Abstract
Autophagy is a cell ‘self-digestion’ pathway involving the synthesis, trafficking and delivery of autophagosomes to lysosomes for degradation. Beclin 1 is a core component of the class III phosphatidylinositol 3-kinase (PI3K-III) complex, which plays an important role in membrane trafficking and restructuring involved in autophagy, endocytosis, cytokinesis and phagocytosis. To date Beclin 1 has largely been characterized in the context of autophagy; it modulates the lipid kinase activity of PI3K-III catalytic unit VPS34, which generates phosphatidylinositol 3-phosphate (PI(3)P), enabling the recruitment of a number of autophagy proteins involved in the nucleation of the autophagosome. Beclin 1 seems to function as an adaptor for recruiting multiple proteins that modulate VPS34. The recent identification of Beclin 1 protein modifications has shed light on its regulation in autophagy, and the discovery of non-autophagy functions of Beclin 1 has expanded our view of Beclin 1's involvement in tissue homeostasis and human diseases.
Keywords: Beclin 1, Atg6, Vps30, VPS34, PIK3C3, Phosphatidylinositol 3-kinase, Phosphatidylinositol 3-phosphate, UVRAG, Vps38, Atg14L, RUBICON, Bif-1, Bcl-2, Vps15, p150, Autophagy, Endocytosis, Phagocytosis, Apoptosis, Neurodegeneration, Phosphorylation, Membrane trafficking, Alzheimer's disease, BH3, Coiled coil, Pathobiology
Introduction
Autophagy is a catabolic, intracellular membrane trafficking pathway exhibited in all eukaryotes from yeast to mammals [1] and misregulation of autophagy in humans is linked to multiple diseases including neurodegenerative diseases [2]. It is one of two major degradative pathways in mammalian cells and is characterized by the formation of double-membrane vesicles called autophagosomes that fuse with the lysosome for degradation. Autophagy generates the means for survival by degrading cytosolic proteins and whole organelles and recycling these back to the cytosol as amino acids and macromolecules. Autophagy is induced in response to nutrient deprivation as its most evolutionarily conserved function, but autophagy induction is also triggered by cellular stress and the accumulation of protein aggregates and damaged organelles, especially in higher organisms [3]. As a result, autophagy maintains cellular homeostasis by clearing the cell of misfolded or long-lived proteins and damaged parts. This is especially important in the brain; recent genetic and molecular evidence shows that neurons rely on basal autophagy to ward off intracellular aggregate accumulation and resulting neurotoxicity [4].
The action of the autophagy-regulating class III PI3-kinase (PI3K-III), also known as VPS34 or PIK3C3, and its Beclin 1-containing complex is required for nucleation of the phagophore and in turn, the protective functions of autophagy [5]. This lipid kinase complex catalyzes the phosphorylation of phosphoinositides to produce Phosphatidylinositol(3)P (PI(3)P) [6] and inhibition of PI3K-III (treatment with 3-methyladenine or wortmannin for example) and subsequent decrease of PI(3)P inhibits autophagy [7]. Through PI(3)P production, the Beclin 1-VPS34 complex enables the recruitment of and provides a platform for important autophagy proteins involved in autophagosome biogenesis such as the mammalian homolog of Atg18, WIPI2 [8] and the omegasome protein DFCP1 [9]. Beclin 1-VPS34 complexes are also required for proper maturation of autophagosomes [10].
Beclin 1 (BECN1) was the first-described mammalian autophagy protein [11] and the first-identified mammalian homolog of an essential yeast autophagy gene (Atg6) [12]. Not only was Beclin 1 shown to be a positive regulator of autophagy, but also the gene BECN1 is a haplo-insufficient tumor suppressor [12],[13],[14] that shows reduced expression or is monoallelically deleted in several cancers [15]. Beclin 1 is required for embryonic development, as Becn1 knock-out mice die early during embryogenesis [13]. The death is earlier and distinct from Atg5 and Atg7 knock-out mice, which survive until the perinatal stage but cannot survive the starvation period after the switch from placental derived nutrients [16].
