Abstract
Cluster analysis methods seek to partition a data set into homogeneous subgroups. It is useful in a wide variety of applications, including document processing and modern genetics. Conventional clustering methods are unsupervised, meaning that there is no outcome variable nor is anything known about the relationship between the observations in the data set. In many situations, however, information about the clusters is available in addition to the values of the features. For example, the cluster labels of some observations may be known, or certain observations may be known to belong to the same cluster. In other cases, one may wish to identify clusters that are associated with a particular outcome variable. This review describes several clustering algorithms (known as “semi-supervised clustering” methods) that can be applied in these situations. The majority of these methods are modifications of the popular k-means clustering method, and several of them will be described in detail. A brief description of some other semi-supervised clustering algorithms is also provided.
Keywords: cluster analysis, high-dimensional data, semi-supervised methods, machine learning
The objective of cluster analysis is to partition a data set into a group of subsets (i.e. “clusters”) such that observations within a cluster are more similar to one another than observations in other clusters. For a more detailed discussion, see Hastie et al. 1 or Gordon 2.
Traditional clustering methods are unsupervised, meaning that there is no outcome measure and nothing is known about the relationship between the observations in the data set. However, in many situations one may wish to perform cluster analysis even though an outcome variable exists or some preliminary information about the clusters is known. For example, an e-mail classification procedure may seek to characterize the properties of “spam” e-mails. Suppose a large database of e-mails is available, a small subset of which has already been classified as “spam” or “not spam.” One may wish to identify clusters in this data set such that one cluster consists primarily of “spam” and the other cluster consists primarily of “not spam.” Or in a genetic study of cancer, one may wish to identify genetic clusters that can be used to determine the prognosis of cancer patients. Such clusters would only be of interest if they were associated with the outcome of interest, namely patient survival.
Clustering methods that can be applied to partially labeled data or data with other types of outcome measures are known as semi-supervised clustering methods (or sometimes as supervised clustering methods). They are examples of semi-supervised learning methods, which are methods that use both labeled and unlabeled data3–6. This review will briefly describe several semi-supervised clustering methods that can be applied to different types of partially labeled data sets. The review will focus primarily on variations of k-means clustering, since most existing semi-supervised clustering methods are modified versions of k-means clustering. However, a brief description of some semi-supervised hierarchical clustering methods will also be provided.
Traditional (Unsupervised) Clustering Methods
This section will briefly describe two of the most common traditional cluster analysis methods, namely k-means clustering and hierarchical clustering.
K-Means Clustering
K-means clustering is one of the most popular cluster analysis methods. It is generally applied to data sets where all the variables are quantitative and the distance between observations is measured using the squared Euclidean distance, which is defined as follows:
| (1) |
Here xi and xi′ are observations from a data set with p features, and xij represents the value of the jth feature for observation i. The k-means clustering algorithm attempts to assign each observation to a cluster to minimize the following objective function:
| (2) |
In the above expression, K represents the number of clusters, and Ci represents the cluster to which observation i is assigned, where 1 ≤ Ci ≤ K. This objective function is also known as the “within-cluster sum of squares” or WCSS. Note that (2) can be written as:
where nk is the number of observations in cluster k and x̄kj is the mean of feature j in cluster k.
Several k-means clustering algorithms have been proposed to minimize (2) 7–10. However, each algorithm uses some variation of the following strategy:
Randomly assign each observation to an initial cluster.
For each feature j and cluster k, calculate x̄kj, the mean of feature j in cluster k.
- Assign each observation i to a new cluster Ci as follows:
Repeat steps 2 and 3 until the algorithm converges.
The above algorithm is guaranteed to converge, but it may converge to a local minimum. Hence, it is advisable to repeat the algorithm multiple times with different initial clusters and choose the set of clusters that produces the minimum WCSS. For a more detailed discussion of k-means clustering and several variations of the k-means algorithm see Hastie et al. 1.
