Summary
Background
Several chromosomal regions have been identified using family-based linkage analysis to contain genes contributing to the development of asthma and allergic disorders. One of these regions, chromosome 2q32-q33, contains a gene cluster containing CFLAR, CASP10 and CASP8. These genes regulate the extrinsic apoptosis pathway utilized by several types of immune and structural cells that have been implicated in the pathogenesis of asthma.
Objective
To assess the role of genetic variation in CFLAR, CASP10 and CASP8 in asthma and related phenotypes in individuals of diverse ethnic backgrounds.
Methods
We tested 26 single nucleotide polymorphisms (SNPs) in the CFLAR, CASP10 and CASP8 gene cluster for association with asthma and related phenotypes in African-American, non-Hispanic whites, and Hispanic case–control populations (cases, N = 517, controls, N = 644).
Results
Five CASP10 SNPS were associated with forced expiratory volume in 1 s (FEV1)/forced expiration volume capacity (FVC) in the African-American subjects with asthma (P = 0.0009–0.047). Nine SNPs, seven in CASP10 and two in CASP8, were also associated with the degree of bronchial hyperresponsiveness (BHR) (as determined by PC20) in race-specific analysis, predominately in the Non-Hispanic white cases. Two SNPs, rs6750157 in CASP10 and rs1045485 in CASP8 were modestly associated with asthma in the African-American (P = 0.025) and Hispanic (P = 0.033) populations, respectively.
Conclusion
These data suggest a role for CASP10 as a potential modifier of the asthma phenotype, specifically with measures of airway obstruction and BHR.
Keywords: asthma, bronchial hyperresponsiveness, CASP10, chromosome 2q, FEV1/FVC
Introduction
Asthma is a respiratory disorder that develops due to the interaction of specific genetic variants and environmental exposures. It is characterized by bronchial inflammation, bronchial hyperresponsiveness (BHR), intermittent airway obstruction, and atopy. Analysis of airway biopsies and bronchioalveolar lavage (BAL) samples from individuals with asthma have supported the role of T helper type II (Th2) cytokines such as IL-4, IL-5, IL-9, and IL-13 [1] as well as other inflammatory cells that include mast cells, eosinophils, fibroblasts, and airway epithelial cells [2, 3]. The large number of diverse cellular elements and signaling molecules involved in asthmatic and allergic inflammation supports the value of genetic approaches to understand and dissect this complex inflammatory process.
Genetic linkage studies have previously identified chromosomal regions that contain genes involved in the development of asthma and related phenotypes [4]. One such region is chromosome 2q32-q33, where there is evidence for linkage to asthma in a Hispanic population [5], and to atopic phenotypes in Hutterite [6], German [7], and Dutch populations [8, 9]. Evidence for linkage to this same region to forced expiratory volume in 1 s (FEV1)/forced expiration volume capacity (FVC) has also been observed in the same Dutch population [10], and a nearby region (2q36) has shown evidence for linkage with pulmonary function in families ascertained by a proband with early onset chronic obstructive pulmonary disease (COPD) [11]. Interestingly, a cluster of genes containing caspase 8 (CASP8), caspase 10 (CASP10), and CASP8 and FADD-like apoptosis regulator (CFLAR) falls within the LOD-1 confidence interval for the linkages with asthma in Hispanics and with total serum IgE levels, eosinophilia, and FEV1/FVC in the Dutch. Because of the localization of multiple asthma-related phenotypes and the relevant biological function of the caspase gene cluster, additional analysis of this chromosomal segment is warranted.
Mutations in CASP10 have been shown to cause or are related to autoimmune lymphoproliferative syndrome, type II (ALPS2), a condition in which abnormal lymphocyte and dendritic cell homeostasis causes chronic, non-malignant adenopathy and splenomegaly as well as autoimmune disorders [12]. Inactivating mutations in CASP10 have also been identified in patients with non-Hodgkin lymphoma [13]. Since mutations in the coding region of this gene have been shown to be associated with severe immune diseases, it is possible that more subtle changes in gene expression could contribute to the more complex immune abnormalities observed in asthma.