Beclin 1 is a scaffold/adaptor protein with three prominent domains: Bcl-2-homology-3 (BH3), coiled coil (CCD) and evolutionarily conserved (ECD) (from N- to C-terminal) (Figure 1A). Beclin 1 was first characterized as a Bcl-2 binding protein [11] and early studies found this interaction to be autophagy-inhibitory, though the precise mechanisms are still unclear. The CCD provides a platform for many important Beclin 1 protein interactions including those with UVRAG, Atg14L (also named Barkor) and RUBICON [17]. VPS34 binds Beclin 1 at its ECD [18], which was recently described through a crystallization study as a novel sort of membrane binding domain [19]. The Beclin 1-VPS34 Class III PI3K complex is known to exist in multiple forms with distinct functions. The recruitment of various regulators through their physical interaction with the scaffolding protein Beclin 1 and the subsequent modulation of VPS34 activity is an essential regulatory mechanism for many VPS34-dependent cellular processes [20]. Yet the molecular details of such regulation as mediated by the physical interaction between Beclin 1 and various modulators is not well understood. In the following section we will describe in detail the different Beclin 1 binding proteins and resulting complexes. The specific interaction between Beclin 1 and its binding partners regulate a vast array of biological and pathophysiological conditions such as development, pathogen infection, heart disease and neurodegeneration [3].
Figure 1. Important modulation of Beclin 1 protein structure and Beclin 1's role in various cellular pathways.
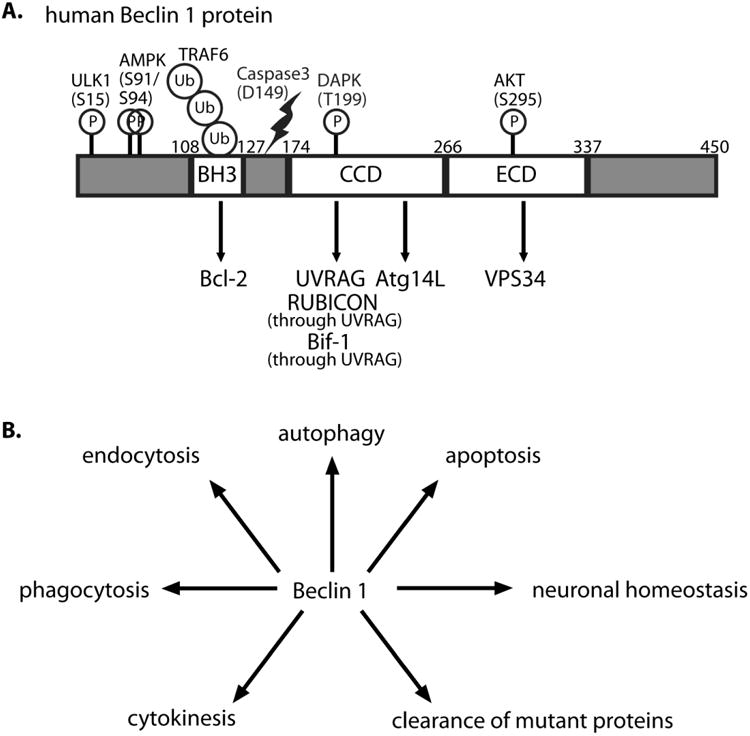
A. The protein structure of human Beclin 1 protein including its BH3, CCD, and ECD domains. The regions of Beclin 1 for binding Bcl-2, UVRAG, Atg14L, VPS34, RUBICON, and Bif-1 are shown with arrowed lines. Reported phosphorylation sites [44-47], a ubiquitination site [50] and a Caspase 3 cleavage site [53] are shown.
B. Beclin 1 is reported to be involved in a vast array of cellular processes including autophagy, apoptosis, and endocytosis.