The k-means clustering algorithm requires one to choose the number of clusters K. Several methods have been proposed for choosing K. One common method is the “gap statistic” proposed by Tibshirani et al. 11. Let Wk be the WCSS (2) when K = k. It is simple to verify that Wk will always decrease as k increases, so one cannot simply choose the value of K that minimizes WK. The motivation for the gap statistic is the following: Let K* denote the true value of K. If k < K*, then at least one cluster produced by the k-means algorithm is actually two separate clusters, and so Wk+1 should be significantly smaller than Wk. On the other hand, if k > K*, then at least two clusters produced by the k-means algorithm are actually a single cluster, so Wk−1 should be only slightly larger than Wk. Thus, the gap statistic seeks to identify the smallest K such that Wk does not decrease significantly for k > K.
Formally, the gap statistic is defined to be
The expected value E [log(Wk)] is calculated under a suitable reference distribution. One common choice of a reference distribution is a multivariate uniform distribution with the same range as the data set of interest. In this case, this expected value may be estimated by sampling from this (uniform) reference distribution. Tibshirani et al. 11 estimate the number of clusters K as follows:
where sk is the estimated standard deviation of E [log(Wk)]. The idea is that when k ≥ K* then Gk+1 ≈ Gk, so one may estimate K* by choosing the minimum k such that Gk+1 ≈ Gk.
A number of other methods have been proposed for choosing the number of clusters K12–14. See the aforementioned references for details of these methods.
Hierarchical Clustering
K-means clustering is an example of what are known as partitional clustering methods, which partition a data set into a fixed number of disjoint subgroups. In contrast, hierarchical clustering groups data points into a series of clusters in a tree-like structure. At each level of the tree, clusters are formed by merging clusters at the next lower level of the tree. Thus, each data point forms a singleton cluster at the bottom level of the tree, and the top level of the tree consists of a single cluster containing all of the data points.
There are a wide variety of different methods for hierarchical clustering. This review will briefly describe a few of the most common hierarchical clustering methods, although many other hierarchical clustering methods have been proposed. See Hastie et al. 1 for more information (including descriptions of several other hierarchical clustering methods).
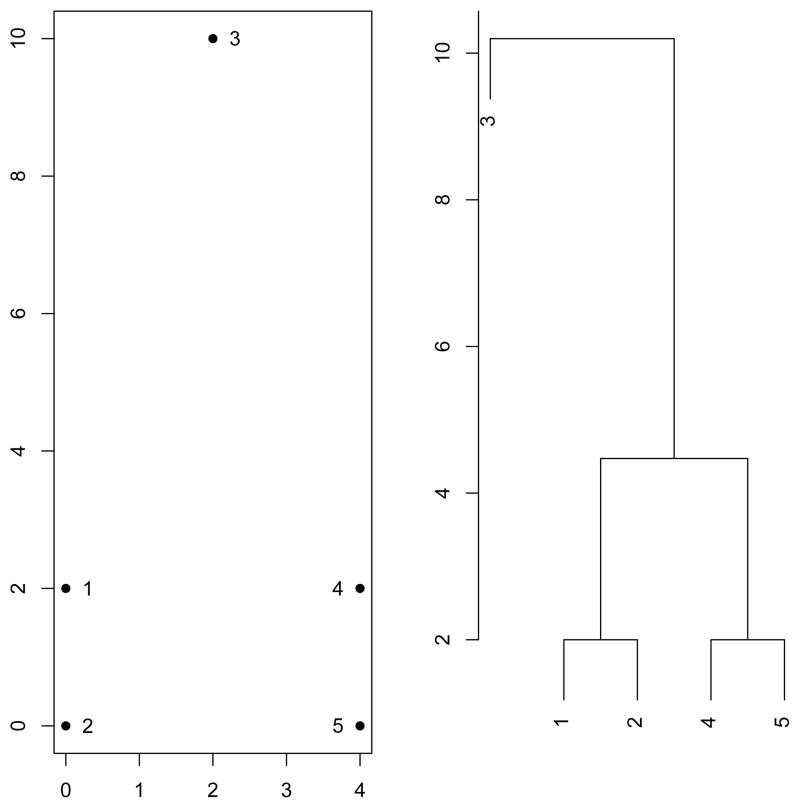
One of the most common hierarchical clustering methods is agglomerative hierarchical clustering. Agglomerative hierarchical clustering methods start with the set of individual data points and merge the two “most similar” points into a cluster. At each step of the procedure, the two “most similar” clusters (which may be individual data points) are merged until all of the data points have been merged into a single cluster. See Figure 1 for an illustration of agglomerative hierarchical clustering.
