Loss of endothelial layer in blood vessel by injury exposes subendothelial extracellular cellular matrix proteins such as collagen, platelets subsequently adhere to collagen-bound von Willebrand factor and other matrix components [1, 2]. Upon platelet adhesion, thromboxane A2 is synthesized from arachidonic acid by cyclooxygenase-1 in platelets and is released. In addition, granules containing various agonists including ADP are released [3]. Platelet agonists are also released from injured cells or generated by the proteolysis during coagulation [4]. These molecules bind to their receptors on platelet surfaces and induce intracellular signaling resulting in platelet aggregation, a critical events in achieving haemostasis to prevent further bleeding at the site of the injury. A platelet adhesion molecule integrin αIIbβ3, a heterodimeric type I transmembrane protein consisting of α and β subunit, is a key component of the platelet aggregation [5]. When activated by the intracellular signaling, the integrin become highly adhesive toward its ligand fibrinogen in the circulating blood stream. Since the fibrinogen has multiple binding sites for the integrin, their interaction can crosslink platelets to facilitate aggregation. In this way, integrin αIIbβ3 plays a vital role in hemostasis.
Proper integrin regulation prevents integrin αIIbβ3 from engaging their ligand and thus avoid the formation of intravascular thrombi under normal conditions. Inappropriate platelet aggregation, usually on vessels involved by atherosclerosis, can be dangerous. Uncontrolled formation of thrombus can block blood flow and cause tissue infarction. Therefore, the affinity of integrin αIIbβ3 is tightly regulated so that the integrin is kept in low affinity state (inactive) and enters a high affinity state (active) only when and where needed. Many structural studies suggest that there are at least two different conformations of extracellular domain corresponding to each affinity state [6-9]. The integrin seems to be inactive when the extracellular domain is folded as V-shape with the ligand binding head piece facing the membrane (Fig. 1A), while it becomes highly adhesive toward its ligand when the extracellular domain is extended (Fig. 1B). These conformational changes can be induced by intracellular signaling, so called “inside-out” signaling [10]. In this review, we will describe how the inside-out signaling can be transmitted to extracellular domain to make such conformational changes in extracellular domain, as well as many intracellular molecular players involved in the signaling. This review will focus on our current understanding, and we apologize for any omissions that this focus engenders.
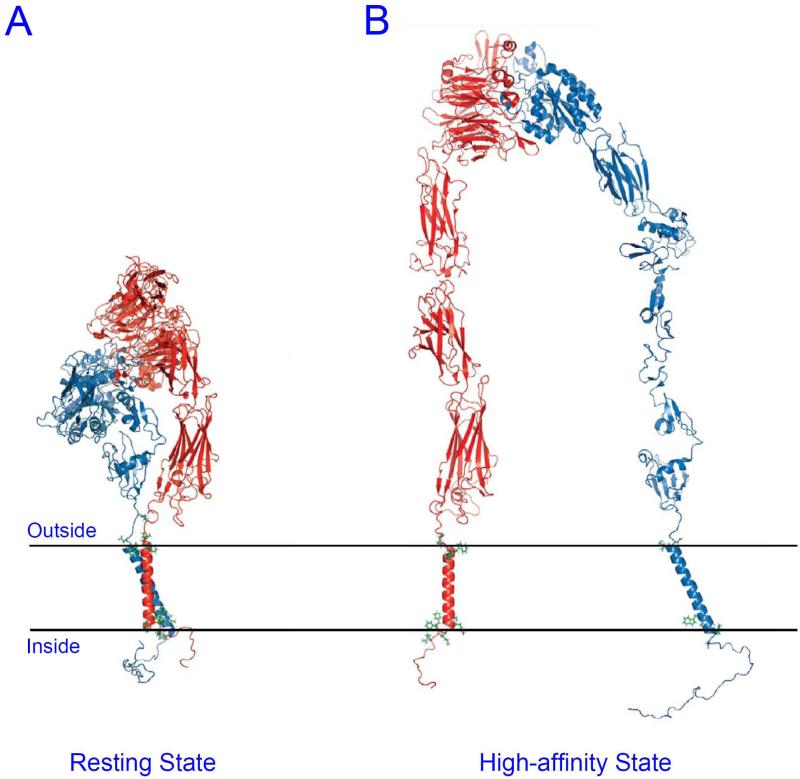
Figure 1. Structural model of proposed integrin αIIbβ3 affinity states.