Multiple Beclin 1-VPS34 complexes
In yeast there are two types of Atg6-Vps34 complexes with distinct functions: Complex I is autophagy-specific and Complex II controls the vacuolar protein sorting (Vps) pathway [21]. Both Complex I and II contain the shared components Vps34, Atg6/Vps30 (the yeast homologue of Beclin 1), and the regulatory/membrane-anchoring protein Vps15. Complex I contains Atg14 as a unique factor for autophagy, while Complex II includes Vps38, which is required for the Vps pathway. Mammalian homologs p150 (PIK3R4), VPS34 (PIK3C3) and Beclin 1 remain as conserved core components in the PI3K-III complex. However, more than two Beclin 1-VPS34 complexes exist in mammals and these often have multiple function that are not well defined [17]. By recruiting different binding partners and forming distinct PI3K-III complexes, Beclin 1 modulates PI3K-III activity and thus regulates autophagy at multiple stages including nucleation and maturation (of note, Vps34 is the only class III phosphoinositide 3 kinase in mammals [22]). Multiple recent studies including those from our lab have uncovered three main Beclin 1-VPS34 complexes in mammals. The Beclin 1-Atg14L complex [23],[24] and the Beclin 1-UVRAG complex [25],[26] are likely to be conserved with the yeast Atg6-Vps34 Complex I and Complex II, respectively. In addition, our studies demonstrate that the Beclin 1-Atg14L and Beclin 1-UVRAG interactions are of high affinity, representing stable VPS34 complexes. It is thought that the competition of Beclin 1-Atg14L versus Beclin 1-UVRAG leads to two mutually exclusive complexes [27]. Recently we showed that Beclin 1 dimerizes in an anti-parallel state that is metastable compared to Beclin 1-UVRAG or Beclin 1-Atg14L heterodimers [28]. This result suggests that the modulation of the homodimer-heterodimer transition by these partners may contribute to the molecular mechanisms that control the formation of various Beclin 1-VPS34 subcomplexes. Atg14L contains a unique ER membrane targeting domain and is thought to recruit autophagy machinery to the ER after induction [29]. Both Atg14L and UVRAG are thought to contribute to autophagosome maturation: Atg14L by sensing autophagosomal membrane curvature [30] and UVRAG by complexing with the C-Vps complex [31].
A third Beclin 1-VPS34 complex contains UVRAG as well as RUBICON, which is a negative regulator of autophagy [23],[24]. RUBICON also interacts with the small GTPase Rab7 [32], which may explain its inhibitory effect on autophagosome maturation. The protein Bif-1, also called SH3GLB1 or SH3-domain GRB2-like endophilin B1 is required for autophagy and interacts with Beclin 1 through a direct interaction with UVRAG [33]. Bif-1 is required for autophagosome membrane expansion and curvature through its N-BAR membrane curvature domain that binds the tethering factor ARF-GAP [34]. Bif-1 may have roles in non-autophagy membrane trafficking pathways, such as fission of Golgi carriers [6]. Recently Bif-1 was shown not to act on autophagy initiation but to regulate degradative endocytic traffic and in turn, autophagosome maturation [35].
Another important Beclin 1 binding protein is AMBRA1, a positive regulator of autophagy and a requirement for neural tube development [36]; it is also required for starvation-induced autophagy [37]. Interestingly, AMBRA1 has recently been implicated in mitochondrial dynamics and degradation [38],[39]. Beclin 1 also plays a role in apoptosis through its binding to the anti-apoptotic protein Bcl-2 [40]. Bcl-2 negatively regulates Beclin 1 by sequestering it away from its VPS34 kinase complex partners and thereby preventing autophagy [41]. Beclin 1 may not only be important for the balance of autophagosome formation and maturation, but may also play a role in modulating cell death versus survival by interacting with factors in the apoptosis pathways [42]. Through its differential binding to the various partners described above, the adaptor molecule Beclin 1 may act as a switch between the complexes in order to achieve the regulation of autophagy in a highly coordinated manner in mammals. Beclin 1 acts, therefore, as a master regulator of PI3K-III activity that controls autophagosome nucleation and maturation by toggling between the Atg14L, the UVRAG complex and the RUBICON complex [17].
Post-translational modifications of Beclin 1 in autophagy control
Recent research shows that Beclin 1 is modified post-translationally by phosphorylation, ubiquitination and cleavage allowing Beclin 1 to fine-tune PI3K-III activity and autophagy in response to various intracellular and extracellular signals (Figure 1A) [43]. Beclin 1 was first proposed to be a phospho-protein in a paper that showed DAPK phosphorylates Beclin 1 [44]. The phosphorylation resulted in autophagy activation due to disruption of the inhibitory Beclin 1-Bcl-2 interaction. More recently, Akt phosphorylation of Beclin 1 was demonstrated; loss of this phosphorylation increased autophagy and inhibited Akt-mediated tumorigenesis [45]. Most recently, demonstration of phosphorylation of Beclin 1 (as well as VPS34) by AMPK provides some idea of the mechanism of autophagy induction after glucose withdrawal [46]. Similarly, phosphorylation of Beclin 1 by the autophagy-initiating kinase ULK1 responded to amino acid starvation by enhancing the autophagy-promoting activity of the Beclin 1-Atg14L complex [47]. This is conserved to C. elegans Atg6 and of interest therapeutically, a Beclin 1 phosphomimic mutant greatly stimulated autophagy [47]. A second study directly connects the ULK1 complex with Beclin 1 complexes: novel autophagy regulators SCOC and FEZ1 were shown to complex with both ULK1 and UVRAG [37]. Collectively these studies not only shed light on the cross-talk between two autophagy kinase complexes (ULK1 protein kinase and VPS34 lipid kinase) essential for phagophore initiation, but also illustrate how the balance between autophagosome initiation and maturation could be mediated by toggling between Beclin 1 and UVRAG [48].