Figure 1.

This figure illustrates how hierarchical clustering would partition a simple data set. In the first two steps, the two pairs of adjacent points would each be combined into a single cluster. In the third step, these two clusters would be combined into a larger cluster. In the final step, the remaining point would be combined to this cluster. All the data points are now combined into a single cluster, so the algorithm terminates.
In order to apply the hierarchical clustering algorithm described above, one must define how the pair of “most similar” clusters is chosen. Note that for hierarchical clustering it is not sufficient to define a dissimilarity (or distance) measure between pairs of points; one must also define a dissimilarity measure between pairs of clusters. Many different dissimilarity measures have been proposed for hierarchical clustering, but the most commonly used methods start by defining a dissimilarity measure between pairs of points. The Euclidean distance defined in (1) is a common choice, but other dissimilarity measures are possible. For example, when clustering DNA microarray data, is it common to define the dissimilarity measure between two points to be 1 − ρ, where ρ is the Pearson correlation coefficient between the two points.15
Once a dissimilarity measure between two points has been defined, there are several ways to define distances between clusters. Two common dissimilarity measures are known as “single linkage” and “complete linkage.” Let C1 and C2 denote the indices of the elements in two clusters. In other words, i ∈ C1 if and only if data point xi is contained in the first cluster. Also, let d(xi, xi′) be the dissimilarity between data points xi and xi′. Then the single linkage dissimilarity between the two clusters is defined to be
and the complete linkage dissimilarity is defined to be
Other dissimilarity measures between clusters can also be used. For example, one could define the dissimilarity between two clusters to be the average dissimilarity between the elements of the two clusters:
where n1 and n2 are the number of data points in clusters 1 and 2, respectively. Each such dissimilarity measure between clusters has certain advantages and disadvantages. See Hastie et al. 1 for details.
As noted earlier, the results of hierarchical clustering may represented as a binary tree. Each node of the tree represents a cluster. (In particular, the root node is the topmost cluster which contains all of the data points, and each terminal node corresponds to a singleton “cluster” consisting of a single data point.) This tree structure can be represented in a graphical form known as a dendogram. It is customary to plot the dendogram such that the height of each node in the tree corresponds to the dissimilarity between the two clusters that were merged to form the cluster. See Figure 2 for an example of a dendogram of a simple data set.
Figure 2.
Hierarchical clustering was applied to the five data points plotted in the left panel. The resulting dendogram is shown on the right panel. Note that point 3 is much more distant from (and hence dissimilar to) the remaining four points. Thus, the height of the node where point 3 is merged to the remaining points is higher than the height of the other nodes in the graph.
Semi-Supervised Clustering Methods
We will now briefly outline several semi-supervised clustering methods. These methods will be organized according to the nature of the known outcome data. First, we will consider the simplest case, namely the case where the data is partially labeled. In other words, the cluster assignments are known for some subset of the observations. We will then consider the case where some sort of relationship between the features is known, and finally the case where one seeks to identify clusters associated with a particular outcome variable.
Partially Labeled Data
In some situations, the cluster assignments may be known for some subset of the data. The objective is to classify the unlabeled observations in the data to the appropriate clusters using the known cluster assignments for this subset of the data.
In a certain sense, this problem is equivalent to a supervised classification problem, where the objective is to develop a model to assign observations in a data set to one of a finite set of classes based on a training set where the true class labels are known. However, traditional supervised classification methods may be inefficient when only a small subset of the data is labeled. For example, if one wishes to classify web pages into a discrete number of groups, one can easily collect millions of unlabeled observations, but classifying any given observation requires human intervention (and hence is likely to be slow). Similarly, if one wishes to develop a method to classify e-mails as “spam” or “not spam,” then one can easily collect numerous unlabeled observations, but the proportion of labeled observations will be much smaller. For these types of problems, conventional supervised classification methods may be inefficient since they typically do not use unlabeled data to build the classification algorithm. Thus, the vast majority of the available data will not be used. In these situations, one can often build more accurate classification rules by combining both labeled and unlabeled data. See Blum and Mitchell 3 or Joachims 4 for a more detailed discussion and examples.
