Abstract
The serotonin (5-hydroxytryptamine, 5-HT) 5-HT2 G protein-coupled receptor (GPCR) family consists of types 2A, 2B, and 2C that share ~75% transmembrane (TM) sequence identity. Agonists for 5-HT2C receptors are under development for psychoses, whereas, at 5-HT2A receptors, antipsychotic effects are associated with antagonists—in fact, 5-HT2A agonists can cause hallucinations and 5-HT2B agonists cause cardiotoxicity. It is known that 5-HT2A TM6 residues W6.48, F6.51, and F6.52 impact ligand binding and function, however, ligand interactions with these residues at the 5-HT2C receptor has not been reported. To predict and validate molecular determinants for 5-HT2C-specific activation, results from receptor homology modeling, ligand docking, and molecular dynamics (MD) simulation studies were compared with experimental results for ligand binding and function at wild type and W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptors.
Keywords: Aromatic Interactions, Serotonin 5-HT2C, GPCR, W6.48, F6.51, F6.52, homology modeling, docking, molecular dynamics, drug design
1. Introduction
The serotonin (5-hydroxytryptamine, 5-HT) 5-HT2 G protein-coupled receptor (GPCR) family consists of types 2A, 2B, and 2C that share ~75% transmembrane (TM) sequence identity and same second messenger signaling [1]. Ligands that activate 5-HT2C receptors are under development for psychoses, whereas, at 5-HT2A receptors, antipsychotic effects are associated with antagonists—in fact, 5-HT2A agonists can cause hallucinations and 5-HT2B agonists cause cardiotoxicity [2,3]. Accordingly, 5-HT2C agonists should be highly specific to avoid potentially adverse clinical outcomes, however, TM sequence similarity and identical signaling pathways among the 5-HT2 GPCRs presents a challenge for development of 5-HT2C agonist drugs.
GPCRs are ubiquitous signaling proteins that share a 3-dimensional (3D) structure consisting of a bundle of seven transmembrane α-helices (TMH), connected by alternating intracellular and extracellular loops, with the N-terminus in the extracellular domain and C-terminus in the intracellular domain. Nearly half of currently-marketed drugs target GPCRs, albeit, GPCR structure and function are not well-understood. For example, although over 900 GPCRs are known, crystal structures are reported only for the following: bovine rhodopsin (bRho) [4–8], opsin, [9, 10], human A2A adenosine receptor bound to an antagonist [11], turkey β1 adrenoceptor [12], human β2 adrenoceptor (β2AR) in an inactive state [13–15], β2AR in a nanobody-stabilized active-state [16], β2AR in complex with an irreversible agonist [17, 18], human dopamine D3 receptor in complex with an agonist [19], human H1 receptor in a complex with an antagonist at 3.1 Å (PDB code 3RZE) [20]. Structural information for particular GPCRs significantly aides drug design targeting the receptor, for example, by providing information regarding ligand–receptor interactions that take place deep inside the orthosteric binding pocket [21]. To date, there are no 3D crystal structures for 5-HT2A and 5-HT2C GPCRs to assist antipsychotic drug design targeting these receptors, however, the related human 5-HT2B, as well as, 5-HT1B structures were reported during the preparation of this manuscript [22, 23].
In the absence of GPCR crystal structures, computational-based homology modeling methods have been used to generate the desired target structures. To validate GPCR receptor models and associated ligand docking studies, experimental studies that measure ligand binding and function at wild type (WT) vs. point-mutated GPCRs are commonly employed. For example, modeling studies suggest ligand binding at serotonin 5-HT2 and other aminergic neurotransmitter GPCRs is facilitated by a critical ionic interaction between a positively charged amine moiety of the ligand and the carboxylate of the fully-conserved aspartate residue D3.32 of the receptor [24–28]. Experimental validation of the proposed D3.32–ligand interaction at 5-HT2C GCPRs recently was reported in studies involving mutation of the 5-HT2C D3.32 residue to alanine (D3.32A), which abolished detectable binding of the 5-HT2C radioligand [3H]-mesulergine [29]. In addition to mutagenesis studies, computational ligand docking results using a homology-based GPCR model ultimately are validated by the solved GPCR crystal structure. For example, drug design results using a human histamine H1 GPCR model built by homology to the β2Ar crystal structure recently were validated using an H1 model built according to the subsequently released human histamine H1 GPCR structure (pdb code 3RZE) [30]; results showed analogous predictions in ligand binding affinities for the two models, as anticipated given the close structural correlation between the homology-based and crystal structure-based H1 models, and, as validated by experimental mutagenesis studies. Accordingly, it is anticipated that computational drug design studies using a 5-HT2C receptor model built by homology to the β2AR and validated by experimental mutagenesis studies will yield fruitful drug discovery results, even in the absence of a 5-HT2C crystal structure [29, 31, 32].