Ubiquitination is an alternative means of Beclin 1 modulation. TRAF6 K63-linked ubiquitination of Beclin 1 was shown to induce autophagy [49]; suggesting a possible feedback loop, the authors also demonstrated that A20 inhibits autophagy by deubiquitinating Beclin 1, thereby inhibiting the autophagy-activating ubiquitination by TRAF6. It was shown separately that the TRAF6 addition of K63-linked Ubiquitin is in Beclin 1's BH3 domain [50]. NEDD4 also polyubiquitinates Beclin 1, which leads to its degradation thereby inhibiting autophagy [51]. Conversely, it was show that Beclin 1 is deubiquitinated by USP10 and USP13 and adding complexity, Beclin 1 itself controlled the protein stabilities of USP10 and USP13 by regulating their deubiquitinating activities, in turn regulating the levels of tumor suppressor p53 [52].
Furthermore, proteolytic cleavage of Beclin 1 adds to its regulation and thus controls its activities in cells [10]. Beclin 1 is subject to Caspase 3 cleavage [53] and it was recently shown that this proteolytic cleavage depleted Beclin 1 in the brains of patients with Alzheimer's disease, contributing to adverse pathology [54]. Beclin 1 is also a substrate of Calpain cleavage [55]. These data contribute to an essential and growing knowledge of how Beclin 1, a non-enzymatic scaffold protein, is essential to so many important cellular pathways.
Potential role of Beclin 1 in endocytic and phagocytic pathways
Accumulating evidence now shows that Beclin 1 is involved in functions beyond autophagy [10]. The endocytic pathway was known to contribute to autophagy [56] but it was recently reported that endocytic traffic itself was disrupted in C. elegans lacking BEC-1 [57] and Drosophila lacking Atg6 [58]. The presence of Atg6/Vps30 in the CVT-directing yeast Vps34 Complex II and Beclin 1 in the mammalian homolog complex containing VPS34 and UVRAG, which is linked to degradation, implicates Beclin 1 in non-autophagy membrane traffic. In mammalian cells, however, the role of Beclin 1 in endocytosis is controversial [59],[35]. It was previously shown that VPS34 is an effector of the small GTPase Rab5 [60] and that Rab5 binds to the Beclin 1-VPS34 complex in order to form autophagosomes [61]. Given the role of Rab5 in early endosome traffic and endosome maturation [62], it remains possible that Beclin 1 may function in the Rab5-VPS34 mediated endocytic pathway. Very recently, the regulation of endocytosis and autophagosome-lysosome fusion by Beclin 1-VPS34 was demonstrated to be critical in highly specialized podocytes, contributing to important kidney homeostasis [63]. Alternatively, Beclin 1 may regulate fusion of endosomes and autophagosomes (amphisome formation). In addition, it was recently shown that Beclin 1 is required for autophagosome fusion to lysosomes [64] strengthening the idea of a strong relationship between the autophagic and endocytic pathways [65], perhaps mediated by Beclin 1.
Beclin 1 also plays a role in phagocytosis. It was shown that Beclin 1-dependent PI(3)K activity and not Atg5-dependent or -independent autophagy is required for phagocyte engulfment apoptotic cells [66]. Additional evidence showed that during phagosome maturation, the interaction of Vps34 with Rab5 and dynamin was important for the engulfment of apoptotic cells [67], most likely through production of PI(3)P on the phagosome membrane. Interestingly, it was reported that Beclin 1 was involved in the clearance of apoptotic cells during embryonic development by regulating the expression of a phospholipid other than PI(3)P, phosphatidylserine (PS) [68]. Last, Beclin 1 is implicated in cytokinesis: the Beclin 1-UVRAG complex mediates the cell division process [35] and it is suggested that this occurs in an autophagy-independent manner [69].