Basu et al. 16 developed a generalization of k-means clustering (which they called “constrained k-means”) for the situation where class labels are known for a subset of the observations. Once again, we let xi and xi′ be observations from a data set with p features, and xij represents the value of the jth feature for observation i. Suppose further that there exists subsets S1, S2, …, SK of the xi’s such that xi ∈ Sk implies that observation i is known to belong to cluster k. (Here K denotes the number of clusters, which is also assumed to be known in this case.) Let |Sk| denote the number of xi’s in Sk. Also let . The algorithm proceeds as follows:
- For each feature j and cluster k, calculate the initial cluster means as follows:
- Assign each observation i to a new cluster Ci. If xi ∈ S, then let Ci = Sk, where xi ∈ Sk. Otherwise let
(3) For each feature j and cluster k, calculate x̄kj, the mean of feature j in cluster k.
Repeat steps 2 and 3 until the algorithm converges.
Note that this procedure is identical to the conventional k-means procedure with the exception of the initial cluster assignments (which are generally arbitrary anyway) and step 2. In step 2, labeled observations are always assigned to their known cluster even if they are closer to the mean of another cluster.
The constrained k-means clustering algorithm described above assumes that none of the labeled observations are misclassified. Using the constrained k-means clustering procedure, if a labeled observation is misclassified, this misclassification can never be corrected, since this observation will be assigned to the same cluster in step 2 in every iteration of the algorithm. Thus, Basu et al. 16 recommend an alternative algorithm (which they call “seeded k-means clustering”) that is identical to constrained k-means clustering with the exception of step 2. The seeded k-means clustering algorithm always assigns observations to the nearest cluster using (3) even if the observation is labeled. Thus, if an observation is initially mislabeled, then the mislabeled observation may be corrected if it is closer to the cluster center of a different cluster.
Observe that seeded k-means clustering is identical to conventional k-means clustering with the exception of the first step in the procedure. Thus, seeded k-means clustering is simply conventional k-means clustering that uses the labeled data to help choose the initial cluster centers. A similar approach is used in the supervised sparse clustering method of Gaynor and Bair 17, which is described below.
Methods for clustering partially labeled data can be useful when analyzing DNA microarray data. In a typical microarray experiment, one measures the gene expression levels of p genes for each of n samples, where normally p ≫ n. One may wish to identify clusters of genes with similar expression levels across samples, since the genes in each such cluster may belong to the same biological pathway. If certain genes are known to belong to certain pathways prior to performing the experiment, then the cluster labels for these genes are known. In this situation, one seeks to cluster the remaining genes using the information from the labeled genes. Several clustering methods have been developed for the specific problem of analyzing partially labeled microarray data18–26. These methods are specifically designed for microarray data and will not be described in this review; see the references for details.
Known Constraints on the Observations
We now consider clustering when more complex relationships among the observations are known. In particular, we will consider two types of possible constraints among observations: “Must-link constraints” require that two observations must be placed in the same cluster, and “cannot-link constraints” require that two observations must not be placed in the same cluster. One possible application is when repeated measurements are collected on some subset of the experimental units. In such a situation, one may want to assign all of the repeated measurements of the same experimental unit to the same cluster.
Note that this is a generalization of the problem considered in the previous section, where the cluster assignments are known for a subset of the features. In that situation, for each feature j that is known to belong to cluster k, one may impose a must-link constraint between feature j and all other features known to belong to cluster k and a cannot-link constraint between feature j and features known not to belong to cluster k. Numerous methods have been proposed for solving the problem of constrained clustering. This review will briefly describe a few of the most commonly used methods, and references for numerous other methods are listed below. Also see Basu et al. 27 for a more detailed description of various algorithms for constrained clustering.
Wagstaff et al. 28 proposed the following algorithm (with they called “COP-KMEANS”) for solving clustering problems given this type of constraint:
Randomly assign each observation to an initial cluster.
For each feature j and cluster k, calculate x̄kj, the mean of feature j in cluster k.
-
Assign each observation i to a new cluster Ci as follows:
where
Dik = {k : no constraints are violated when observation i is assigned to cluster k}
Repeat steps 2 and 3 until the algorithm converges. The algorithm fails if Dik = ∅ for any i at any step of the procedure.