(A) In resting state, α and β TMDs interact with each other, and the extracellular domains folded with the ligand binding site facing cell membrane. (B) In high affinity state, α and β TMDs are separated, and the extracellular domains are extended.
Disrupting the Transmembrane Domain (TMD) interaction between α and β subunits induces integrin activation
Earlier mutational study suggested that there is an electrostatic interaction in the membrane-proximal region of cytoplasmic domain (Fig. 2A), between Arg995 of αIIb and Asp723 of β3 [11]. Charge reversal (e.g. αIIb(R995D), β3(D723R)) of either residue induces integrin activation but combined charge reversals of both residues makes the integrin inactive again. This observation led to a hypothesis that an interaction between cytoplasmic tails of integrin αIIb and β3 subunits can keep the integrin inactive and breaking the interaction can activate integrins. In addition to the charged residues, many other activating mutations are also found in the membrane proximal regions. For example, mutating two Phe in the membrane proximal region of αIIb (Fig. 2A) into Ala makes the integrin constitutively active [11]. Since the membrane proximal regions of integrins contain unusual hydrophobic patches which may be embedded in membrane together with the transmembrane domain (Fig. 2A), those activating effects induced by mutations in the regions were interpreted that membrane embedding of those membrane proximal region may be important for regulating integrin affinity [12].
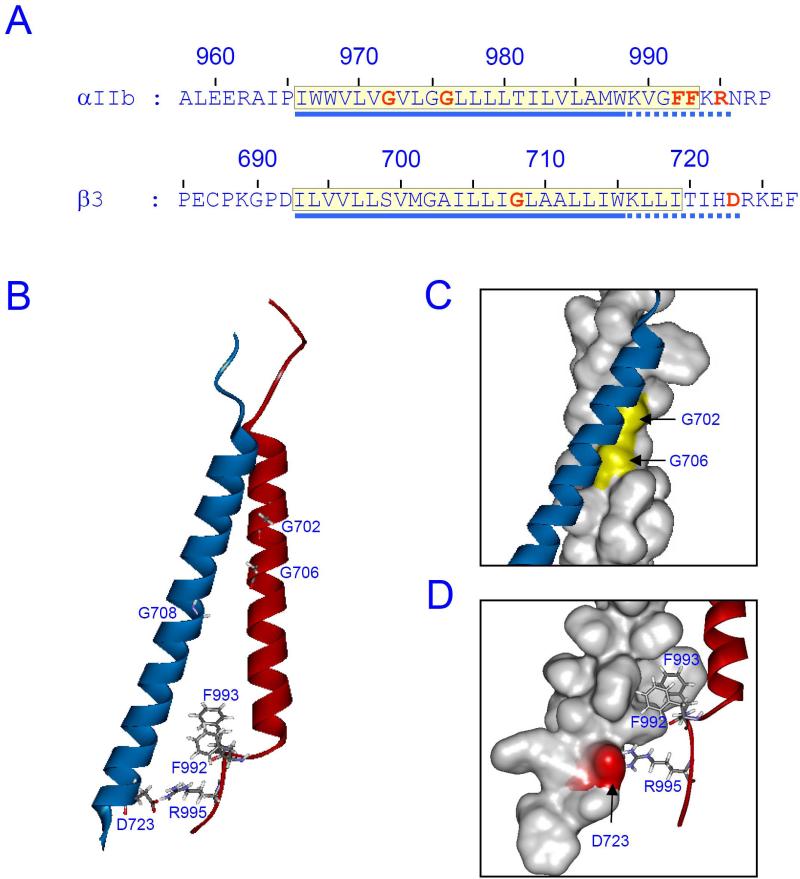
Figure 2. Integrin αIIbβ3 TMD structure.
(A) Amino acid sequences of integrin αIIbβ3 around TMD. TMD regions are highlighted in yellow box. Amino acid residues mentioned in the text are indicated in red. The conventional TMD regions and the membrane proximal regions are indicated with blue solid and dotted lines, respectively. (B) NMR structure of integrin αIIb (red) and β3 (blue) (PDB entry 2k9j) is shown. The amino acid residues indicated in (A) are shown as stick models. (C) The αβ TMD interaction through GXXXG motif, or outer membrane clasp, is highlighted. Surface of αIIb TMD is shown with glycines in the GXXXG motif indicated as yellow. (D) The αβ TMD interaction through the Asp723-Arg995, or inner membrane clasp, is highlighted. Surface of β3 TMD with the Asp723 indicated as red.
