Abstract
Mice can recognize one another by individually characteristic body scents that reflect their genetic constitution at the extremely polymorphic major histocompatibility complex (MHC) of genes on chromosome 17. Reproductive behavioral manifestations of this sensory communication system include MHC-related mating preferences and neuroendocrine responses that affect preimplantation pregnancy and arise from the MHC-related scent of alien males. We have shown previously that mice can be trained in a Y maze to distinguish the scents of urine of congeneic mice that differ genetically only at the MHC. By means of an automated olfactometer, we now show that rats also can similarly distinguish the urinary scents of MHC congeneic mice. Thus, the mode of individual recognition that depends on scents determined by MHC genes can operate across species barriers.
Full text
PDF
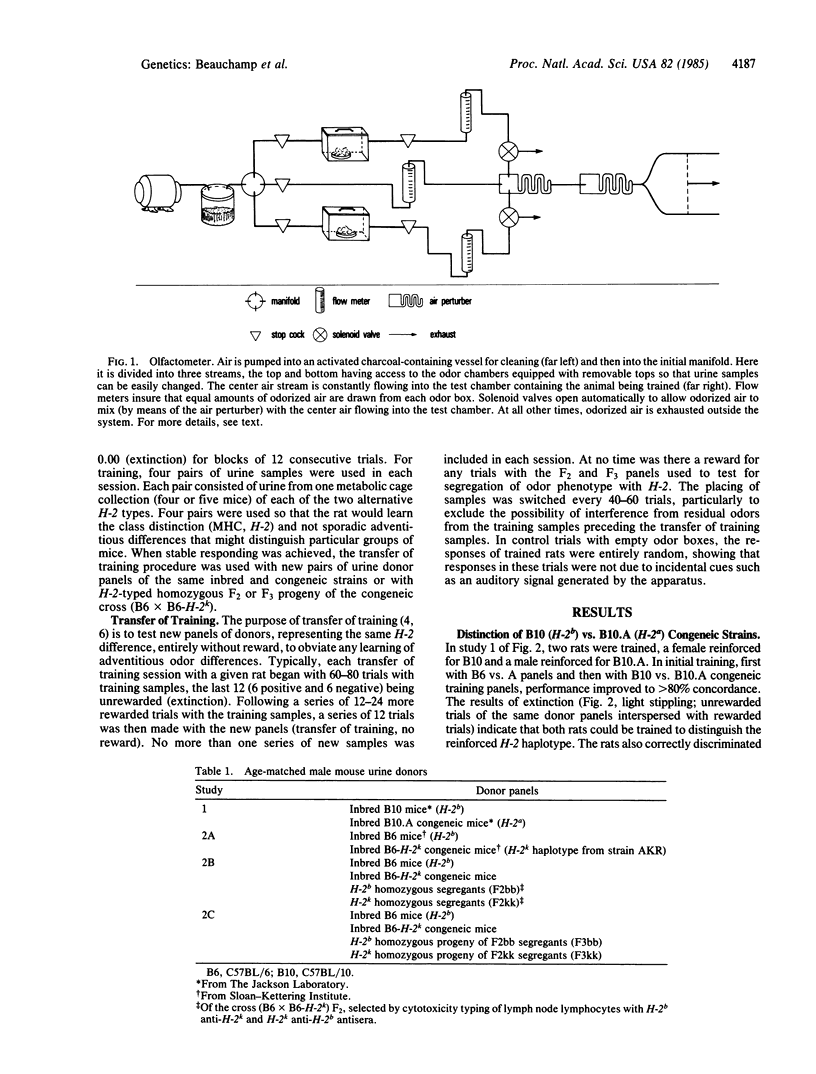

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyse E. A., Beauchamp G. K., Yamazaki K. Critical review: the sensory perception of genotypic polymorphism of the major histocompatibility complex and other genes: some physiological and phylogenetic implications. Hum Immunol. 1983 Apr;6(4):177–183. doi: 10.1016/0198-8859(83)90090-3. [DOI] [PubMed] [Google Scholar]
- Slotnick B. M., Born W. S. Transfer of a sex odor discrimination by rats. Physiol Behav. 1979 Sep;23(3):589–591. doi: 10.1016/0031-9384(79)90061-1. [DOI] [PubMed] [Google Scholar]
- Slotnick B. M., Ptak J. E. Olfactory intensity-difference thresholds in rats and humans. Physiol Behav. 1977 Dec;19(6):795–802. doi: 10.1016/0031-9384(77)90317-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Yamazaki K., Beauchamp G. K., Bard J., Thomas L., Boyse E. A. Distinctive urinary odors governed by the major histocompatibility locus of the mouse. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5817–5820. doi: 10.1073/pnas.78.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K., Beauchamp G. K., Bard J., Thomas L., Boyse E. A. Chemosensory recognition of phenotypes determined by the Tla and H-2K regions of chromosome 17 of the mouse. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7828–7831. doi: 10.1073/pnas.79.24.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K., Beauchamp G. K., Egorov I. K., Bard J., Thomas L., Boyse E. A. Sensory distinction between H-2b and H-2bm1 mutant mice. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5685–5688. doi: 10.1073/pnas.80.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K., Beauchamp G. K., Wysocki C. J., Bard J., Thomas L., Boyse E. A. Recognition of H-2 types in relation to the blocking of pregnancy in mice. Science. 1983 Jul 8;221(4606):186–188. doi: 10.1126/science.6857281. [DOI] [PubMed] [Google Scholar]
- Yamazaki K., Boyse E. A., Miké V., Thaler H. T., Mathieson B. J., Abbott J., Boyse J., Zayas Z. A., Thomas L. Control of mating preferences in mice by genes in the major histocompatibility complex. J Exp Med. 1976 Nov 2;144(5):1324–1335. doi: 10.1084/jem.144.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K., Yamaguchi M., Baranoski L., Bard J., Boyse E. A., Thomas L. Recognition among mice. Evidence from the use of a Y-maze differentially scented by congenic mice of different major histocompatibility types. J Exp Med. 1979 Oct 1;150(4):755–760. doi: 10.1084/jem.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]