Note that COP-KMEANS is identical to conventional k-means clustering with the exception of step 3. COP-KMEANS assigns each observation to the nearest cluster such that no constraints are violated (whereas conventional k-means clustering assigns each observation to the nearest cluster without considering the constraints).
One potential drawback of the COP-KMEANS algorithm is the fact that it requires that no constraints are violated. In some situations, one may wish to allow for the possibility that some constraints may be violated if there is a strong evidence that a particular constraint is incorrect. Thus, Basu et al. 29 proposed a method (which they call “PCKmeans”) that solves the problem of identifying clusters given a set of must-link and cannot-link constraints on the observations that allows some constraints to be violated. PCKmeans seeks to minimize a modified version of the objective function (2) that is defined as follows: Let observations (xi, xi′ ) ∈
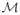 if there is a must-link constraint between observations i and i′, and let (xi, xi′) ∈
if there is a must-link constraint between observations i and i′, and let (xi, xi′) ∈
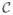 if there is a cannot-link constraint between observations i and i′. Then PCKmeans minimizes the following objective function:
if there is a cannot-link constraint between observations i and i′. Then PCKmeans minimizes the following objective function:
| (4) |
Here li,i′ is a user-defined penalty for violating a must-link constraint between observations i and i′ and is the penalty for violating a cannot-link constraint between i and i′. See Basu et al. 29 for details of the PCKmeans algorithm for minimizing (4).
The methods described above modify an existing clustering method (namely k-means clustering) such that the constraints are satisfied. Thus, such methods are sometimes referred to as “constraint-based methods” in the literature6,30. In contrast, “distance-based methods” (or “metric-based methods”) use an existing clustering method but modify the metric used to measure the “distance” between a pair of observations such that the constraints are satisfied. For example, rather than using the simple Euclidean distance (1), one may use an alternative distance metric such that two observations with a “must-link constraint” will necessarily have a lower distance between them 31–43. Moreover, other constraint-based methods have been proposed 44–48, and still other methods combine both of these approaches into a single model6,30. Other forms of constrained clustering are also possible, such as clustering on graph data49,50. These methods will not be described further in this review; see the original references for details.
Thus far we have also assumed that the constraints on the observations were specified when the data was collected. In some situations, the data analyst may have the opportunity to select some subset of the observations and impose constraints on this subset. For example, suppose the objective is to cluster a large set of text documents based on the frequency of selected words that appear in the documents. One may manually examine any given pair of documents to determine if they should be classified to the same cluster (and hence imposing either a must-link constraint or a cannot-link constraint). Suppose a researcher looked up the titles of three documents and determined that two of the documents were romance novels for teenagers and the third document was an article from a medical journal. In this case, the researcher would impose a must-link constraint between the two novels and a cannot-link constraint between each novel and the journal article. However, there is a cost associated with making such a determination, so typically one may only analyze a small subset of the observations. In such a situation, it is advantageous to choose this subset to maximize the information about the clusters.
Basu et al. 29 describe a variant of PCKmeans (called “active PCKmeans”) that chooses a subset of the observations on which to impose constraints such that the accuracy of the clustering algorithm is maximized. They show that this method outperforms the generic PCKmeans algorithm for this type of problem. For other methods for constraint selection in this situation, see Greene and Cunningham 51 or Mallapragada et al. 52.
Semi-Supervised Hierarchical Clustering
The majority of existing semi-supervised clustering methods are based on k-means clustering or other forms of partitional clustering. Comparatively few semi-supervised hierarchical clustering methods have been proposed 53. This is partly due to the fact that the problem must be formulated differently for hierarchical clustering. As noted earlier, most semi-supervised partitional clustering methods utilize either partially labeled data or known constraints (e.g. “must-link” or “cannot-link” constraints) on the observations. It is more difficult to define such constraints for hierarchical clustering, since hierarchical clustering links all observations in a data set at some level of the clustering hierarchy. Thus, a “must-link” constraint will always be satisfied at some level of the hierarchy and likewise a “cannot-link” constraint will always be violated.