Activating mutations of integrin αIIbβ3 are also found in transmembrane domains (TMDs). Interestingly, most of the activating mutations found were substitutions of Gly resides in both αIIb (Gly972, Gly976) and β3 TMDs (Gly708) with amino acids containing bulky side chain, such as Leu, Ile, or Asn [13-15]. Especially, two Gly in αIIb TMD are part of a GXXXG motif, a motif important in TMD helix-helix association [16] in other transmembrane proteins. Therefore, the α-β TMD packing guided by a groove formed by those Gly was also considered important to keep the integrin inactive. However, this finding is not consistent with other previous mutational and structural study. For example, TMDs and membrane proximal regions of both αIIb and β3 were considered to form continuous helices [17] and the packing of TMDs via those Gly residues makes TMDs to cross each other [18]. The crossing angle between the TMDs, predicted by the structural studies[18], does not permit the known αIIb Arg995-β3 Asp723 electrostatic interaction (at least 10Å apart in the structure). Nevertheless, mutation either in the αIIb GXXXG motif or in the αIIb or β3 membrane proximal regions disrupts the αIIbβ3 TMD-interaction [19], suggesting simultaneous formation of both the TMD packing (later termed as outer membrane clasp or OMC) and the membrane-proximal region interaction (later termed as inner membrane clasp or IMC) are required to maintain an inactive integrin. Therefore, a structural explanation of these activating mutations is of great interest.
Recently, the NMR structure of integrin αIIbβ3 TMD complex embedded in lipid bilayer was solved (Fig. 2B) [20]. The structure shows that αIIb and β3 TMDs pack together via the Gly residues with a vertical αIIb TMD and tilted β3 TMD, which forms the OMC (Fig. 2C). β3 TMD and the membrane proximal region make a continuous helix as expected. The hydrophobic amino acid patch in β3 membrane-proximal region is inserted into membrane as a part of TMD and makes long and tilted helix. In contrast, αIIb TMD makes short and straight helix that ends at Gly991. The membrane proximal region next to the Gly991 turns toward β3, which enables the two Phe (Phe992 and Phe993) outside of the transmembrane helix to be embedded into membrane (Fig 2D) generating an unusual transmembrane domain. In addition, the turning of membrane proximal region places the Arg995 residue close to Asp723 of β3, thus allowing formation of IMC (Fig 2D). Thus, the structure of αIIbβ3 TMD complex, explains how all the mutations discussed above can activate integrin.
Talin binding to the integrin β taildisrupts the TMD interaction
Talin, a 270kDa cytoplasmic protein, can bind to integrin β tails through its 50kDa head domain [21, 22] and has multiple actin binding sites in its 220kDa rod domain [11, 23, 24]. Over-expression of the integrin-binding head domain in cells strongly activates β3 [21], β1 [25] and β2 integrins [26, 27]. Genetic ablation of talin is embryonic lethal due to defects in cell migration at gastrulation [28]. Tissue specific deletion of talin in cells of the megakaryocyte lineage caused severe defects in platelet aggregation and agonist-stimulated integrin activation. Consequently mice that have lost talin in megakaryocytes and platelets have prolonged bleeding time and experience frequent pathological bleeding [29, 30]. Conditional knockout of talin in macrophages result in defective phagocytosis and β2 integrin mediated adhesion [26]. Thus talin is an important regulator of integrin activation.