Early mutagenesis and homology-based (bacterial rhodopsin) molecular modeling studies of the 5-HT2A GPCR indicate aromatic amino acids (Ballesteros numbering [33, 34]) W6.48, F6.51, and F6.52 in TMH 6, which are highly conserved across aminergic neurotransmitter GPCRs and present in 5-HT2C, affect ligand binding and functional activity [35–37]. Using a 5-HT2C receptor model built by homology to the β2-AR [29, 31, 32], hydrophobic and/or aromatic interactions were proposed between 5-HT2C residues W6.48, F6.51, and F6.52, and the novel 5-HT2C agonist/5-HT2A/2B inverse agonist, (2S, 4R)-(−)-trans-4-phenyl-N,N-dimethy-1,2,3,4-tetrahydronaphthalene-2-amine (PAT) [38]. The present work reports novel 5-HT2C GPCR homology molecular modeling, ligand docking, molecular dynamics (MD) simulations, and pharmacological results using point-mutated receptors to delineate the role of 5-HT2C residues W6.48, F6.51, and F6.52 on binding and function of the endogenous agonist 5-HT, as well as, PAT, another 5-HT2C agonist/5-HT2A/2B inverse agonist, (2S, 4R)-(−)-trans-4-(3'-bromophenyl)-N,N-dimethy-1,2,3,4-tetrahydronaphthalene-2-amine (3'-Br-PAT), and a 5-HT2A/2B/2C inverse agonist, (2S, 4R)-(−)-trans-4-cyclohexyl-N,N-dimethy-1,2,3,4-tetrahydronaphthalene-2-amine (CAT). The W6.48, F6.51, and F6.52 aromatic residues are fully-conserved among 5-HT2 receptors and fully- or semi-conserved among several other aminergic neurotransmitter GPCRs, thus, results here are expected to provide a molecular basis for design of 5-HT2C-specific agonist ligands for development as novel antipsychotic drugs.
2. Theoretical methods
2.1. Homology Modeling
The crystal structure of the β2AR/T4-lysozyme chimera (PDB entry 2RH1), [15] was used as template to build the human 5-HT2C homology model. A full description of the methods is described elsewhere [31]. Briefly, the 5-HT2C native sequence was aligned to the β2AR sequence using ClustalW multiple sequence alignment [39, 40]. Other structures (e.g., inverse agonist carazolol, T4-lysozyme, and cholesterol molecules) present in the β2AR/T4-lysozyme chimera crystal structure were deleted. Point mutations were performed as needed and the gaps were analyzed, followed by the appropriate sequence additions and deletions to match the 5-HT2C receptor amino acid sequence. TMHs were built using the Biopolymer module of Sybyl-X 1.2 [41] and the crude model of the unbound receptor was minimized using the Powell method implemented in Sybyl with Tripos force field [42] and AMBER charges [43], followed by equilibration in a 1-palmitoyl-2-oleyl-sn-glycero phosphatidyl choline (POPC) bilayer [44]. The system was relaxed using the Tripos force field to a gradient 0.05 Kcal/Å mol, prior to MD simulations in the POPC membrane. MD simulation conditions were time run 5 μs, time step 1 fs, with snapshots collected every 5 fs. Other parameters were the NVT canonical ensemble, 300 K temperature, Boltzmann initial velocities, and non-bonded cutoff set at 8 Å. Constraints for alpha carbons in the TM domains were employed. Subsequently, the constraints were removed for a 1000 ps MD simulation run. The final unbound wild-type 5-HT2C homology model was obtained from the median structure after clustering analysis of the frames from the last 10 ps. of the MD simulation, and optimized using the Tripos force field to a convergence of 0.05 Kcal/Å.mol. The W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptor models used the same optimization and MD simulation parameters as described above for the WT receptor model.
2.2. Ligands
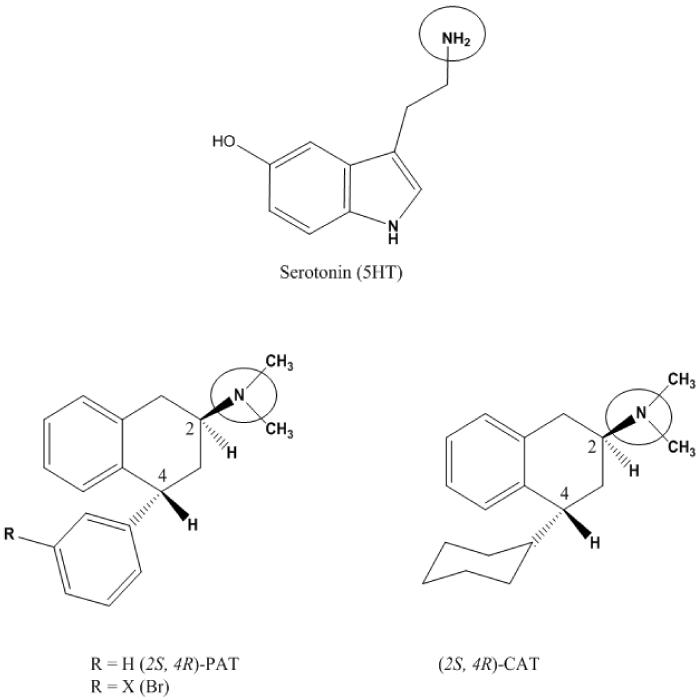
The ligands (5-HT, PAT, 3'-Br-PAT, CAT; structures in Figure 1) were built as monocations (protonated amines) using HyperChem 8.0 [45] and optimized using PM3 model Hamiltonian to a gradient of 0.01 Kcal/ Å mol. Synthesis and absolute configuration (based on X-ray crystal structure) of PAT, 3'-Br-PAT and CAT (Figure 1) are reported elsewhere [46, 47] and 5-HT was purchased from was purchased from Alfa Aesar (Ward Hill, MA).
