interface attacks; that is, a frequent interface is less likely to hit topologically critical elements of the network. This makes the network more resistant to failures.
CONCLUSIONS
We proposed a new network representation (P2IN), which introduces the structures of protein interfaces into the protein interaction networks (PINs). In addition to providing the binary information of whether two proteins interact with each other, the protein interface and interaction network (P2IN) also provides information on the structure of the complex that they form. This representation allows us to propose a new attack strategy of hitting edges between protein pairs that interact via structurally similar interfaces rather than nodes.
We generated the signaling network of the p53 P2IN and tested its robustness to various attacks. Both node and edge attacks are performed. The interface attack is found to be as destructive as hub node attacks; however, it is not as harmful as distributed attack that targets maximal edges. A drug that disturbs a frequent interface type may be as destructive as a drug targeting a high degree protein, suggesting the usefulness of considering the frequency of interface motifs during drug development. We discovered that some drugs (Aminopurvalanol, PD-0332991, CHEBI:792519, CHEBI:792520 and Fisetin) binding to CDK6, disrupt its interaction with CDKN2D. We applied our interface attack strategy to this case and found that drugs blocking this interface may also affect the interaction between CDK4 and CDKN2D. CDK4 also appears an off-target for drugs binding to CDK6. This example illustrates the promise in our strategy as a first step in indentifying potential off-target drug hits. Finally, we provided a case study of a comparison between node and interface attacks. Challenging next steps are accounting for molecular flexibility. Proteins are highly dynamic, and structure-based drug discovery requires detailed structural treatment to uncover transient pockets which are unlikely to be observed in the static crystal snapshots and rigid docking. Nonetheless, systems-wide outcome involving possible off-targets of a drug is an important consideration, and eventually would need to be integrated with detailed structural investigation in attempts to forecast potential side effects. Here, our concept of interface attack exploits structural motifs. It is inspired by network pharmacology, an emerging paradigm in drug discovery.
MATERIALS AND METHODS
Constructing Protein Interface and Interaction Network (P2IN)
The first step of building a P2IN is to gather raw data of protein interactions and their 3D structures. Protein interactions are collected from the literature and databases; the 3D structure of the interfaces is obtained from application of PRISM 68,69. There may be more than one possible template interface for one interaction pair; in such a case, there is more than one possible binding site between two proteins. All possibilities are considered, and every matching interface template is included in the interface and interaction networks. Proteins whose interface sites cannot be predicted by the PRISM server are discarded. This decreases the number of proteins and interactions.
PRISM predicts interfaces for two target proteins, which have 3D structures in the PDB. However, the interaction data gathered from the literature are UniProt 98 IDs. Thus, target (UniProt) proteins should be mapped to PDB IDs, and these will constitute the input for PRISM. The main problem in this phase is that a protein may have multiple PDB IDs or its 3D structure might not be known. To overcome this inconsistency, proteins that do not have matching PDB chains are discarded and all PDB counterparts of a protein are taken into account while constructing the network.
P53 P2IN
We studied the interactions between the proteins in the p53 signaling pathway. The list of proteins that are involved in this pathway was compiled from the literature 70 and databases by Tuncbag et al.50. Among these proteins, 85 had 3D structures in the PDB. The interaction and interface data is obtained from PRISM predictions. We used 1037 template interfaces that were extracted from the PDB 99 for the prediction process. The resulting interface predictions with energies lower than -10 are accepted.
PRISM predicted 251 interactions among 81 proteins and there are 46 different interface structures in the network. The number of proteins dropped from 85 to 81, since PRISM did not infer interactions for some proteins. If we were to link each protein in the network to other proteins, we would end up with ~3300 edges. PRISM infers 251 interactions out of those 3300 possibilities and 41% of those predictions are already known. Furthermore, in the generated p53 P2IN, there are 15 PPIs, which have PDB structures in complex form. PRISM was able to predict 13 of those interfaces correctly (Table SI2).
Docking Parameters
For adding polar hydrogens, assigning Gasteiger charges and drawing grid boxes AutoDockTools 1.5.491 was used. Binding affinities were calculated with AutoGrid version 4. Lamarckian genetic algorithm (trials of 50 dockings, population size of 150, and maximum number of generations of 27000) was used to do the docking experiments using AutoDock 4.0 91.
Clustering Algorithm
We partitioned the network according to the “Affinity Propagation” algorithm 94 with the help of Clustermaker plugin 100 of Cytoscape 101.