Talin head domain contains a FERM domain, (band 4.1, Ezrin, Radixin, and Moesin homology domain), which is usually further divided into F1, F2 and F3 subdomains [31]. Talin head has an extra N-terminal F0 subdomain [32]. The integrin binding PTB (phospho-tyrosine binding) domain is located within F3 and is sufficient to activate β3 integrin [33]. What features in talin made it unique in its ability to activate integrin amongst the FERM domain containing or PTB domain containing proteins? Comparing to other FERM domain proteins, talin has two integrin binding sites: a strong binding site to the β3- W739DTANNPLY747 sequence involving mainly the S5 strand of the F3 domain and a hydrophobic pocket formed by R358, A360, and Y377 of F3 (membrane distal (MD) interaction) [34, 35], and a weak binding site to the β3-H722DRKEFAKFEEER726 involving the loop between S1-S2 strand (membrane proximal (MP) interaction) [34]. This weak interaction between talin F3 and the integrin MP interaction site depends upon the presence of the strong talin-integrin MD interaction. In addition to two integrin binding sites, the talin F2 and F3 domain also have multiple positively charged residues on its surface that can interact with the negatively charged phospholipids and potentially favor the right configuration of talin-integrin interaction [34, 36]. A patch of basic residues in talin F1 domain has also been identified [32]. A recent crystal structure of talin head domain revealed a novel extended arrangement of talin F0, F1, F2 and F3 domains, which is different from the typical compact cloverleaf structure observed in most FERM domains [37]. Thus, the unique extended structure of the talin FERM domain allows simultaneous contact of the basic residues on F1, F2, and F3 with lipids [37].
Since we have detailed understanding about the structural mechanism of how integrin activated by disruption of TM and cytoplasmic domain interactions, the obvious question to ask is how these unique features of talin-integrin interactions lead to disruption of integrin cytoplasmic and TM interactions? Talin is recruited to the plasma membrane by intracellular signals such as Rap1 and RIAM (Rap1 interacting adaptor molecule) [38-41]. At the plasma membrane, talin will then engage with β integrin cytoplasmic tails. Whether talin is actively recruited to integrin β tail by some signaling events or it simply encounters integrin through an efficient two dimensional diffusion once at the plasma membrane is still unclear. Upon engaging with integrin, talin interacts with β integrin cytoplasmic tail through its strong MD binding site. Any mutation to integrin or talin that interrupts this strong site interaction would abolish integrin activation by talin [34, 35]. Following this strong site interaction, the weak talin-β tail MP interaction and talin-membrane interaction occurs. These sequential interactions are supported by the observation that talin can still bind to soluble β integrin cytoplasmic tail even when the weak interaction site is disrupted in a system where the membrane is absent [34]. Whether the weak talin-β3 MP interaction and the talin-membrane interaction occur sequentially or simultaneously is not clear.
The engagement of talin with plasma membrane and with two β integrin cytoplasmic tail binding sites as well as the restricted topology of β transmembrane and cytoplasmic domain by lipid bilayer probably puts the talin-β integrin complex in a precise configuration poised to trigger the activation of integrin. Mutations in β integrin or in talin that disrupt the weak talin-β cytoplasmic interaction site have small or no effects on the affinity of talin for the β integrin cytoplasmic tail but markedly impair integrin activation [34]. Platelets expressing talin with the same point mutation that disrupts talinintegrin β3 MP weak interaction site have defects in integrin activation and platelet aggregation [42]. Mutations in F1, F2 or F3 that interrupt with the talin-lipid interaction also impaired integrin activation by talin [32, 34, 36]. The final definitive proof is provided by an in vitro reconstitution of inside-out activation with purified talin head and integrin nanodiscs. Talin head is sufficient to activate integrins in a purified system only when interactions with membrane and two talin binding sites in β integrin cytoplasmic tail are present [9]. Thus the requirement for the multiple interactions between talin, integrin and membrane is well established by multiple lines of work.
The more interesting question though, is how this configuration of talin, integrin and membrane can trigger integrin activation. There are several possible mechanisms. The first one is that talin cause a motion, possibly tilting, pistoning or lateral motion in integrin β transmembrane and cytoplasmic domain, which disrupt integrin α and β subunit interactions at both IMC and OMC. This hypothesis is supported by a recent molecular dynamics simulation which showed that talin can change the tilting angle of integrin β subunit [43]. Since the tilting angle is important to maintain optimal α and β subunit IMC and OMC, a tilting angle change would disrupt these interactions leading to integrin activation [19, 20]. The second hypothesis is that this precise configuration brings Talin Lys324 close to Asp723 of the integrin β3 subunit, disrupting the Asp-Arg (R995-D723 in αIIbβ3) salt bridge, thus favoring integrin activation. This hypothesis is supported by the observation of talin Lys-integrin β Asp interaction in a crystal structure of talin F2F3-β1D complex [36]. The third hypothesis is that talin might come in between integrin α and β subunit, acting like a wedge to open the integrin α and β TM and cytoplasmic clasps through steric hindrance. This idea is illustrated in Fig 3. When structure of talin head-integrin α and β TM complex is assembled by aligning talin head and α and β TM complex with talin F2F3-β1D complex, we observed a possible clash between talin head and integrin α tails. These three possible mechanisms are not mutually exclusive and further research will be needed to pinpoint the exact mechanism.