The role of Beclin 1 in neurodegeneration and neuroprotection
A recent surge of research has revealed that the cellular process of macroautophagy (involving critical steps such as formation, trafficking, fusion and degradation of autophagosomes) in which Beclin 1 is intimately involved is evolutionarily conserved from yeast to mammals. But the regulation and functional adaptation of autophagy in the cells and tissues of higher organisms are poorly understood, and even more so in the central nervous system (CNS) [4]. Our previous study reported early evidence linking Beclin 1 to excitotoxicity in mouse brains containing a Lurcher mutation in GluRd2 via the PDZ-containing protein nPIST [70]. However, the exact mechanism whereby Beclin 1 mediates autophagy dysfunction that contributes to neurodegeneration is unclear [71]. In addition, closed head injury results in upregulation of Beclin 1 protein perhaps as part of a stress response in the brain including upregulation of autophagy [72].
Emerging evidence demonstrates that autophagy is primarily responsible for the turnover of aggregate-prone proteins or macromolecular complexes. In fact, an increasing body of work has shown the neuroprotective function of Beclin 1 through removal of disease-related proteins in the CNS including APP metabolites [73],[74], α-synuclein [75], Huntingtin [76], and mutant ataxin [77]. In an Alzheimer's disease (AD) mouse model, reduced Beclin 1 expression causes an increase in intraneuronal and extracellular amyloid beta accumulation and accelerated neurodegeneration. Remarkably, AD-associated amyloid pathology in mice can be rescued by re-introduction of Beclin 1, strengthening the idea that it is a potential target protein for AD treatments [73]. Similar results were seen when increased expression of Beclin 1 reduced accumulation of alpha-synuclein and repaired associated neuritic alterations caused by the toxic accumulation of alpha-synuclein in a mouse model for PD [75]. Very recently it was shown that Beclin 1 is depleted in AD disease brains as a result of caspase-3 cleavage [54]. Furthermore, overexpression of Beclin 1 was associated with the clearance of mutant ataxin-3 and neuroprotection in a model for Machado-Joseph disease [78].
These results suggest the therapeutic potential of modulating Beclin 1-associated PI3K-III activity for the treatment of neural proteinopathies. However, whether the neuroprotective role of Beclin 1 is mediated through strictly autophagy-dependent or independent pathways is unclear. Endocytic trafficking is indeed particularly important for synaptic function [79], and, thus, the effects on neurodegenerative disease may reflect both the endocytic and autophagic functions of Beclin 1. Therefore, it is important to dissect molecular mechanism underlying the Beclin 1 protection against neurodegeneration. For instance, what specific Beclin 1-VPS34 subcomplex or Beclin 1 function is involved in the removal of pathological mutants of disease proteins and in turn, neuroprotection remains to be answered. At this time, the function of Beclin 1 in the brain has not been elucidated and brain or neuron type-specific conditional knock-out models should be characterized. In addition, the sequence information in Beclin 1 required for the interaction with various binding partners should be identified for the generation of specific binding mutants of Beclin 1. These mutants can then be used to dissect the specific Beclin 1-VPS34 subcomplex important for neuroprotection in different neuropathological conditions.
Conclusions
Beclin 1 is implicated in multiple biological processes and it is critical to understand Beclin 1's multiple functions in higher organisms as its deregulation has been linked to multiple diseases [3]. Previous thought might have placed primary responsibility on the role of Beclin 1 in autophagy because in mammals it has almost exclusively been characterized in the context of autophagy. Through recruitment of critical effectors such as UVRAG and Atg14L, Beclin 1 coordinates overarching membrane trafficking pathways and cellular functions that are critical to mammalian development and cell viability (Figure 1B). Autophagy maintains cellular homeostasis: for instance, neurons rely on basal levels of autophagy to remove intracellular toxic aggregates and in neurodegenerative disease conditions, autophagy clears disease-associated mutant proteins. Autophagy-enhancing drugs could perhaps be of use as anti-cancer or anti-aging therapies and may even prevent neurodegeneration by clearing toxic aggregates. We must, however, be cautious; autophagy can be cytotoxic when autophagosome synthesis is activated but autophagosome degradation and substrate clearance is impaired [80]. We need highly specific drug targets such as components in the Beclin 1-VPS34 complex, which play a role in various critical pathways including autophagy, endocytosis and apoptosis. Two important lines of study have shown that decreased levels of Beclin 1 is causal in cancer [12] and Alzheimer's disease [73]; the authors attributed these results to Beclin 1's role in autophagy, but in light of recent studies that implicate Beclin 1 in other cellular processes, the mechanisms should be reexamined. Of particular interest, Beclin 1 has been recently linked to multiple neurodegenerative diseases, however, the function of Beclin 1 in the brain has not been elucidated. Nonetheless, characterization of Beclin 1 in the progression of neurodegenerative disorders and other diseases will assist in developing therapies.