Hence, semi-supervised hierarchical clustering methods have considered different types of constraints. For example, Miyamoto and Terami 54 require observations linked by a “must-link” constraint to be clustered together at the lowest possible level of the hierarchy. They further require that observations separated by a “cannot-link” constraint must not be part of the same clustering hierarchy. Thus, rather than identifying a single clustering hierarchy, the method of Miyamoto and Terami 54 returns several clustering hierarchies. A separate hierarchy is produced for each observation that is part of a “cannot-link” constraint. Several related methods have been proposed to perform hierarchical clustering subject to such constraints45,54–56.
Other types of constraints have been proposed for semi-supervised hierarchical clustering. Bade and Nurnberger 57 describe a method for performing hierarchical clustering given a set of “must-link before” constraints, where certain a certain set of observations must be clustered together before they are clustered with other data points.53 develop an alternative method for hierarchical clustering with this type of constraint. Zhao and Qi 58 consider hierarchical clustering with “ordering constraints,” wherein observations must be combined in a certain order. In other words, given an ordering constraint of (x3, x1, x4, x2), observations x1 and x3 must be clustered together before they can be combined into a cluster containing x4, and observations x1, x3, and x4 must be clustered together before they can be combined into a cluster containing x2. Hamasuna et al. 59 define “clusterwise tolerance based pairwise constraints” which define “must-link” and “cannot-link” constraints between pairs of clusters based on a weighted count of the number of such constraints that exist between observations in the clusters. They developed algorithms for implementing several variants of hierarchical clustering subject to this type of constraint59–61.
Most of these methods for semi-supervised hierarchical clustering are very new and little research has been performed on the advantages and disadvantages of the various methods. The development of methods for semi-supervised hierarchical clustering remains an active research area.
Clusters Associated with an Outcome Variable
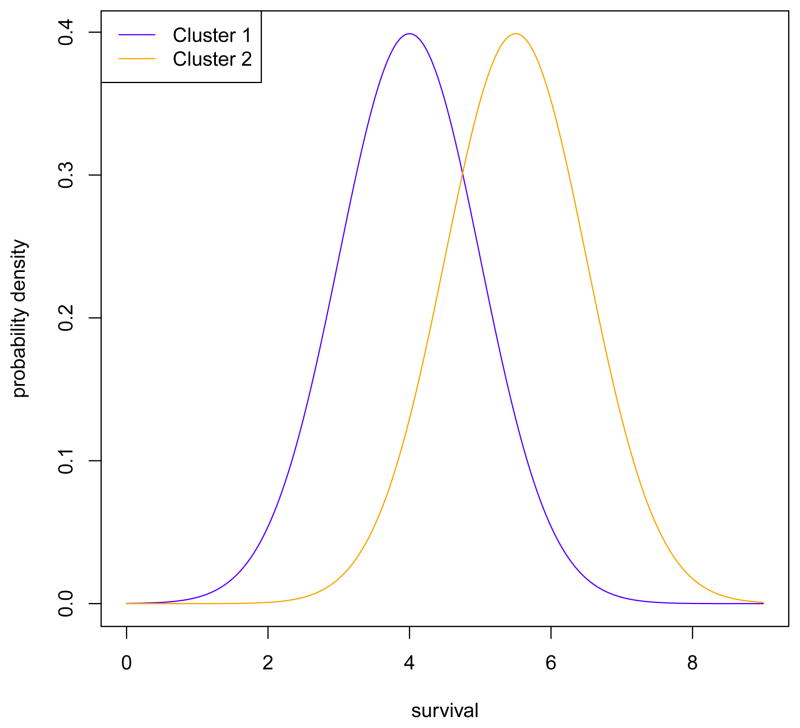
In other situations, one may wish to identify clusters that are associated with a given outcome variable. Typically the outcome variable is a “noisy surrogate”62 for the (unobserved) clusters of interest. For example, in genetic studies of cancer, there may exist subtypes of cancer with different genetic characteristics. Some subtypes may be more likely to metastasize, resulting in a poorer prognosis for patients with these subtypes. In this case these genetic subtypes are unobserved, but the survival times of the patients in the study may be available. A patient who has a “high-risk” subtype is more likely to have a low survival time than a patient who has a “low-risk” subtype, but there is considerable variation within subtypes. It is possible to observe a patient with a “low-risk” subtype and a low survival time (and vice versa). See Figure 3 for an illustration of such a scenario. In this example, patients in cluster 2 have a higher mean survival time than patients in cluster 1, but there is significant overlap in the two groups, so it is not possible to identify the clusters using only the survival times.