In this study, 26 single nucleotide polymorphisms (SNPs) encompassing CASP8, CASP10 and CFLAR were identified through the dbSNP database. Additional CASP10 SNPs in two genomic regions that are conserved between multiple species were identified through DNA sequencing. Single SNP association and haplotype analyses were performed in three ethnically diverse, well-characterized populations with asthma and related phenotypes.
Materials and methods
Population
African-American, non-Hispanic white, and Hispanic (from New Mexico) subjects with asthma were characterized using the NHLBI Collaborative Study on the Genetics of Asthma (CSGA) criteria [14]. Written informed consent for adults or written parental assent for children was obtained from all participants. This study was approved by the Institutional Review Board at Wake Forest University School of Medicine.
Clinical characteristics for each of the three populations used in this study are shown in Table 1. The case population for association studies was assembled from the original CSGA families used for the linkage studies, the probands from CSGA trios, or recruited individually. Since the CSGA families contained affected sibling pairs (each a proband), the ‘case’ proband for this study was selected by choosing the sibling with the most phenotype data available. The controls were generally older than the cases, reducing the probability of undiagnosed asthma among the controls. Other characteristics were similar between the populations, with the exception of total serum IgE levels, which showed some variability. A subject was considered to have asthma if he/she met the following criteria: physician’s diagnosis for asthma, demonstrated BHR or bronchodilator reversibility, and a medical history of at least two of the following symptoms: wheeze, dyspnoea, chest tightness, or cough. Subjects were considered hyperresponsive if their PC20 was ≤ 25 mg/mL methacholine. Because of safety concerns, in children (≤ 18 years of age) with an FEV1 < 70% of predicted or adults with an FEV1 < 60% of predicted, reversibility testing to an inhaled beta agonist was used to demonstrate bronchial liability, defined by an increase in FEV1 of ≥ 15% after bronchodilator inhalation [14].
Table 1.
Characteristics of case–control populations
| African-American
|
Non-Hispanic white
|
Hispanic
|
||||
|---|---|---|---|---|---|---|
| Cases (n = 168) |
Controls (n = 269) |
Cases (n = 233) |
Controls (n = 245) |
Cases (n = 116) |
Controls (n = 130) |
|
| Age, mean±SD | 22.3±12.7 | 32.5±11.3 | 23.9±13.1 | 32.7±11.0 | 17.4±11.7 | 36.8±12.3 |
| % Male | 45.8 | 44.6 | 39.5 | 39.8 | 46.5 | 32.3 |
| IgE, IU/mL (geo. mean) | 204.2±4.1 | 66.1±4.4 | 138.0±4.6 | 15.5±6.8 | 223.9±4.7 | 32.4±4.9 |
| FEV1, % predicted (SD)* | 75.5 (20.6) | ND | 82.6 (16.4) | ND | 95.2 (15.9) | ND |
| Mean FEV1/FVC | 0.7±0.1 | ND | 0.8±0.1 | ND | 0.8±0.1 | ND |
| Mean PC20, mg/mL | 3.5±5.1 | ND | 3.6±5.8 | ND | 3.9±5.9 | ND |
Percent predicted FEV1 values for African-Americans are race-adjusted [14].
ND, not determined.
Cases and controls were characterized by a questionnaire that evaluated asthma and allergy symptoms, health care utilization, and medication use. Controls were recruited separately by public advertisement and had no history of asthma and no first-degree relatives with asthma, but did not undergo further testing. Whole blood was collected for determination of total serum IgE levels and isolation of DNA.
Sequencing and Genotyping
SNPs were selected for genotyping from dbSNP (http://www.ncbi.nlm.nih.gov/SNP) based on allele frequency and putative functional relevance. Two intronic regions of CASP10 spanning base pairs (bp) 201 834 876–201 835 395 and 201 820 881–201 823 623 (NCBI Build 35) were highly conserved between multiple species, indicative of a potential functional element. For the detection of polymorphisms in these regions, resequencing was performed on a panel of subjects including US Non-Hispanic white, African-American, and US Hispanic cases and controls (eight cases and 16 controls from each population). Genotyping was performed using the MassARRAY SNP genotyping system according to the manufacturer’s instructions (Sequenom Inc., San Diego, CA, USA).