Figure 1.
Ligand used in docking studies at WT and point mutated W6.48A, F6.51A, and F6.52A 5-HT2C GPCRs include: serotonin (5HT), PAT, 3'-Br-PAT and CAT. The protonation site is circled.
2.3. Docking
Ligands were pre-positioned in the binding pocket by performing rigid docking with the PatchDock server [48]. The low-energy-high-score solutions were analyzed to select the initial configuration, ensuring the essential interaction between the carboxylate oxygen of receptor residue D3.32 and the ligand protonated amine moiety [18, 20]. The initial ligand-receptor complex configuration was used for flexible ligand docking with Flexidock in Sybyl-x 1.2 [41]. The binding site was defined by assigning residue D3.32 as a definitive binding site interaction point with the ligand protonated amine moiety [24–28], and including residues within a 7 Å radius. Structure preparation was carried out prior to docking studies by assigning AMBER [43] charges for the protein and Gesteiger-Marsili [49] charges for the ligand. Rotatable bonds in the ligand and the side chains of residues defining the receptor putative active site were screened for optimal positioning of the ligand and side chains in the conformational space; remaining residues were frozen during docking. Default FlexiDock parameters were set at 80,000-generation. The best docking solution, according to the highest FlexiDock score, was minimized using the Tripos force field to a gradient 0.05 Kcal/Å mol, prior to molecular dynamics simulation.
2.4. Molecular Dynamics
The selected high-score pose of the docked ligand was subjected to a MD simulation run for 500 ps, with other parameters the same as above, to allow adjustment of the positions of side chains and helices. The final structure of the ligand docked at the receptor model was obtained from the average of last 10 ps of the MD simulation.
2.5. Ligand Affinity and Function Studies
Saturation and competition binding studies were carried out as previously described [29, 38]. The cDNA encoding the human unedited WT 5-HT2C receptor was obtained from UMR cDNA Resource Center (Rolla, MO). The W6.48A, F6.51A and F6.52A point-mutated 5-HT2C receptors were generated by PCR using our previously reported procedures [29, 38]. WT, W6.48A, F6.51A, and F6.52A 5-HT2C receptors were radiolabeled with [3H]-mesulergine (specific activity 92 Ci/mmol; Perkin-Elmer, Waltham, MA) and nonspecific binding was defined with 10 μM mesulergine hydrochloride (Tocris, Ellisville, MO). Ligand affinity (based on competitive displacement of radioligand) is expressed as Ki value by conversion of the IC50 data using the equation Ki = IC50/1 + L/KD, where L is the concentration of radioligand having affinity KD [50]. Comparisons of Ki values were performed using two-way ANOVA with Bonferroni's post-hoc test. Differences were considered statistically significant when the p-value was less than 0.05.
Functional activity was measured as phospholipase C (PLC) activation and [3H]-inositol phosphates (IP) formation in HEK cells transiently expressing WT, W6.48A, F6.51A, or F6.52A 5-HT2C receptors, as previously reported in detail [29, 38]. Resulting data were analyzed using the nonlinear regression algorithms in Prism, with the one-site model providing the best fit. Data is expressed as mean percentage of basal control [3H]-IP formation, with potency expressed as concentration required to stimulate (EC50) [3H]-IP formation by 50% ± SEM (n ≥ 3). Comparisons of potency values (EC50) were performed using Student's t-test.
3. Results and Discussion
3.1 Homology Model of 5-HT2C Receptor
Sequence alignment (performed with ClustalW) of the WT of 5-HT2A, 5-HT2B, and 5-HT2C receptors with the β2AR template structure allowed visualization of the conserved TMHs 3–7 (Table I; conserved residues are in bold print and reference residues are labeled according to Ballesteros standard GPCR nomenclature [33, 34]). The very close similarity of the 5-HT2 TMHs demands careful analysis of the configuration of the orthosteric pocket, particularly with regard to residues W6.48, F6.51, and F6.52.
Table I.