Figure 3.
This figure shows an example of a situation where an (observed) outcome variable (namely survival) is a “noisy surrogate” for two unobserved clusters. Suppose there are two subtypes of cancer, and patients with the first subtype (cluster) tend to have lower survival than patients with the second subtype. However, there is considerable overlap in the distribution of the survival times, so while a patient with a low survival time is more likely to be in cluster 1, it is not possible to assign each patient to cluster based only on their survival time.
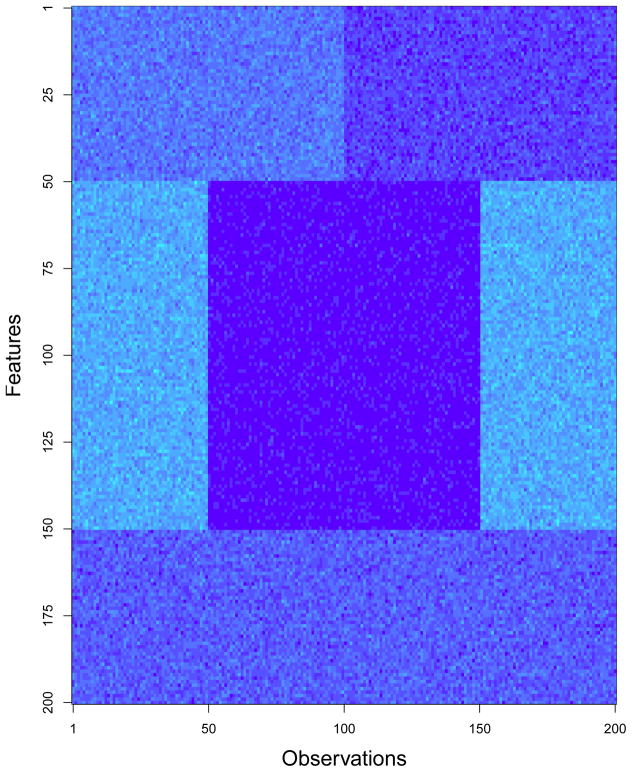
Since conventional clustering methods do not use the values of an outcome variable, they may fail to identify clusters associated with the outcome and instead identify clusters unrelated to the outcome. Figure 4 shows an example of a situation where a specialized clustering method is needed to identify clusters associated with an outcome variable of interest. In this situation, features 1–50 form clusters that are associated with the outcome variable and features 51–150 form clusters that are unrelated to the outcome variable. Conventional clustering methods will nevertheless identify the clusters defined by features 51–150, since the distance between the centers of these clusters is greater than the distance between the centers of the clusters defined by features 1–50. Thus, special methods are needed to identify the clusters of interest (i.e. the clusters defined by features 1–50) in this scenario.
Figure 4.
This figure shows an example of a data set where two different sets of clusters exist and only one cluster is associated with the outcome of interest. In the above figure, darker shades of blue correspond to higher values of the features and lighter shades of blue correspond to lower values. Suppose that observations 1–100 have a disease of interest and observations 101–200 are controls. In this case we would be interested in identifying the clusters formed by features 1–50. However, conventional clustering algorithms will identify the clusters formed by features 50–150, since the distance between the centers of these two clusters is greater than the distance between the centers of the clusters formed by features 1–50.
Despite the importance of this problem, relatively few methods have been proposed for identifying clusters associated with an outcome variable. Methods exist for identifying secondary clusters for data sets similar to the data shown in Figure 4 (see for example Nowak and Tibshirani 63), but these methods also do not use information from the outcome variable to identify the secondary clusters. One of the earliest methods for identifying clusters associated with an outcome variable is the “supervised clustering” method of Bair and Tibshirani 62, which proceeds as follows:
For each feature in the data set, calculate a test statistic Tj for testing the null hypothesis of no association between the jth feature and the outcome variable. If the outcome variable is binary (i.e. case versus control), Tj may be a t-statistic. If the outcome variable is continuous, Tj may be a t-statistic for testing the null hypothesis that the regression coefficient for predicting the outcome based on feature j is equal to 0. If the outcome variable is a right-censored survival time, Tj may be the corresponding test statistic from a Cox proportional hazards model.