Genetic Analysis
Tests for Hardy–Weinberg Equilibrium (HWE) were assessed using chi-square tests, as determined by SnpA-nalyzer 1.1 (http://www.fhcrc.org/labs/kruglyak/Downloads/). Pairwise marker–marker linkage disequilibrium (LD) was assessed using r2 and plotted using Haploview [15].
Asthma was analyzed as a binary trait using logistic regression models performed using PROC LOGISTIC in SAS statistical software (SAS Institute, Cary, NC, USA). Measures of pulmonary function (FEV1/FVC, percent of predicted FEV1, and percent of predicted FVC) and BHR (PC20) were analyzed as continuous variables, in the cases only, using linear regression models (PROC GLM). P-values were calculated based on a two degree-of-freedom test for genotypic association. Because race is potentially a confounding factor, interactive effects between each polymorphism and race were examined in the tests described above. If the interaction term was not significant, results are presented for the combined racial populations (for associated SNPs only). Otherwise, analyses stratified by race were performed.
Haplotype analysis was performed using a score test developed by Schaid [16], as implemented by the haplo.stats program (http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm//www.mayo.edu/statgen). Statistical differences in overall haplotype frequencies were tested for association with all outcomes. Specific haplotype effects were examined if the test for the overall frequency distribution was significant (P≤0.05).
Results
Sequencing of the two evolutionarily conserved CASP10 genomic regions identified 11 SNPs, two of which had a minor allele frequency (MAF) >5% in each of the sequenced populations (rs10188598 and rs13396523). These two SNPs were included in the genotyping. Allele frequencies and sequences for all of the SNPS identified can be found in Table S1 in the online data supplement.
Twenty-six SNPs were genotyped and analyzed in the African-American, non-Hispanic white and Hispanic case–control populations (Table 2). As expected, LD patterns differed between the three populations with two to three haplotype blocks calculated using the method described by Gabriel et al. [17] (Fig. 1).
Table 2.
Single nucleotide polymorphisms (SNPs) and minor allele frequencies in each population
| Gene | SNP ID | SNP # | Reference Allele | African-American
|
Non-Hispanic whites
|
Hispanic