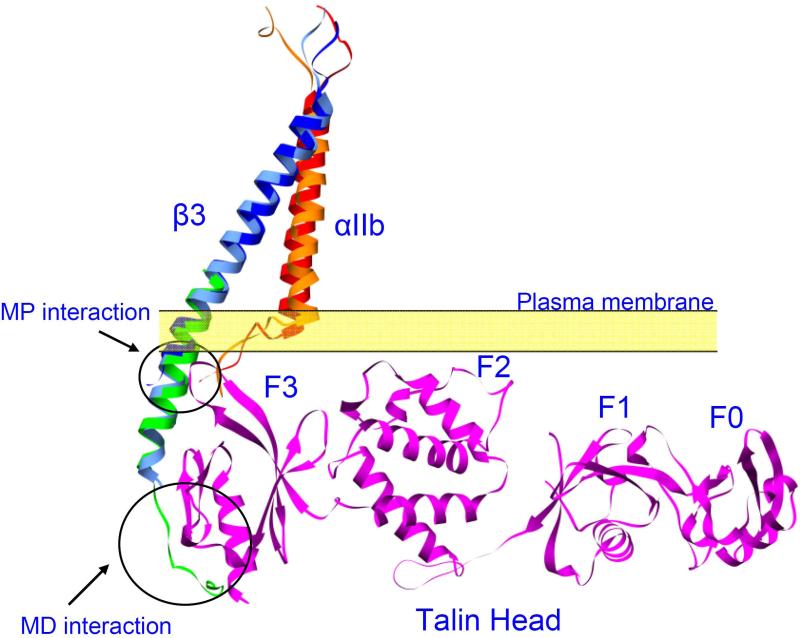
Figure 3. Binding of talin to β3 tail might result in steric clash between talin and αIIb.
Two αIIbβ3 TMD structure was depicted in the figure. Shown in red and blue are the integrin αII and β3 TMD from the recent NMR structure (PDB entry 2k9j) [20]. Shown in orange and cyan are the αIIb and β3 TMD from the recent Rosetta structure [62]. Talin head domain (PDB entry 3IVF) was shown in magenta. The structure of the whole complex was assembled by first aligning a β1D-Talin F2F3 (PDB entry 3G9W. β1D cytoplasmic domain in green) complex structure to the β3, and then aligning the talin head structure to the talin F2F3 (not shown) structure. Membrane distal (MD) and membrane proximal (MP) interactions were indicated. Talin head overlaps with αIIb in the Rosetta structure, indicating potential steric hinderance.
Kindlin is an important regulator of integrin
Recently, another family of integrin binding proteins, kindlins, has been shown to be important for proper integrin regulation. There are three kindlin orthologues in mammals: kindlin-1, 2 and 3. Genetic ablation of kindlin-2 or 3 are embryonic lethal in mice [44, 45]. Loss of functional kindlin-3 in platelets results in severe bleeding, defective platelet aggregation and platelet integrin activation [44]. Similarly, defects in β2 integrin activation and the integrin-mediated adhesion were reported in leukocytes that have lost functional kindlin-3 [46, 47]. Knock out of kindlin-1 and 2 inhibited β1 integrin activation and resulted in defective cell attachment [45, 48]. In human, loss of functional kindlin-3 has been shown to be the cause of a subset of leukocyte adhesion deficiency III (LAD III, LAD Iv) patients [47, 49, 50]. Mutations and truncations in kindlin-1 are the cause of Kindler's Syndrome [48, 51]. Thus, the importance of kindlin in integrin regulation is well supported by current literatures.