Many questions remain to be answered: what triggers differential Beclin 1 complexing with VPS34 and other components, how does Beclin 1 regulate specific functions of VPS34, how do Beclin 1 post-translational modifications affect its function, and what are the non-autophagy functions of Beclin 1? Multiple lines of evidence have linked altered expression of Beclin 1 to several major human diseases such as cancer, infectious diseases and, in particular, neurodegenerative disorders, but the exact mechanism underlying the influence of Beclin 1 levels in disease progression is unknown. It is thus imperative to understand the precise molecular mechanism whereby Beclin 1 regulates VPS34 functions in autophagy and other membrane trafficking pathways, the disruption of which may underlie the pathogenesis of multiple diseases.
Footnotes
Compliance with Ethics Guidelines: Conflict of Interest: Nicole C. McKnight and Zhenyu Yue declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent: This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Recently published papers of particular interest have been highlighted as:
• Of importance
•• Of major importance
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKnight NC, Mizushima N, Yue Z. The Cellular Process of Autophagy and Control of Autophagy in Neurons. In: Yue Z, Chu CT, editors. Autophagy of the Nervous System. Singapore: World Scientific Publishing Co. Pte. Ltd.; 2012. pp. 3–35. [Google Scholar]
- 5.Yue Z, Zhong Y. From a global view to focused examination: understanding cellular function of lipid kinase VPS34-Beclin 1 complex in autophagy. J Mol Cell Biol. 2010;2:305–307. doi: 10.1093/jmcb/mjq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 9.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M, Vandenabeele P. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8:6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- 11.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 13.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends in cell biology. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 19•.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. The crystal structure of Beclin 1's ECD unveils a novel class of membrane-binding domain that prefers cardiolipin-enriched lipids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He C, Levine B. The Beclin 1 interactome. Current opinion in cell biology. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. The EMBO journal. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 26.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662. doi: 10.1038/ncomms1648. Beclin 1's CCD structure was revealed through crystalization. It binds itself to form a homodimer with an imperfect dimer interface; in turn, the more-stable Beclin 1-Atg14L or Beclin 1-UVRAG heterodimer interactions and favored. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. Atg14L binds directly to autophagosomes through its BATS (Barkor/Atg14L autophagosomes targeting sequence) domain and Atg14L senses autophagosomal membrane curvature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell death and differentiation. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 37•.McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. The EMBO journal. 2012;31:1931–1946. doi: 10.1038/emboj.2012.36. Three novel modulators of mammalian starvation-induced autophagy, SCOC, FEZ1 and WAC, were identified. SCOC and FEZ1 complex with both the ULK1 kinase complex and UVRAG, linking the two autophagy-essential kinase complexes together. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. The EMBO journal. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. AMBRA1 binds to BCL-2 and competes with binding to Beclin 1, perhaps regulating both autophagy and apoptosis. AMBRA1 is recruited to Beclin 1 during starvation-induced autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell death and differentiation. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Molecular cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abrahamsen H, Stenmark H, Platta HW. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: Tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS letters. 2012;586:1584–1591. doi: 10.1016/j.febslet.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 44.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO reports. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. Beclin 1 is a phosphorylation target of Akt, which is an autophagy-inhibiting process by increasing Beclin 1 binding to 14/3/3 and the vimentin intermediate filament complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress andautophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. AMPK phosphorylates both VPS34 and Beclin 1 (at S91/S94) in response to nutrient starvation and this differential phosphorylation is dictated by Atg14L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activatingVPS34 lipid kinase. Nature cell biology. 2013 doi: 10.1038/ncb2757. Amino acid withdrawal induces ULK1 phosphorylation of Beclin 1 (at S15) which activates autophagy by promoting the activity of Atg14L-containing VPS34 complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirth M, Joachim J, Tooze SA. Autophagosome formation-The role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013 doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik SA, Orhon I, Morselli E, Criollo A, Shen S, Marino G, BenYounes A, Benit P, Rustin P, Maiuri MC, et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene. 2011;30:3918–3929. doi: 10.1038/onc.2011.104. [DOI] [PubMed] [Google Scholar]