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | ||||
| CFLAR | rs2110728 C/T | 1 | T | 0.34 | 0.36 | 0.18 | 0.18 | 0.14 | 0.17 |
| rs2041767 C/T | 2 | T | 0.08* | 0.07 | 0.00 | 0.00 | 0.01 | 0.00 | |
| rs4487072 C/T | 3 | T | 0.42 | 0.37 | 0.21 | 0.20 | 0.16 | 0.17 | |
| rs719125 C/A | 4 | A | 0.43 | 0.39 | 0.21 | 0.20 | 0.17 | 0.17 | |
| rs2268791 C/G | 5 | G | 0.49 | 0.46 | 0.48 | 0.50 | 0.38 | 0.41 | |
| rs1594 C/T | 6 | T | 0.50 | 0.45 | 0.49 | 0.49 | 0.36 | 0.39 | |
| CASP10 | rs2098355 A/G | 7 | G | 0.25* | 0.22* | 0.05 | 0.03 | 0.05 | 0.03 |
| rs3900115 G/A | 8 | A | 0.50 | 0.40 | 0.48 | 0.48 | 0.37 | 0.42 | |
| rs3731714 G/A | 9 | A | 0.05 | 0.05 | 0.31 | 0.28 | 0.19 | 0.23 | |
| rs10166093 A/G | 10 | G | 0.29 | 0.31 | 0.04 | 0.03 | 0.05 | 0.03 | |
| rs6750157 C/T | 11 | T | 0.19 | 0.22* | 0.00 | 0.00 | 0.02 | 0.01 | |
| rs6435068 T/C | 12 | C | 0.33 | 0.34 | 0.04 | 0.03 | 0.04 | 0.03 | |
| rs10188598 A/G | 13 | G | 0.20 | 0.15 | 0.50 | 0.47 | 0.33 | 0.39 | |
| rs13396523 T/C | 14 | C | 0.28 | 0.31* | 0.04 | 0.03 | 0.05 | 0.03 | |
| rs6435069 A/G | 15 | G | 0.32 | 0.33 | 0.47 | 0.49 | 0.62 | 0.57 | |
| rs3851975 A/C | 16 | C | 0.30 | 0.32* | 0.05 | 0.03 | 0.04 | 0.02 | |
| rs3851976 A/G | 17 | G | 0.20 | 0.20 | 0.05 | 0.04* | 0.05 | 0.04 | |
| CASP8 | rs3769825 T/C | 18 | C | 0.36 | 0.34 | 0.46 | 0.45* | 0.42 | 0.45 |
| rs3754935T/G | 19 | G | 0.23 | 0.22 | 0.08 | 0.06 | 0.06 | 0.03 | |
| rs3769823 C/T | 20 | T | 0.38 | 0.39 | 0.32 | 0.35* | 0.41 | 0.47 | |
| rs2349070 C/A | 21 | A | 0.18 | 0.22 | 0.26 | 0.28 | 0.42 | 0.45 | |
| rs3754934 G/T | 22 | T | 0.23 | 0.20 | 0.07 | 0.06 | 0.05 | 0.03 | |
| rs3817578 G/A | 23 | A | 0.19 | 0.22 | 0.06 | 0.06 | 0.05 | 0.03 | |
| rs1045485 G/C | 24 | C | 0.06 | 0.07 | 0.11 | 0.13 | 0.08 | 0.03 | |
| rs1045487 G/A | 25 | A | 0.22 | 0.21 | 0.06 | 0.05 | 0.04 | 0.03 | |
| rs2141331 C/T | 26 | T | 0.17 | 0.21 | 0.27 | 0.30 | 0.44 | 0.46 | |
Indicates deviation from expected values of Hardy–Weinberg equilibrium (P<0.05).
ND, not determined.
Fig. 1.
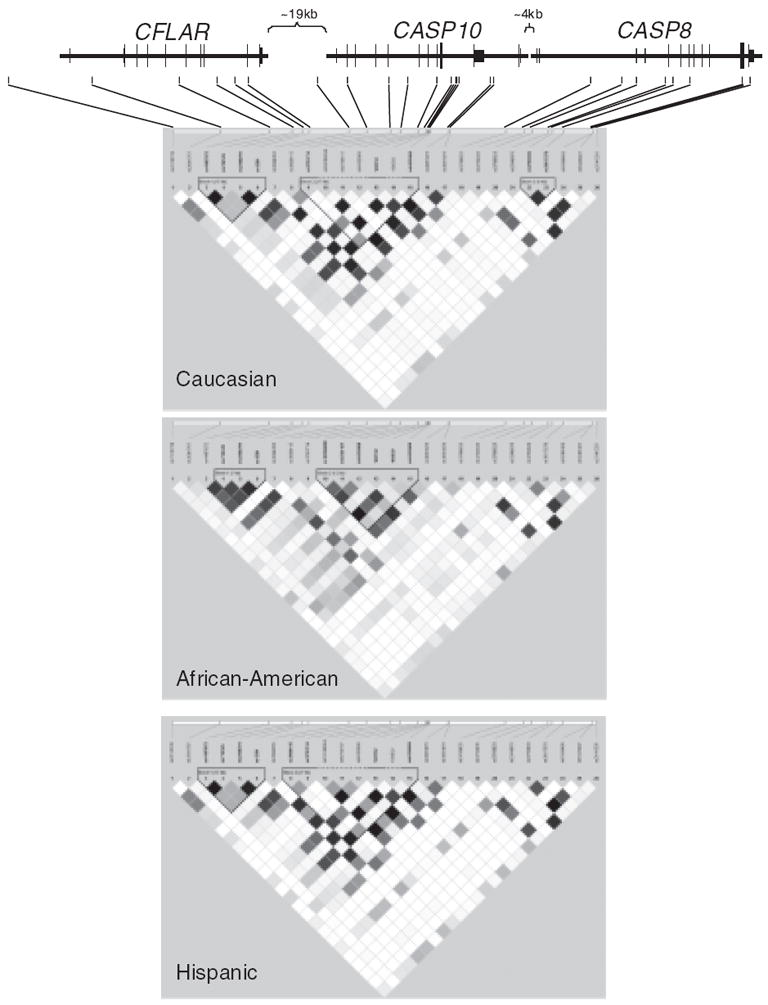
Linkage disequilibrium (LD) pattern of CFLAR, CASP10 and CASP8 in non-Hispanic white (top), African-American (middle), and Hispanic (bottom) populations generated by HaploView [15]. LD is represented by r2 values, with black boxes indicating an r2 value ≥0.90. Haplotype blocks were defined by the Confidence Interval algorithm [17].