Partial alignment of 5-HT2 and β2AR GPCRs sequences using ClustalW, showing transmembrane helices TMH 3,5,6,7 highlighted in yellow. Conserved residues are indicated in bold. Reference residues are labeled according to Ballesteros nomenclature [33, 34]

|
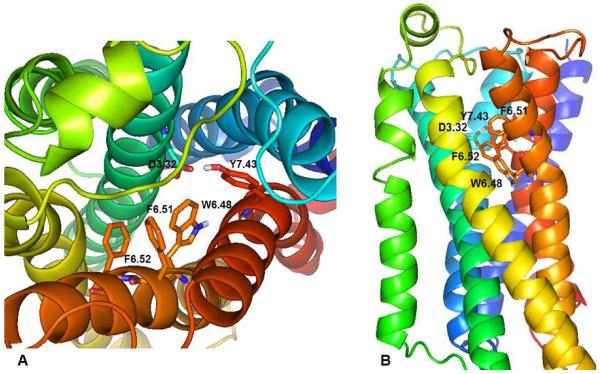
The alignments were verified using conserved residue sequences among 5-HT2 GPCRs and the β2AR in the TMHs (Table I). These sequences are: GNXLVI motif in TMH 1, with reference residue N1.50 (not shown), TNYF and SLAXAD motifs in TMH 2, with reference residue D2.50 (not shown), DVL, TASI, and DRY motifs in TMH 3, with essential reference residue D3.32, reference residue W4.50 in TMH 4, FXXPLXIM motif in TMH 5, with reference residue P5.50, WXPFFIXXNI motif in TMH 6 (that includes the W6.48, F6.51, F6.52 aromatic residues in this study), with reference residue P6.50, and WIGY and NPLXY motifs in TMH 7, with reference residue P7.50. The β2AR has been used to build homology models of several related aminergic neurotransmitter GPCRs, including, our previously reported 5-HT2C receptor homology model that was validated by Ramachandran analysis, as well as, mutagenesis and ligand binding pharmacological studies [29–32]. Figure 2, produced with PyMOL 1.3 [51], shows two views of the TMH bundle of the unbound 5-HT2C receptor. The TMHs are spectrum color coded, blue for TMH 1 through red for TMH 7. The orthosteric ligand binding pocket is located in the upper-third part of the TMH bundle, involving residues in TMHs 3, 5, 6, and 7. Residue D3.32 in TMH 3 (forms the essential ionic interaction with the ligand protonated ligand amine moiety), residues W6.48, F6.51, F6.62 in TMH 6, and residue Y7.43 in TMH 7 are labeled. Similar to the cognate 5-HT2A receptor [35–37], the 5-HT2C aromatic residues W6.48, F6.51, and F6.52 form the putative ligand binding pocket.
Figure 2.
5-HT2C Trans-membrane helices (TMH) bundle, color coded from blue for TMH 1 through red for TMH 7. A: TMH view from the extracellular domain. B: TMH across the membrane; extracellular domain is on top and intracellular at the bottom. The binding site is in the upper 1/3 of the TM bundle. Essential residue D3.32 and aromatic residues W6.48, F6.51, F6.52, and Y7.43 are displayed
3.2. Ligand–Receptor Interactions
3.2.1 Mesulergine
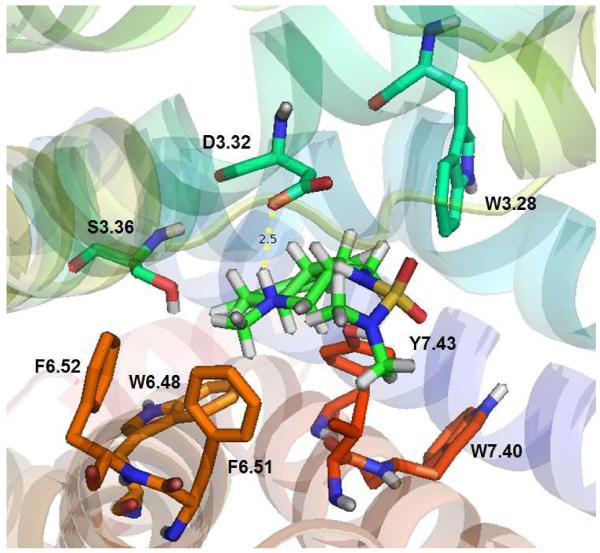
After MD simulations in the POPC lipid bilayer, the equilibrated 5-HT2C structure was used in ligand docking studies. The WT 5-HT2C receptor radioligand [3H]-mesulergine retains affinity for W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptors [52], thus, it was predicted that critical interactions between mesulergine and these TMH 6 residues would not be apparent. Indeed, as shown in Figure 3, no significant interactions between mesulergine and 5-HT2C residues W6.48, F6.51, and F6.52 were observed. These results provide a parsimonious explanation for [3H]-mesulergine radiolabeling of W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptors, and, validate the use of [3H]-mesulergine in the competition displacement studies here using the point-mutated receptors.
Figure 3.
Mesulergine docked at WT 5-HT2C receptor. No close interactions with W6.48, F6.51, and F6.52 are observed. Residues are displayed in spectrum color coded scheme indicating the TMH, i.e., orange for W6.48, F6.51, and F6.52, in TMH 6 and red for W7.40, Y7.43 in TMH 7.