Compared to the detailed mechanistic understanding of integrin activation by talin, we are still at the beginning stage of understanding the mechanism of integrin regulation by kindlin. Kindlin has a FERM domain. But the F2 subdomain is separated into two halves by a PH domain in the middle. Deletion of the PH domain reduced kindlin's capacity to synergize with talin head in activating integrin αIIbβ3 [52]. Giving the usual property of PH domain as a lipid bilayer interacting partner, this result would indicate that the contact between kindlin and membrane might also plays a role in kindlin function. The consensus from current literature is that kindlin binds to the membrane distal NPxY motif while talin binds to the membrane proximal NPxY motif [44, 52, 53]. Similar to talin, kindlin binds to integrin through its F3 subdomain [44]. What structural features gave kindlin the specificity to the membrane distal rather than the membrane proximal NPxY motif is not clear. Mutations in kindlin or integrin β tail that disrupt kindlin-integrin interaction inhibited the capacity of kindlin to synergize with talin head in integrin activation [52, 53]. Thus the interaction between kindlin and integrin is required for kindlin function, at least for its function to synergize with talin head in activating αIIbβ3. However, rather than synergizing with talin head in activation α5β1 integrin, over-expression of kindlins inhibited the capacity of talin head to activate α5β1 [53]. This observation is incompatible with the simple mechanism that kindlin and talin bind to integrin β tail and co-activate integrins. Instead, a hypothesis that kindlin might function as a scaffold has been proposed. A couple of recent reviews summarized possible mechanisms of kindlin function [10, 54].
Other signaling molecules that may contribute to integrin activation
Talin and kindlin are the two most well documented integrin regulators, supported by data from genetically engineered mice, human patients, biochemical studies, structural studies and cell biological studies. Other molecules have also been implicated in the regulation of integrin, most notably integrin linked kinas (ILK). Conditional knock-out of ILK in platelets caused modest defects in platelet integrin activation and platelet aggregation [55]. Loss of ILK expression in CHO cells inhibited activation of a chimeric integrin αIIbα6Bβ3, which is constitutively active but sensitive to cytoplasmic integrin regulators [56]. Current literature sheds very little light on the mechanism for ILK function in integrin regulation. ILK was reported to interact with integrins by some labs [57], but others have not been able to detect the interaction [58]. ILK also displays interactions with kindlin in yeast-2-hybrid system [59]. Thus it is possible that ILK function in integrin regulation is associated with kindlin function. Another kindlin binding protein, migfilin, can weakly activate integrin when over-expressed in cells. Migfilin was proposed to exhibit its integrin activation effect by displacing a known integrin inhibitor, filamin [60]. However, genetic ablation of migfilin in mice produced no obvious phenotype and had no effects on cell adhesion, cell spreading, and integrin activation [61]. The question then is whether there are any paralogues of migfilin that might have compensated its function.
Future developments
The regulation of integrin is important for haemostasis, proper leukocyte function and endothelial cell function. Thus, this continues to be an area of intense interest. In the past several years, remarkable progress in idenfitying the key players and understanding their mechanism of action has been achieved. Nevertheless, important unsanswered questions, such as the mechanism whereby kindlins support integrin activation, remain. The significance of integrin activation in thrombosis and inflammation also suggests that this extraordinary progress in the basic understanding of this process may lead to “translational moments” in which new therapies can emerge.