FEV1/FVC, a measure of airway obstruction, was associated with five SNPs in CASP10 in African-Americans and one in Hispanics (Table 3). In both populations, the minor allele was generally associated with lower FEV1/FVC compared with the major allele, and this trend appeared to be additive.
Table 3.
Single nucleotide polymorphisms (SNPs) associated with forced expiratory volume in 1 s (FEV1)/forced expiration volume capacity (FVC) in CASP10
| Gene | SNP ID | SNP No. | Geno | African-American
|
Non-Hispanic white
|
Hispanic
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean (FEV1/FVC) | P-value | N | Mean (FEV1/FVC) | P-value | N | Mean (FEV1/FVC) | P-value | ||||
| CASP10 | rs10166093 | 10 | AA | 77 | 0.77 | 0.0033 | 179 | 0.75 | NS | 85 | 0.81 | NS |
| AG | 55 | 0.71 | 18 | 0.74 | 9 | 0.75 | ||||||
| GG | 11 | 0.65 | ||||||||||
| rs6750157 | 11 | CC | 97 | 0.76 | 0.0009 | 195 | 0.75 | NS | 93 | 0.81 | NS | |
| CT | 47 | 0.70 | 1 | 0.76 | 3 | 0.77 | ||||||
| TT | 3 | 0.55 | ||||||||||
| rs6435068 | 12 | TT | 68 | 0.77 | 0.026 | 176 | 0.76 | NS | 83 | 0.81 | NS | |
| CT | 60 | 0.72 | 17 | 0.75 | 7 | 0.78 | ||||||
| CC | 15 | 0.69 | ||||||||||
| rs13396523 | 14 | TT | 79 | 0.77 | 0.0014 | 180 | 0.75 | NS | 82 | 0.81 | NS | |
| CT | 56 | 0.71 | 17 | 0.74 | 9 | 0.75 | ||||||
| CC | 11 | 0.65 | ||||||||||
| rs6435069* | 15 | AA | 64 | 0.73 | 0.047 | 51 | 0.76 | NS | 13 | 0.80 | NS | |
| AG | 63 | 0.73 | 104 | 0.76 | 46 | 0.80 | ||||||
| GG | 17 | 0.81 | 41 | 0.73 | 36 | 0.81 | ||||||
| rs3851975 | 16 | AA | 71 | 0.76 | NS | 175 | 0.75 | NS | 90 | 0.81 | NS | |
| AC | 60 | 0.71 | 18 | 0.74 | 8 | 0.75 | ||||||
| CC | 10 | 0.69 | ||||||||||
| rs3851976 | 17 | AA | 99 | 0.74 | NS | 181 | 0.75 | NS | 86 | 0.81 | 0.0038 | |
| AG | 39 | 0.75 | 15 | 0.74 | 11 | 0.73 | ||||||
| GG | 6 | 0.67 | 1 | 0.85 | ||||||||
Significant for race×gene interaction (P<0.05).
NS, not significant.
Associations with BHR, as measured by PC20, were observed with seven SNPs in CASP10 (P = 0.0003–0.0024), predominately in non-Hispanic white cases (Table 4). The phenotype trend was similar between non-Hispanic whites and Hispanics, with individuals homozygous for the major allele having higher PC20 values, indicating less BHR. Marginal associations (P = 0.016–0.043) were also observed with two SNPs in CASP8 – one in non-Hispanic whites and one in Hispanics (Table 4). The association data for all SNPs with these two phenotypes is available in supplementary Tables S2 and S3. Only two SNPs were associated with asthma – rs6750157 in CASP10 in African-Americans (P = 0.025) and rs1045485 in CASP8 in Hispanics (P = 0.033) (Table S4).