3.2.2 Serotonin (5-HT)
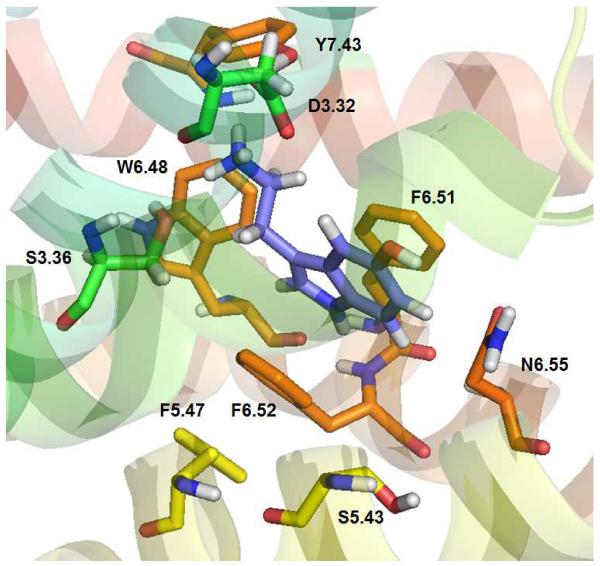
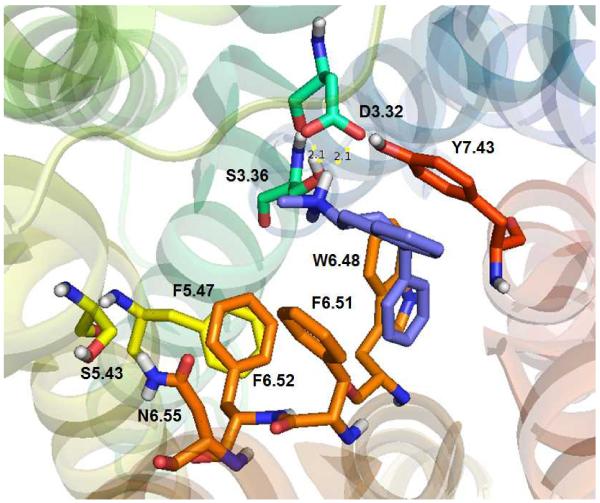
Previously, it has been reported that 5-HT docks to the WT 5-HT2C receptor binding pocket in two low energy configurations [29]. The lowest energy configuration of 5-HT is shown docked to the WT 5-HT2C receptor model here in Figure 4. The 5-HT indole moiety docks parallel to the aromatic ring of 5-HT2C residue F6.51, with the distance between the two entities being 3.5 Å, suggesting, π–π interactions. 5-HT also docks nearly parallel to the F6.52 residue, at a longer distance (4.2 Å) as compared to the interaction with F6.51, but, within the likelihood of π–π interactions. These results suggest an expected detrimental effect regarding 5-HT affinity at the F6.51A point-mutated 5-HT2C receptor and a more moderate negative effect on affinity at the F6.52A 5-HT2C receptor, as compared to the WT receptor, and, indeed, this is the case (see Table II). The W6.48 residue is in the binding pocket, however, the indol moiety of W6.48 is close to the terminal CH2 of 5HT ethylamino (2.5 Å) and far from the indol system of 5-HT (6.2 Å) and cannot form π–π interactions, consequently, affinity of 5-HT for the W6.48A point-mutated 5-HT2C receptor is not expected to be largely different than its affinity at the WT receptor, and, indeed, this is the case (see Table II). Similar results are obtained for the other low energy pose of 5-HT [29].
Figure 4.
5-HT docked at the WT 5-HT2C receptor. The indole moiety in 5-HT is parallel to aromatic ring of F6.51 (3.5 Å) and F6.52 (4.2 Å); W6.48 also is in the binding pocket but not able to form π–π interactions with 5-HT.
Table II.
Ligand affinities (Ki ± SEM; nM) of test ligands at WT and point-mutated 5-HT2C GPCRs
| Test Ligand | WT 5-HT2C | W6.48A 5-HT2C | F6.51A 5-HT2C | F6.52A 5-HT2C |
|---|---|---|---|---|
| 5-HT | 7.5 ± 3.5 | 37.2 ± 10.9 | 4036 ± 1339 | 1059 ± 637.8 |
| PAT | 28.5 ± 9.8 | 2042 ± 830.4 | 172.1 ± 47.1 | 70.18 ± 43.1 |
| 3'-Br-PAT | 11.2 ± 6.0 | 1566 ± 182.5 | 372.9 ± 0.0 | 23.1 ± 4.0 |
| CAT | 8.3 ± 2.5 | 460.1 ± 17.1 | 35.1 ± 3.4 | 15.5 ± 8.4 |
3.2.3. PAT
Figure 5 shows PAT docked at the WT 5-HT2C receptor, with likely π–π stacking interactions occurring between the PAT 4-pheny moiety and the 5-HT2C W6.48 indole group (3.8 Å). The PAT tetrahydronaphthalene aromatic ring docks nearly parallel to the 5-HT2C F6.51 phenyl group, at 4.4 Å distance and the 4-phenyl ring of PAT can form T-stacking interactions with F6.52 aromatic ring, 4.5 Å. Although the 5-HT2C F6.52 residue appears to form part of the binding pocket, distance 5.3Å from PAT, and the 4-phanyl ring of PAT is not parallel to the aromatic ring of F6.52 precluding π–π stacking, however, F6.52 likely contributes to the hydrophobicity of the binding pocket. It is noted that F6.52 appears to be involved in π–π interaction with F5.47 in TMH 5, likely, contributing to stabilization of the binding pocket. The interactions observed at the WT 5-HT2C receptor model are, of course, abolished at the W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptor models, accordingly, it is expected that PAT affinity would be diminished at the W6.48A, F6.51A, and, perhaps F6.52A point-mutated 5-HT2C receptors—and, indeed this is the case (see Table II).
Figure 5.