References
- 1.Cruz MA, Chen J, Whitelock JL, Morales LD, Lopez JA. The platelet glycoprotein Ib-von Willebrand factor interaction activates the collagen receptor alpha2beta1 to bind collagen: activation-dependent conformational change of the alpha2-I domain. Blood. 2005;105:1986–91. doi: 10.1182/blood-2004-04-1365. [DOI] [PubMed] [Google Scholar]
- 2.Savage B, Almus-Jacobs F, Ruggeri ZM. Specific synergy of multiple substrate-receptor interactions in platelet thrombus formation under flow. Cell. 1998;94:657–66. doi: 10.1016/s0092-8674(00)81607-4. [DOI] [PubMed] [Google Scholar]
- 3.Kahner BN, Shankar H, Murugappan S, Prasad GL, Kunapuli SP. Nucleotide receptor signaling in platelets. J Thromb Haemost. 2006;4:2317–26. doi: 10.1111/j.1538-7836.2006.02192.x. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Delaney MK, O'Brien KA, Du X. Signaling during platelet adhesion and activation. Arterioscler Thromb Vasc Biol. 2010;30:2341–9. doi: 10.1161/ATVBAHA.110.207522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller BS, Shattil SJ. The GPIIb/IIIa (integrin alphaIIbbeta3) odyssey: a technology-driven saga of a receptor with twists, turns, and even a bend. Blood. 2008;112:3011–25. doi: 10.1182/blood-2008-06-077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries MJ, Mould AP. Structure. An anthropomorphic integrin. Science. 2001;294:316–7. doi: 10.1126/science.1066240. [DOI] [PubMed] [Google Scholar]
- 7.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–11. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 8.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294:339–45. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–73. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes PE, Diaz-Gonzalez F, Leong L, Wu C, McDonald JA, Shattil SJ, Ginsberg MH. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J Biol Chem. 1996;271:6571–4. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 12.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–16. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain helix packing stabilizes integrin alphaIIbbeta3 in the low affinity state. J Biol Chem. 2005;280:7294–300. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- 14.Luo BH, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci U S A. 2005;102:3679–84. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Mitra N, Gratkowski H, Vilaire G, Litvinov R, Nagasami C, Weisel JW, Lear JD, DeGrado WF, Bennett JS. Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science. 2003;300:795–8. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 16.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–9. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 17.Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas T, Plow E, Qin J. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–97. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Metcalf DG, Gorelik R, Li R, Mitra N, Nanda V, Law PB, Lear JD, Degrado WF, Bennett JS. A push-pull mechanism for regulating integrin function. Proc Natl Acad Sci U S A. 2005;102:1424–9. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim C, Lau TL, Ulmer TS, Ginsberg MH. Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009;113:4747–53. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. Embo J. 2009;28:1351–61. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274:28071–4. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 22.Patil S, Jedsadayanmata A, Wencel-Drake JD, Wang W, Knezevic I, Lam SC. Identification of a talin-binding site in the integrin beta(3) subunit distinct from the NPLY regulatory motif of post-ligand binding functions. The talin n-terminal head domain interacts with the membrane-proximal region of the beta(3) cytoplasmic tail. J Biol Chem. 1999;274:28575–83. doi: 10.1074/jbc.274.40.28575. [DOI] [PubMed] [Google Scholar]
- 23.Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, Emsley J, Barsukov IL, Roberts GC, Critchley DR. Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem. 2010;285:29577–87. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, Grossmann JG, Liu H, Putz NS, Roberts GC, Volkmann N, Hanein D, Barsukov IL, Critchley DR. The structure of the C-terminal actin-binding domain of talin. Embo J. 2008;27:458–69. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouaouina M, Lad Y, Calderwood DA. The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate beta1 and beta3 integrins. J Biol Chem. 2008;283:6118–25. doi: 10.1074/jbc.M709527200. [DOI] [PubMed] [Google Scholar]
- 26.Lim J, Wiedemann A, Tzircotis G, Monkley SJ, Critchley DR, Caron E. An essential role for talin during alpha(M)beta(2)-mediated phagocytosis. Mol Biol Cell. 2007;18:976–85. doi: 10.1091/mbc.E06-09-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–5. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 28.Monkley SJ, Zhou XH, Kinston SJ, Giblett SM, Hemmings L, Priddle H, Brown JE, Pritchard CA, Critchley DR, Fassler R. Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev Dyn. 2000;219:560–74. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1079>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Petrich BG, Marchese P, Ruggeri ZM, Spiess S, Weichert RA, Ye F, Tiedt R, Skoda RC, Monkley SJ, Critchley DR, Ginsberg MH. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J Exp Med. 2007;204:3103–11. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieswandt B, Moser M, Pleines I, Varga-Szabo D, Monkley S, Critchley D, Fassler R. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J Exp Med. 2007;204:3113–8. doi: 10.1084/jem.20071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Jr., Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–2. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- 32.Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, Gingras AR, Grossmann JG, Roberts GC, Calderwood DA, Critchley DR, Barsukov IL. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. Embo J. 2010;29:1069–80. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calderwood DA, Yan B, de Pereda JM, Alvarez BG, Fujioka Y, Liddington RC, Ginsberg MH. The phosphotyrosine binding-like domain of talin activates integrins. J Biol Chem. 2002;277:21749–58. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 34.Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID. Structural basis of integrin activation by talin. Cell. 2007;128:171–82. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 36.Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. Embo J. 2009;28:3623–32. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elliott PR, Goult BT, Kopp PM, Bate N, Grossmann JG, Roberts GC, Critchley DR, Barsukov IL. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–99. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem. 2009;284:5119–27. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, Boussiotis VA, Ginsberg MH, Shattil SJ. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J Cell Biol. 2008;181:1211–22. doi: 10.1083/jcb.200803094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16:1796–806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 41.Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7:585–95. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 42.Haling JR, Monkley SJ, Critchley DR, Petrich BG. Talin-dependent integrin activation is required for fibrin clot retraction by platelets. Blood. 2010;117:1719–22. doi: 10.1182/blood-2010-09-305433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalli AC, Wegener KL, Goult BT, Anthis NJ, Campbell ID, Sansom MS. The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure. 2010;18:1280–8. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–30. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 45.Montanez E, Ussar S, Schifferer M, Bosl M, Zent R, Moser M, Fassler R. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–30. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang HV, Sperandio M, Fassler R. Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat Med. 2009;15:300–5. doi: 10.1038/nm.1921. [DOI] [PubMed] [Google Scholar]
- 47.Malinin NL, Zhang L, Choi J, Ciocea A, Razorenova O, Ma YQ, Podrez EA, Tosi M, Lennon DP, Caplan AI, Shurin SB, Plow EF, Byzova TV. A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat Med. 2009;15:313–8. doi: 10.1038/nm.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ussar S, Moser M, Widmaier M, Rognoni E, Harrer C, Genzel-Boroviczeny O, Fassler R. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 2008;4:e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svensson L, Howarth K, McDowall A, Patzak I, Evans R, Ussar S, Moser M, Metin A, Fried M, Tomlinson I, Hogg N. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat Med. 2009;15:306–12. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuijpers TW, van de Vijver E, Weterman MA, de Boer M, Tool AT, van den Berg TK, Moser M, Jakobs ME, Seeger K, Sanal O, Unal S, Cetin M, Roos D, Verhoeven AJ, Baas F. LAD-1/variant syndrome is caused by mutations in FERMT3. Blood. 2009;113:4740–6. doi: 10.1182/blood-2008-10-182154. [DOI] [PubMed] [Google Scholar]
- 51.Mas-Vidal A, Minones-Suarez L, Toral JF, Mallo S, Perez-Oliva N. A novel mutation in the FERMT1 gene in a Spanish family with Kindler's syndrome. J Eur Acad Dermatol Venereol. 2011;24:978–9. doi: 10.1111/j.1468-3083.2009.03554.x. [DOI] [PubMed] [Google Scholar]
- 52.Ma YQ, Qin J, Wu C, Plow EF. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J Cell Biol. 2008;181:439–46. doi: 10.1083/jcb.200710196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harburger DS, Bouaouina M, Calderwood DA. Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J Biol Chem. 2009;284:11485–97. doi: 10.1074/jbc.M809233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–9. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 55.Tucker KL, Sage T, Stevens JM, Jordan PA, Jones S, Barrett NE, St-Arnaud R, Frampton J, Dedhar S, Gibbins JM. A dual role for integrin-linked kinase in platelets: regulating integrin function and alpha-granule secretion. Blood. 2008;112:4523–31. doi: 10.1182/blood-2008-03-148502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honda S, Shirotani-Ikejima H, Tadokoro S, Maeda Y, Kinoshita T, Tomiyama Y, Miyata T. Integrin-linked kinase associated with integrin activation. Blood. 2009;113:5304–13. doi: 10.1182/blood-2008-07-169136. [DOI] [PubMed] [Google Scholar]
- 57.Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC, Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–6. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 58.Wickstrom SA, Lange A, Montanez E, Fassler R. The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! Embo J. 2010;29:281–91. doi: 10.1038/emboj.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–97. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- 60.Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, Gupta S, Wu C, Plow EF, Qin J. Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009;284:4713–22. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moik DV, Janbandhu VC, Fassler R. Loss of migfilin expression has no overt consequences on murine development and homeostasis. J Cell Sci. 2011;124:414–21. doi: 10.1242/jcs.075960. [DOI] [PubMed] [Google Scholar]
- 62.Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Mol Cell. 2009;34:234–49. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]