Table 4.
Single nucleotide polymorphisms (SNPs) associated with PC20 in CASP10 and CASP8
| Gene | SNP ID | SNP # | Genotypes | African-American
|
Non-Hispanic white
|
Hispanic
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean PC20 | P-value | N | Mean PC20 | P-value | N | Mean PC20 | P-value | ||||
| CASP10 | rs2098355* | 7 | AA | 56 | 3.18 | NS | 183 | 3.87 | 0.0011 | 87 | 4.27 | NS |
| AG | 44 | 3.87 | 18 | 1.24 | 10 | 2.32 | ||||||
| GG | 11 | 3.05 | ||||||||||
| rs3731714 | 9 | GG | 109 | 3.73 | NS | 99 | 3.12 | NS | 66 | 3.53 | 0.013 | |
| GA | 9 | 2.90 | 89 | 4.51 | 32 | 5.21 | ||||||
| AA | 22 | 2.05 | 2 | 4.07 | ||||||||
| rs10166093* | 10 | AA | 58 | 3.60 | NS | 193 | 3.84 | 0.0005 | 89 | 4.25 | NS | |
| AG | 50 | 3.75 | 19 | 0.88 | 9 | 2.33 | ||||||
| GG | 9 | 3.05 | ||||||||||
| rs6435068* | 12 | TT | 52 | 3.10 | NS | 191 | 3.99 | 0.0004 | 87 | 4.13 | NS | |
| TC | 52 | 4.25 | 19 | 0.88 | 8 | 1.88 | ||||||
| CC | 12 | 2.56 | ||||||||||
| Rs13396523* | 14 | TT | 61 | 3.59 | NS | 194 | 3.81 | 0.0003 | 86 | 4.14 | NS | |
| TC | 51 | 3.68 | 18 | 0.83 | 9 | 2.33 | ||||||
| CC | 9 | 3.05 | ||||||||||
| rs3851975* | 16 | AA | 54 | 2.93 | NS | 189 | 3.92 | 0.0006 | 94 | 4.14 | NS | |
| AC | 52 | 4.81 | 19 | 0.88 | 8 | 2.58 | ||||||
| CC | 9 | 2.54 | ||||||||||
| rs3851976* | 17 | AA | 77 | 3.02 | NS | 196 | 3.85 | 0.0024 | 92 | 4.11 | NS | |
| AG | 37 | 4.66 | 16 | 0.89 | 9 | 2.33 | ||||||
| GG | 4 | 6.70 | 1 | 0.33 | ||||||||
| CASP8 | rs3769825 | 18 | TT | 41 | 3.98 | NS | 40 | 2.00 | 0.016 | 34 | 3.52 | NS |
| CT | 60 | 3.38 | 100 | 4.12 | 48 | 4.35 | ||||||
| CC | 10 | 5.24 | 63 | 3.80 | 19 | 3.39 | ||||||
| rs1045485 | 24 | GG | 106 | 3.75 | 0.043 | 172 | 3.25 | NS | 88 | 4.02 | NS | |
| GC | 14 | 1.58 | 37 | 5.08 | 15 | 3.78 | ||||||
| CC | 4 | 1.99 | ||||||||||
Significant for race×gene interaction (P<0.05).
NS, not significant.
Haplotypes for the entire CASP10 locus (SNPs 7–17) were generated for each population to evaluate FEV1/FVC and PC20 in the cases. In the non-Hispanic white cases, the CASP10 haplotype GGGGCCACACG (frequency = 4%) was associated with lower PC20 (P = 0.0004). This haplotype alone contains each of the alleles associated with lower PC20 individually, and none of the other three haplotypes observed in cases contained any associated allele, making it unclear whether the observed association is due to the individual SNPs or the haplotype. In the African-American cases, two CASP10 haplotypes were associated with FEV1/FVC. The GAGGTCACACA haplotype (13%) was associated with a lower FEV1/FVC (P = 0.001), and all of the alleles individually associated with the lower FEV1/FVC were contained in this haplotype. The AGGACTATAAG haplotype (14%) was associated with a higher ratio FEV1/FVC (P = 0.014), and all of the single SNP alleles associated with higher values were present on this haplotype.