PAT docked at WT 5-HT2C receptor. The PAT 4-phenyl moiety is able to form π–π interactions with the indole moiety of W6.48. The aromatic ring of the PAT tetrahydronapthyl moiety docks nearly parallel to the phenyl ring of F6.51, at 4.4 Å distance, and the 4-phenyl group of PAT can form T-stacking interactions with F6.51, whereas F6.52 is farther away (5.3 Å).
3.2.4. 3'-Br-PAT
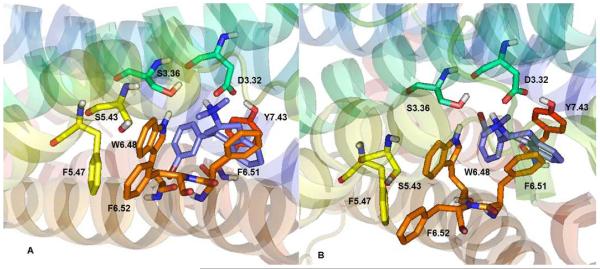
As shown in Figure 6, the 3'-Br-PAT ligand was considered in two low energy poses, with the 4-(3'-bromo)-phenyl moiety oriented toward TMH6 (panel A) or TMH 3 (panel B). Similar to PAT, the 3'-Br-PAT 4-(3'-bromo)-phenyl moiety docks nearly parallel to the 5-HT2C W6.48 indole moiety, facilitating π–π interaction (Panel A), while the aromatic part of the tetrahydronapthyl system interacts with the receptor F6.51 phenyl group—at the W6.48A and F6.51A 5-HT2C models, these interactions are lost. Affinity of 3'-Br-PAT is greatly diminished at the W6.48A and F6.51A point-mutated 5-HT2C receptors in comparison to the WT receptor (see Table II), as would be expected based on the modeling results here. Meanwhile, consistent with the lack of direct interactions observed between 3'-Br-PAT and the WT 5-HT2C receptor model (Figure 5), there is no significant difference in affinity of 3'-Br-PAT at the WT and F6.52A 5-HT2C receptors Table II).
Figure 6.
3'-Br-PAT docked at WT 5-HT2C receptor with 4-(3-bromophenyl) moiety oriented toward TMH 6 (pose 1, panel A) or TMH 3 (pose 2, panel B). Similar to PAT (Figure 5), the 4-(3'-bromo)-phenyl moiety participates in apparent π–π interactions with the W6.48 indole moiety, and, the aromatic part of the tetrahydronapthyl system interacts with F6.51; 6.52 is too far from the ligand for direct interactions.
3.2.5. CAT
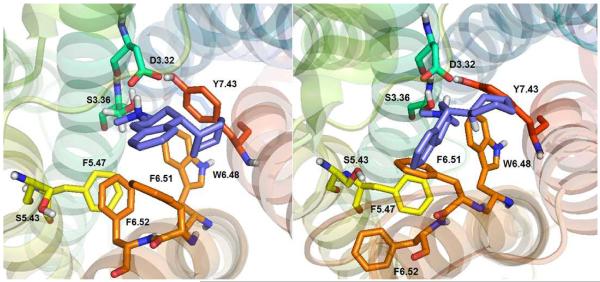
Figure 7 shows CAT docked to the WT 5-HT2C receptor. During MD simulations the CAT cyclohexyl ring underwent conformation change from the chair to semi-chair to twisted boat configuration. On average, the CAT cyclohexyl moiety docked in the chair and twisted boat conformations, as shown in Figures 7A and 7B, respectively. The cyclohexyl moiety, of course, cannot form π–π interactions with the W6.48, F6.51, and F6.52 aromatic residues that apparently still form the binding pocket, but, hydrophobic interactions are possible. Thus, it was observed that CAT docks close to W6.48 and F6.51 (4.5 and 4.1 Å), however, F6.62 was more distant, 8.1 Å. These results suggest that CAT affinity would be diminished at the W6.48A and F6.51A point-mutated 5-HT2C receptors in comparison to the WT receptor, with less affinity differences expected at the F6.52A receptor—these predictions are borne out by the experimental data in Table II.
Figure 7.
(2S, 4R)-CAT docked at WT 5-HT2C receptor. Two main poses were considered, chair (Panel A), and twisted boat (Panel B).
3.3. Ligand Affinity Results at WT and W6.48A, F6.51A, and F6.52A 5-HT2C Receptors
Ligand affinities from radio-receptor assays are shown in Table II. Experimental results indicate a large impact of the F6.51A mutation on 5-HT binding, with less effect observed for the F6.52A point-mutation, and the W6.48A mutation had no significant effect. In contrast to results for 5-HT, the W6.48A and F6.51A point-mutations had large negative effects on binding of PAT and 3'-Br-PAT, and less but significant negative impact on binding of CAT, whereas, the F6.52A point-mutation did not negatively impact binding. All in all, the experimental results closely reflect the computational molecular modeling results and clearly validate the docking studies, including, the noteworthy difference in orientation the endogenous agonist 5-HT assumes in the binding pocket as compared to the novel PAT- and CAT-type ligands.