Mapping the experimentally validated PRISM interface predictions of p53 pathway on the Kohn's MIM
Kohn's MIM has some nodes that do not have a protein counterpart, or some nodes correspond to multiple proteins. Before constructing the PIN, we updated nodes in Kohn's MIM by removing or expanding some of them (Table SI3). If a node was replaced with multiple proteins, the number of interactions automatically increased. We searched the String database for validating the new edges and picked the ones which were coming from experiments or databases. For example, the “CDK4-6” node corresponds to three proteins (CDK4 – CDK5 – CDK6). In the original map there was an interaction between “CDK7” and “CDK4-6”. The “CDK7” interactions with CDK4 and CDK5 are validated, but not with CDK6. The full list of interactions can be found in Table SI4.
Robustness Measures
AIGL is the sum of the inverses of all shortest paths, divided by the number of possible node combinations. The definition is given in Equation 1. The notation used is as follows:
ℓ = average geodesic length
n = number of nodes
i, j = proteins
dij = distance between proteins i and j
If there is no path connecting nodes i and j, the distance between them is set to infinity. Some studies use the average geodesic length but we preferred to use AIGL. Even after several attacks, AIGL will not be equal to infinity, because if there is no navigable route between i and j, .
| (Eq. 1) |
GCS is the number of nodes in the network's largest connected sub-graph and it may give important clues about the collapsing mechanism of network under attacks.
Supplementary Material
Table 2.
AutoDock91 Results Results given in terms of the lowest binding energy of the largest conformational clusters are in the first two rows. The RMSD values of superimpositions of the best poses of each drug molecule docked to CDK4 compared to CDK6 are in the last row.
| PD-0332991 | Fisetin | Aminopurvalanol | CHEBI: 792520 | CHEBI: 792519 | |
|---|---|---|---|---|---|
| CDK4 | −8.22 kcal/mol | −7.59 kcal/mol | −5.97 kcal/mol | −7.55 kcal/mol | −6.51 kcal/mol |
| CDK6 | −8.05 kcal/mol | −6.75 kcal/mol | −7.69 kcal/mol | −6.81 kcal/mol | −6.18 kcal/mol |
| RMSD | 0.57 Å | 0.68 Å | 0.89 Å | 1.83 Å | 1.92 Å |
ACKNOWLEDGEMENTS
This work has been supported by TUBITAK (Research Grant Numbers: 109T343 and 109E207) and Turkish Academy of Sciences (TUBA). This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Table SI1 provides PRISM interaction predictions for p53 network, Table SI2 shows the correctness status of PRISM predictions for 15 interactions which have available PDB structures. A full list of proteins in Kohn's MIM can be found in Table SI3. Lastly, in Table SI4 the list of updated interactions of Kohn's MIM is given. Figure SI1 highlights the CDK6 binding site (one chain of the CDK6-CDKN2D interface) on CDK6-drug complexes present in PDB, and shows the overlap between the drugs and the binding site. Table SI5 contains the RMSD values of CDK6 structures we used in this study. Figure SI2 depicts the hotspots of CDK4 and the drugs docked on CDK4, while Figure SI3 depicts the hotspots of CDK6 and the drugs docked on CDK6. Figure SI4 provides the superimposition of pockets of CDK6 (cyan) and CDK4 (dark blue) using VMD visualization tool. Figure SI5 shows the change in drugs’ conformations when the pockets of CDK6 (dark blue), CDK4 (red) are superimposed. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Xie L, Bourne PE. Structure-based systems biology for analyzing off-target binding. Curr Opin Struct Biol. 2011;21:189–199. doi: 10.1016/j.sbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mestres J, Gregori-Puigjane E, Valverde S, Sole RV. Data completeness--the Achilles heel of drug-target networks. Nat Biotechnol. 2008;26:983–984. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- 3.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 4.Pujol A, Mosca R, Farres J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol Sci. 2010;31:115–123. doi: 10.1016/j.tips.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, Salzberg AC, Huang ES. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 6.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, Zheng S, Li Z, Li H, Jiang H. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–W614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byvatov E, Fechner U, Sadowski J, Schneider G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J Chem Inf Comput Sci. 2003;43:1882–1889. doi: 10.1021/ci0341161. [DOI] [PubMed] [Google Scholar]
- 9.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 10.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 11.Scheiber J, Jenkins JL, Sukuru SC, Bender A, Mikhailov D, Milik M, Azzaoui K, Whitebread S, Hamon J, Urban L, Glick M, Davies JW. Mapping adverse drug reactions in chemical space. J Med Chem. 2009;52:3103–3107. doi: 10.1021/jm801546k. [DOI] [PubMed] [Google Scholar]