Discussion
CFLAR, CASP10, and CASP8 form a gene cluster in a genomic region on chromosome 2q with consistent evidence for linkage in multiple studies of asthma and related phenotypes. In the present study, 26 SNPs were selected throughout this cluster and association studies were conducted for asthma and associated phenotypes in three ethnically diverse case–control populations. The results demonstrate that both SNPs and haplotypes from this region, predominantly in CASP10, contribute to phenotypes that are closely associated with asthma. The most consistent associations observed were with phenotypes associated with asthma expression instead of disease susceptibility. FEV1/FVC, a measure of pulmonary obstruction, was associated with five SNPs in CASP10 in the African-American population. PC20, a measure of BHR, was significantly associated with CASP10 SNPs in the non-Hispanic white population, with a trend towards a similar effect in the Hispanic population. Haplotype analysis was consistent with the single SNP analysis, but did not shed light on which SNPs may be of primary importance.
Airway obstruction is related to increased airway responsiveness in asthma [18]. In addition, increased airway responsiveness is related to longitudinal decline in pulmonary function [19]. Therefore it is not surprising that we observed association with measurements of these specific phenotypes simultaneously. Our measure of airway obstruction in this population, FEV1/FVC, was associated with SNPs in CASP10 in the African-American population. The lack of association in the non-Hispanic white and Hispanic groups is most likely due to the lower MAF of 4% in these populations compared with 30% in the African-American group. Replication of these SNPs in larger populations will be helpful in evaluating the significance of this finding.
For PC20, significant association was observed predominately with CASP10 SNPs in the non-Hispanic white population. A similar trend was also observed in the Hispanic population (carriers of the minor allele have lower PC20 values), but the same SNPs were not significantly associated. This is probably due to the smaller number of Hispanic cases, and therefore less power in this population. This trend was much more difficult to detect in the African-American population, which may be a reflection of the ascertainment of cases in this population. To be classified as a ‘case,’ individuals were required to be hyperresponsive (PC20 ≤25 mg/mL methacholine) or show 15% reversibility of FEV1 post-bronchodilator. Individuals with baseline FEV1 values ≤70% (children under 18 years of age) or ≤60% (over 18 years of age) were not tested for BHR but were included based on reversibility data only. More African-American cases (26.2%) were included based on reversibility compared with non-Hispanic whites (5.6%) and Hispanics (8.6%). Thus, fewer African-Americans were available for the PC20 analysis.
With a few exceptions, the same SNPs associated with FEV1/FVC in African-Americans were associated with PC20 in the non-Hispanic White population, indicating the importance of these SNPs in the expression of asthma. These SNPs were in LD with each other, limiting our ability to determine which single SNPs or combinations may have potential function effects. Only one of the associated SNPs codes for an amino acid change (rs1045485 in CASP8), and there were no coding polymorphisms in CASP10 associated with either FEV1/FVC or PC20. All other associated SNPs were located in introns or upstream of the first exon. Bioinformatic analysis reveals that all of these SNPs alter the consensus-binding site of a transcription factor, and inappropriate regulation of this gene may alter its expression (data not shown). Also, one of the SNPs from the sequencing in the conserved regions (rs13396523) was associated with both FEV1/FVC and PC20. Further tests will be required to elucidate if any of these SNPs cause changes in transcription factor binding or regulation of this gene. It is also possible that associations may be due to another variation in LD with the associated alleles, which would also contribute to the different associations in each ethnic group.
Because of the variable LD between racial groups, we did not provide a formal adjustment for multiple comparisons. However, using a simple Bonferroni correction and adjusting for 26 independent tests (i.e. 26 SNPs), the adjusted P-value would be 0.0019. Two SNPs remain significant for FEV1/FVC in the African-American group and five SNPs remain significant for PC20 in the non-Hispanic white group for this adjusted P-value.