3.4. Ligand Function at WT and W6.48A, F6.51A, and F6.52A 5-HT2C Receptors
The ligands 5-HT, PAT, 3'-Br-PAT and CAT were incubated with HEK cells expressing WT, F6.51A, F6.52A or W6.48A 5-HT2C receptors and effects on receptor-mediated second messenger signaling (PLC activity and IP formation) were measured. For the 5-HT2C agonist ligands (i.e, 5-HT, PAT, 3'-Br-PAT), effects on second messenger signaling at the point-mutated in comparison to WT 5-HT2C receptors reflected differences in binding, i.e., point-mutations that resulted in decreased ligand affinity also resulted in decreased potency and efficacy to activate PLC/IP signaling (data not shown). Unexpectedly, however, CAT, which is an inverse agonist at the WT 5-HT2C receptor, demonstrated (partial) agonist activity at the W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptors (data not shown). As this dramatic change in pharmacological activity for CAT went beyond the scope of the current work, it was decided to follow-up on this phenomenon in another report.
4. Conclusions
The computational and β2AR-based homology molecular modeling results for 5-HT, PAT, 3'-Br-PAT, and CAT were translated and validated experimentally in binding and functional studies using WT and W6.48A, F6.51A, and F6.52A point-mutated 5-HT2C receptors. Thus, computational approaches to drug design targeting GPCRs can be an accurate tool in the drug discovery and development process. Important information learned from the ligand docking and MD simulations conducted here (validated experimentally) includes the different binding modes noted for the endogenous agonist 5-HT in comparison to PAT, 3'-Br-PAT and CAT. Notably, 5-HT docked far from W6.48, precluding π–π or other interactions with this residue, which, is in contrast to computational and experimental results regarding 5-HT interaction with W6.48 at the 5-HT2A receptor [33]. The results for 5-HT suggest that an agonist ligand can dock differently at 5-HT2A vs. 5-HT2C receptors, despite the close sequence similarity between the two receptors. Thus, theoretically, it should be possible to design 5-HT2C-specific agonists for development as antipsychotic drugs. Indeed, PAT is a serendipitous example of a 5-HT2C-specific agonist that is, in fact, an inverse agonist at 5-HT2A (and 5-HT2B) receptors [38]. Future 5-HT2A molecular modeling and mutagenesis studies using PAT-type ligands analogous to the studies here for 5-HT2C receptors are expected to provide molecular insight into rational design of 5-HT2C-specific agonists, as well as, 5-HT2A-specific antagonists for development of antipsychotic drugs. Likewise, molecular modeling studies based on the very recently reported 5-HT2B crystal structure [22, 23] are expected to aide design of drugs that avoid activation of this receptor that is deleterious to cardiac valve function [1]. Also noteworthy from the ligand docking and MD simulations conducted here (validated experimentally) is the finding that ligand functional activity (i.e., agonism vs. inverse agonism) apparently can be determined by single amino acids in aminergic GPCRs. Thus, point-mutations of three different 5-HT2C residues (W6.48, F6.51, and F6.52) involved (more or less) in the binding of CAT also changed its function from an inverse agonist to a (partial) agonist. Results here from MD simulations lead to the hypothesis that the TMH 6 aromatic amino acids are impacted by conformational changes of the CAT cyclohexyl moiety that lead to stabilization of agonist vs. inverse agonist conformations of the 5-HT2C receptor—future work will assess this possibility. Thus, homology-based GPCR molecular modeling studies coupled with experimental studies can lead to molecular understanding of GPCR function, as well as structure, for drug design purposes.
Acknowledgements
NIH RO1 DA023928, DA030989, MH081193.
References
- [1].Kroeze WK, Kristiansen K, Roth BL. Curr. Top. Med. Chem. 2002;2:507–528. doi: 10.2174/1568026023393796. [DOI] [PubMed] [Google Scholar]
- [2].Jensen NH, Cremers TI, Sotty F. Scientific World Journal. 2010;10:1870–1885. doi: 10.1100/tsw.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bubar MJ, Cunningham KA. Prog. Brain. Res. 2008;172:319–346. doi: 10.1016/S0079-6123(08)00916-3. [DOI] [PubMed] [Google Scholar]
- [4].Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:793–745. [Google Scholar]