- 12.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 13.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62:479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 15.Weisel M, Proschak E, Schneider G. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J. 2007;1:7. doi: 10.1186/1752-153X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13:323–30. 307–308. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 17.Xie L, Bourne PE. A robust and efficient algorithm for the shape description of protein structures and its application in predicting ligand binding sites. BMC Bioinformatics. 2007;8(Suppl 4):S9. doi: 10.1186/1471-2105-8-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S, Okuno Y, Tsujimoto G, Mamitsuka H. A probabilistic model for mining implicit ‘chemical compound-gene’ relations from literature. Bioinformatics. 2005;21(Suppl 2):ii245–ii251. doi: 10.1093/bioinformatics/bti1141. [DOI] [PubMed] [Google Scholar]
- 19.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for pharmacogenomics. Clin Pharmacol Ther. 2009;86:183–189. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24:i232–i240. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanishi Y, Kotera M, Kanehisa M, Goto S. Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics. 2010;26:i246–i254. doi: 10.1093/bioinformatics/btq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Defranchi E, Schalon C, Messa M, Onofri F, Benfenati F, Rognan D. Binding of protein kinase inhibitors to synapsin I inferred from pair-wise binding site similarity measurements. PLoS One. 2010;5:e12214. doi: 10.1371/journal.pone.0012214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy J, Cyert MS. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signal. 2009;2:re9. doi: 10.1126/scisignal.2100re9. [DOI] [PubMed] [Google Scholar]
- 25.Agoston V, Csermely P, Pongor S. Multiple weak hits confuse complex systems: a transcriptional regulatory network as an example. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71:051909. doi: 10.1103/PhysRevE.71.051909. [DOI] [PubMed] [Google Scholar]
- 26.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 27.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 28.Chene P. Drugs targeting protein-protein interactions. ChemMedChem. 2006;1:400–411. doi: 10.1002/cmdc.200600004. [DOI] [PubMed] [Google Scholar]
- 29.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 30.Gao M, Skolnick J. Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc Natl Acad Sci U S A. 2010;107:22517–22522. doi: 10.1073/pnas.1012820107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fry DC, Vassilev LT. Targeting protein-protein interactions for cancer therapy. J Mol Med (Berl) 2005;83:955–963. doi: 10.1007/s00109-005-0705-x. [DOI] [PubMed] [Google Scholar]
- 32.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 33.Whitty A, Kumaravel G. Between a rock and a hard place? Nat Chem Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 34.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug Discov Today. 2009;14:155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem. 2006;13:2607–2625. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 36.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuritzkes D, Kar S, Kirkpatrick P. Fresh From The Pipeline; Maraviroc. Nature Reviews Drug Discovery. 2008;7:15–16. [Google Scholar]
- 38.Domling A. Small molecular weight protein-protein interaction antagonists: an insurmountable challenge? Curr Opin Chem Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]
- 39.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 40.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nature reviews. Genetics. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 41.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabasi AL, Oltvai ZN, Osterman AL. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol. 2009;5:e1000601. doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gursoy A, Keskin O, Nussinov R. Topological properties of protein interaction networks from a structural perspective. Biochem Soc Trans. 2008;36:1398–1403. doi: 10.1042/BST0361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keskin O, Nussinov R. Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure. 2007;15:341–354. doi: 10.1016/j.str.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O. Architectures and functional coverage of protein-protein interfaces. J Mol Biol. 2008;381:785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 48.Mashiach E, Nussinov R, Wolfson HJ. FiberDock: a web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Res. 2010;38:W457–W461. doi: 10.1093/nar/gkq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mashiach E, Nussinov R, Wolfson HJ. FiberDock: Flexible induced-fit backbone refinement in molecular docking. Proteins. 2010;78:1503–1519. doi: 10.1002/prot.22668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuncbag N, Keskin O, Nussinov R, Gursoy A. Fast and accurate modeling of protein-protein interactions by combining template-interface-based docking with flexible refinement. Proteins. 2012;80:1239–1249. doi: 10.1002/prot.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keskin O, Tsai CJ, Wolfson H, Nussinov R. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keskin O, Ma B, Rogale K, Gunasekaran K, Nussinov R. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up Systems Biology approach. Phys Biol. 2005;2:S24–S35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- 53.Xie L, Li J, Bourne PE. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haupt VJ, Schroeder M. Old friends in new guise: repositioning of known drugs with structural bioinformatics. Brief Bioinform. 2011;12:312–326. doi: 10.1093/bib/bbr011. [DOI] [PubMed] [Google Scholar]