In summary, we report associations with SNPs in CASP10 with measurements of airway obstruction (FEV1/FVC) and BHR (PC20). Because of the strong LD between these SNPs, it is unclear which specific SNPs or haplotypes are ultimately important, or if they all play a particular role. Additional replication studies and functional evaluation of CASP10 and its contribution to apoptosis in immune or airway cells are required to further understand the mechanisms responsible for the persistent inflammation, injury, and remodelling characteristic of asthma.
Supplementary Material
Additional Supporting Information may be found in the online version of this article:
Table S1. Minor Allele Frequency and Sequence of novel CASP10 SNPs.
Table S2. All Association Results for FEV1/FVC in Asthma Cases.
Table S3. All Association Results for PC20 in Asthma Cases.
Table S4. SNPs Associated with Asthma in CASP8 and CASP10.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Acknowledgments
We would like to thank all participants of the study. We would also like to thank Lilly Zheng and Jonathan Clark for their technical assistance as well as Dr Annette Hastie and Dr James Zangrilli for sharing their experience and knowledge. This work was supported by the Collaborative Study on the Genetics of Asthma Grant U01 HL49602.
References
- 1.Krug N, Madden J, Redington A, et al. T-cell cytokine profile evaluated at the single cell level in BAL and blood in allergic asthma. Am J Respir Cell Mol Biol. 1996;14:319–26. doi: 10.1165/ajrcmb.14.4.8600935. [DOI] [PubMed] [Google Scholar]
- 2.Renauld JC. New insights into the role of cytokines in asthma. J Clin Pathol. 2001;54:577–89. doi: 10.1136/jcp.54.8.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddox L, Schwartz DA. The pathophysiology of asthma. Annu Rev Med. 2002;53:477–98. doi: 10.1146/annurev.med.53.082901.103921. [DOI] [PubMed] [Google Scholar]
- 4.Howard TD, Celedon JC. Linkage to asthma and its intermediate phenotypes. In: Postma DS, Weiss ST, editors. Genetics of asthma and chronic obstructive pulmonary disease. New York: Informa Healthcare; 2007. pp. 199–208. [Google Scholar]
- 5.CSGA. A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15:389–92. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 6.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genome-wide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67:1154–62. doi: 10.1016/s0002-9297(07)62946-2. In process citation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wjst M, Fischer G, Immervoll T, et al. A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics. 1999;58:1–8. doi: 10.1006/geno.1999.5806. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Postma DS, Howard TD, et al. Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet. 2000;67:1163–73. doi: 10.1086/321190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koppelman G, Stine O, Xu J, et al. Genome-wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol. 2002;109:498–506. doi: 10.1067/mai.2002.122235. [DOI] [PubMed] [Google Scholar]
- 10.Postma DS, Meyers DA, Jongepier H, Howard TD, Koppelman GH, Bleecker ER. Genome wide screen for pulmonary function in 200 families ascertained for asthma. Am J Respir Crit Care Med. 2005;172:446–52. doi: 10.1164/rccm.200407-864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman EK, Mosley JD, Palmer LJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet. 2002;70:1229–39. doi: 10.1086/340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Zheng L, Lobito A, et al. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 13.Shin MS, Kim HS, Kang CS, et al. Inactivating mutations of CASP10 gene in non-Hodgkin lymphomas. Blood. 2002;99:4094–9. doi: 10.1182/blood.v99.11.4094. [DOI] [PubMed] [Google Scholar]
- 14.Lester LA, Rich SS, Blumenthal MN, et al. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108:357–62. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 18.Boushey HA, Holtzman MJ, Sheller JR, Nadel JA. Bronchial hyperreactivity. Am Rev Respir Dis. 1980;12:389–413. doi: 10.1164/arrd.1980.121.2.389. [DOI] [PubMed] [Google Scholar]
- 19.Rijcken B, Weiss ST. Longitudinal analyses of airway responsiveness and pulmonary function decline. Am J Respir Crit Care Med. 1996;154:S246–S249. doi: 10.1164/ajrccm/154.6_Pt_2.S246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article:
Table S1. Minor Allele Frequency and Sequence of novel CASP10 SNPs.
Table S2. All Association Results for FEV1/FVC in Asthma Cases.
Table S3. All Association Results for PC20 in Asthma Cases.
Table S4. SNPs Associated with Asthma in CASP8 and CASP10.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.