- 51•.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. The Biochemical journal. 2012;441:399–406. doi: 10.1042/BJ20111424. The ubiquitin ligase Nedd4 polyubiquitinates Beclin 1 thereby controlling its stability (degradation). Nedd4 interacts with Beclin 1's C-terminal PY domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. Beclin1 Controls the Levels of p53 by Regulating the Deubiquitination Activity ofUSP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. Spautin-1, a small molecule inhibitor of autophagy, works by promoting the degradation of Beclin 1, stimulated by USP10 and USP13 ubiquitin-specific peptidase inhibition. Beclin 1 also controls the protein stabilities of USP10 and USP13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Rohn TT, Wirawan E, Brown RJ, Harris JR, Masliah E, Vandenabeele P. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer's disease brain. Neurobiology of disease. 2011;43:68–78. doi: 10.1016/j.nbd.2010.11.003. Beclin 1 levels dictate Alzheimer's disease pathology in the brains of Alzheimer's disease patients. Beclin 1 levels are decreased as a result of Caspase 3 cleavage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, Rotiroti D, Morrone LA, Bagetta G, Corasaniti MT. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 57•.Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. The Beclin 1 ortholog BEC-1 is required for the endocytic trafficking in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013 doi: 10.1242/dev.089490. The Beclin 1 ortholog Atg6 is required for the endocytic trafficking in Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. Journal of cell science. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 60.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nature cell biology. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 61.Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. Journal of cell science. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature reviews. Molecular cell biology. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 63.Bechtel W, Helmstadter M, Balica J, Hartleben B, Schell C, Huber TB. The class III phosphatidylinositol 3-kinase PIK3C3/VPS34 regulates endocytosis and autophagosome-autolysosome formation in podocytes. Autophagy. 2013;9 doi: 10.4161/auto.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gladue DP, O'Donnell V, Baker-Branstetter R, Holinka LG, Pacheco JM, Fernandez Sainz I, Lu Z, Brocchi E, Baxt B, Piconne ME, et al. Foot and Mouth Disease Virus non structural protein 2C interacts with Beclin1 modulating virus replication. J Virol. 2012 doi: 10.1128/JVI.01610-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lamb CA, Dooley HC, Tooze SA. Endocytosis and autophagy: Shared machinery for degradation. Bioessays. 2013;35:34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- 66.Konishi A, Arakawa S, Yue Z, Shimizu S. Involvement of Beclin 1 in engulfment of apoptotic cells. The Journal of biological chemistry. 2012;287:13919–13929. doi: 10.1074/jbc.M112.348375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nature cell biology. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 69.Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nature cell biology. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 70.Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35:921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 71.Yue Z, Friedman L, Komatsu M, Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta. 2009;1793:1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diskin T, Tal-Or P, Erlich S, Mizrachy L, Alexandrovich A, Shohami E, Pinkas-Kramarski R. Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma. 2005;22:750–762. doi: 10.1089/neu.2005.22.750. [DOI] [PubMed] [Google Scholar]
- 73.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaeger PA, Pickford F, Sun CH, Lucin KM, Masliah E, Wyss-Coray T. Regulation of amyloid precursor protein processing by the Beclin 1 complex. PloS one. 2010;5:e11102. doi: 10.1371/journal.pone.0011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson's and Lewy body diseases. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:13578–13588. doi: 10.1523/JNEUROSCI.4390-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. The Journal of biological chemistry. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 77.Nascimento-Ferreira I, Santos-Ferreira T, Sousa-Ferreira L, Auregan G, Onofre I, Alves S, Dufour N, Colomer Gould VF, Koeppen A, Deglon N, et al. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain : a journal of neurology. 2011;134:1400–1415. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- 78•.Nascimento-Ferreira I, Nobrega C, Vasconcelos-Ferreira A, Onofre I, Albuquerque D, Aveleira C, Hirai H, Deglon N, Pereira de Almeida L. Beclin 1 mitigates motor and neuropathological deficits in genetic mouse models of Machado-Joseph disease. Brain : a journal of neurology. 2013;136:2173–2188. doi: 10.1093/brain/awt144. Beclin 1 overexpression prevents neuronal dysfunction and neurodegeneration associated with spinocerebellar ataxia type 3. [DOI] [PubMed] [Google Scholar]
- 79.Plowey ED, Chu CT. Synaptic dysfunction in genetic models of Parkinson's disease: a role for autophagy? Neurobiology of disease. 2011;43:60–67. doi: 10.1016/j.nbd.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]