- 55.Perot S, Sperandio O, Miteva MA, Camproux AC, Villoutreix BO. Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Discov Today. 2010;15:656–667. doi: 10.1016/j.drudis.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 57.Erdös P, R. A. On Random Graphs. Publicationes Mathematicae. 1959;6:290–297. [Google Scholar]
- 58.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 59.Holme P, Kim BJ, Yoon CN, Han SK. Attack vulnerability of complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65:056109. doi: 10.1103/PhysRevE.65.056109. [DOI] [PubMed] [Google Scholar]
- 60.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 61.Dartnell L, Simeonidis E, Hubank M, Tsoka S, Bogle ID, Papageorgiou LG. Robustness of the p53 network and biological hackers. FEBS Lett. 2005;579:3037–3042. doi: 10.1016/j.febslet.2005.03.101. [DOI] [PubMed] [Google Scholar]
- 62.Crucitti P, L. V, Marchiori M, Rapisard A. Efficiency of Scale-free Networks: Error and Attack Tolerance. Physica A. 2003:622–642. [Google Scholar]
- 63.ZHANG DM, YIN YP, TAN J, PAN GJ, HE MH. Multiple Partial Attacks on Complex Networks. Chinese Physical Society. 2008;25:769–772. [Google Scholar]
- 64.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 65.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 66.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 67.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077–4085. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 68.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohn KW. Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Molecular biology of the cell. 1999;10:2703–2734. doi: 10.1091/mbc.10.8.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prasad TS, Kandasamy K, Pandey A. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- 72.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolton E, W. Y, Thiessen PA, Bryant SH. Annual Reports in Computational Chemistry. Vol. 4. American Chemical Society; Washington, DC: 2008. PubChem: Integrated Platform of Small Molecules and Biological Activities. [Google Scholar]
- 80.Lu H, Schulze-Gahmen U. Toward understanding the structural basis of cyclin-dependent kinase 6 specific inhibition. J Med Chem. 2006;49:3826–3831. doi: 10.1021/jm0600388. [DOI] [PubMed] [Google Scholar]
- 81.Baughn LB, Di Liberto M, Wu K, Toogood PL, Louie T, Gottschalk R, Niesvizky R, Cho H, Ely S, Moore MA, Chen-Kiang S. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 2006;66:7661–7667. doi: 10.1158/0008-5472.CAN-06-1098. [DOI] [PubMed] [Google Scholar]
- 82.Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho YS, B. M, Brain C, Chen CH, Cheng H, Chopra R, Chung K, Groarke J, He G, Hou Y, Kim S, Kovats S, Lu Y, O'Reilly M, Shen J, Smith T, Trakshel G, Vögtle M, Xu M, Xu M, Sung MJ. 4-(Pyrazol-4-yl)-pyrimidines as Selective Inhibitors of Cyclin-Dependent Kinase 4/6. Journal of Medicinal Chemistry. 2010;53:7938–7957. doi: 10.1021/jm100571n. [DOI] [PubMed] [Google Scholar]
- 84.Lu H, Chang DJ, Baratte B, Meijer L, Schulze-Gahmen U. Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin. J Med Chem. 2005;48:737–743. doi: 10.1021/jm049353p. [DOI] [PubMed] [Google Scholar]
- 85.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brotherton DH, Dhanaraj V, Wick S, Brizuela L, Domaille PJ, Volyanik E, Xu X, Parisini E, Smith BO, Archer SJ, Serrano M, Brenner SL, Blundell TL, Laue ED. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 87.Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 88.Chan FK, Zhang J, Cheng L, Shapiro DN, Winoto A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol Cell Biol. 1995;15:2682–2688. doi: 10.1128/mcb.15.5.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 90.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14:33–8. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 91.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, Toogood PL. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 93.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, Schneider R, Skoblo R, Russell RB, Bourne PE, Bork P, Preissner R. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 95.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Seminars in cancer biology. 2003;13:41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 96.El Baroudi M, Cora D, Bosia C, Osella M, Caselle M. A curated database of miRNA mediated feed-forward loops involving MYC as master regulator. PLoS One. 2011;6:e14742. doi: 10.1371/journal.pone.0014742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- 98.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tuncbag N, Kar G, Gursoy A, Keskin O, Nussinov R. Towards inferring time dimensionality in protein-protein interaction networks by integrating structures: the p53 example. Mol Biosyst. 2009;5:1770–1778. doi: 10.1039/b905661k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, Bader GD, Ferrin TE. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. 2011;12:436. doi: 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.