- [5].Li J, Edwards PC, Burghammer M, Villa C. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- [6].Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- [7].Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Biochemistry. 2001;40:7761–7762. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- [10].Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauβ N, Choe HW, Hofmann KP, Ernst OP. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- [11].Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- [14].Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- [16].Rassmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosembaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–181. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rosembaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Nature. 2011;469:236–242. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Congreve M, Langmea CJ, Mason JS, Marshall FH. J. Med. Chem. 2011;54(13):4283–4311. doi: 10.1021/jm200371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chien EYT, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Science. 2010;330(6007):1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Won Han G, Kobayashi T, Stevens RC, Iwata S. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Congreve M, Andrews SP, Dore AS, Hollenstein K, Hurrell E, Langmead CJ, Mason JS, Ng IW, Tehan B, Zhukov A, Weir M, Marshall FH. J. Med. Chem. 2012;55(5):1898–1903. doi: 10.1021/jm201376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang C, Jiang Y, Ma J, Wu H, Wacker D, Katritch V, Han GW, Liu W, Huang XP, Vardy E, Mc.Corvy JD, Gao X, Zhou EX, Melcher K, Zhang C, Bai F, Yang H, Yang L, Jiang H, Roth BL, Cherezov V, Stevens RC, Xu HE. Science. 2013 doi: 10.1126/science.1232808. DOI: 10.1126/science. 12232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, Mc.Corvy JD, Jiang Y, Chu M, Siu FY, Liu W, Xu FE, Cherezov V, Roth BL, Stevens RC. Science. 2013 DOI: 10.1126/science. 12232808. [Google Scholar]
- [24].Booth RG, Fang L, Huang Y, Wilczynski A, Sivendran S. Eur. J. Pharmacol. 2009;615:1–9. doi: 10.1016/j.ejphar.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bucholtz EC, Brown RL, Tropsha A, Booth RG, Wyrick SD. J. Med. Chem. 1999;42:3041–3054. doi: 10.1021/jm980428x. [DOI] [PubMed] [Google Scholar]
- [26].Kristiansen K, Dahl SG. Eur. J. Pharmacol. 1996;306:195–210. doi: 10.1016/0014-2999(96)00180-x. [DOI] [PubMed] [Google Scholar]
- [27].Kristiansen K, Kroeze WK, Willins DL, Gelber EI, Savage JE, Glennon RA, Roth BL. J. Pharmacol. Exp. Ther. 2000;293:735–746. [PubMed] [Google Scholar]
- [28].Booth RG, Fang L, Wilczynski A, Sivendren S, Sun Z, Travers S, Bruysters M, Sansuk K, Leurs R. Inflam. Res. 2008;57(suppl 1):S43–S44. doi: 10.1007/s00011-007-0621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Canal C, Cordova-Sintjago TC, Villa NY, Fang LJ, Booth RG. Eur. J. Pharmacol. 2011;673:1–12. doi: 10.1016/j.ejphar.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cordova-Sintjago TC, Fang L, Bruysters M, Leurs R, Booth RG. J. Chem. Pharm.Res. 2012;4(6):2937–2951. 2012. [PMC free article] [PubMed] [Google Scholar]
- [31].Cordova-Sintjago T, Villa N, Canal C, Booth R. Int. J. Quant. Chem. 2011;112:140–149. [Google Scholar]
- [32].Cordova-Sintjago T, Sakhuja R, Kondabolu K, Canal CE, Booth RG. Int. J. Quant. Chem. 2012;112(24):3807–3814. doi: 10.1002/qua.24237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SG, Shi L, Gether U, Javitch JA. J. Biol. Chem. 2001;276:29171–7. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- [34].Ballesteros JA, Shi L, Javitch JA. Mol. Pharmacol. 2001;60:1–19. [PubMed] [Google Scholar]
- [35].Roth BL, Shoham M, Choudhary MS, Khan N. Mol. Pharmacol. 1997;52:259–266. doi: 10.1124/mol.52.2.259. [DOI] [PubMed] [Google Scholar]
- [36].Braden MR, Parris JC, Naylor JC, Nichols DE. Mol. Pharmacol. 2006;70:1956–1964. doi: 10.1124/mol.106.028720. 2006. [DOI] [PubMed] [Google Scholar]
- [37].Chambers JJ, Nichols DE. J. Comput. Aided Mol. Des. 2002;16:511–520. doi: 10.1023/a:1021275430021. [DOI] [PubMed] [Google Scholar]
- [38].Booth RG, Fang L, Huang Y, Wilczynsky A, Sivendran S. Eur. J. Pharmacol. 2009;615:1–9. doi: 10.1016/j.ejphar.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. Clustal W and Clustal X version 2.0. [DOI] [PubMed] [Google Scholar]
- [41]. SYBYL-X 1.2, Tripos International, 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA.
- [42].Clark M, Cramer RD, III, van Opdenhosch N. J. Comp. Chem. 1989;10:982–1012. [Google Scholar]
- [43].Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr., Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- [44].Heller H, Schaefer M, Schulten K. J. Phys. Chem. 1993;97:8343–8360. [Google Scholar]
- [45]. HyperChem (TM) Professional 8.0, Hypercube, Inc., 1115 NW 4th Street, Gainesville, Florida 32601, USA.
- [46].Wyrick SD, Booth RG, Myers AM, Owens CE, Kula NS, Baldessarini RJ, McPhail AT, Mailman RB. J. Med. Chem. 1993;36:2542–2551. doi: 10.1021/jm00069a013. [DOI] [PubMed] [Google Scholar]
- [47].Vincek AS, Booth RG. Tetrahedron Lett. 2009;50:5107–5109. doi: 10.1016/j.tetlet.2009.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Nucleic Acids. Res. 2006;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gasteiger J, Marsili M. Tetrahedrom. 1980;36:3219. [Google Scholar]
- [50].Cheng Y, Prusoff WH. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- [51].The PyMOL Molecular Graphics System. Version 1.3 Schrödinger, LLC: [Google Scholar]
- [52].Villa NY, Cordova-Sintjago TC, Sakhuja R, Canal CE, Fang L, Booth RG. Soc. Neurosci. Abstr. 2012;42 [Google Scholar]