Choose a threshold M, and apply k-means clustering to the features for which |Tj| > M. Features with |Tj| ≤ M are discarded and do not affect the cluster assignments.
Although this approach is relatively simple, Bair and Tibshirani 62 show that this method can identify biologically relevant clusters in several data sets. In particular, Bullinger et al. 64 used this method to identify subtypes of acute myeloid leukemia that were associated with patient survival. An advantage of this method is the fact that it performs well even when the data is high-dimensional. Since clustering is performed using only a subset of the features, a high-dimensional data set can be effectively reduced to a data set with fewer features.
This supervised clustering procedure requires the choice of a tuning parameter M, which may be chosen using cross-validation. Also, while the method proposed by Bair and Tibshirani 62 applies k-means clustering to the subset of the features that are most strongly associated with the outcome variable, one could use the same strategy of selecting the features that are most strongly associated with the outcome and then apply hierarchical clustering or an alternative clustering method. Indeed, Koestler et al. 65 propose a method called “semi-supervised recursively partitioned mixture models (RPMM)” that uses this strategy. Semi-supervised RPMM first selects a set of features that are most strongly associated with the outcome variable and then applies the RPMM method of Houseman et al. 66 to this subset of the features. One possible advantage of RPMM over k-means clustering is that RPMM does not require one to choose the number of clusters K. Koestler et al. 65 provide several examples where semi-supervised RPMM produces more accurate results than the supervised clustering method of Bair and Tibshirani 62. However, in other situations semi-supervised RPMM can fail to detect clusters even when such clusters exist; see Gaynor and Bair 17 for examples.
One possible drawback to methods such as supervised clustering and semi-supervised RPMM is the fact that any feature that is discarded after the initial screening step is permanently excluded from the analysis. This is problematic if one wishes to identify the features that differ across clusters, since it is possible for features that differ across clusters to be only weakly associated with the outcome variable, particularly if the association between the clusters and the outcome variable is weak. Indeed, if the association between the clusters and the outcome variable is very weak, supervised clustering and semi-supervised RPMM can fail to identify the correct clusters.
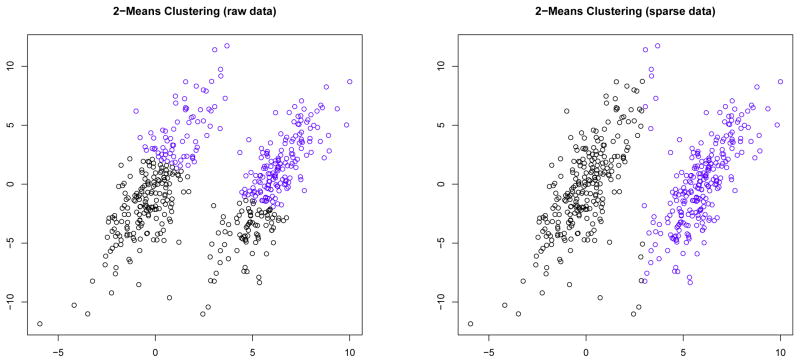
To overcome this difficulty, Gaynor and Bair 17 propose a method called “supervised sparse clustering,” which is a modification of the “sparse clustering” method of Witten and Tibshirani 67. Sparse clustering is an unsupervised clustering method that is useful when the clusters differ with respect to only a subset of the features. See Figure 5 for an example of a data set where sparse clustering produces better results than traditional k-means clustering. In this (two-dimensional) example, the clusters differ with respect to x but not with respect to y. Applying 2-means clustering to both x and y results produces inaccurate results, but applying 2-means clustering only to x identifies the correct clusters.
Figure 5.
In the above figure, there are two clusters such that the cluster means differ with respect to x but not with respect to y. If 2-means clustering is applied to both x and y, then it fails to identify the correct clusters, but 2-means clustering produces satisfactory results when applied only to x.
The following is a brief description of the sparse clustering algorithm of Witten and Tibshirani 67: First, note that minimizing the k-means objective function (2) is equivalent to maximizing
Here each xij is an observation from a data set with n observations and p features that is partitioned into K clusters, where Ci = k if and only if observation i belongs to cluster k and nk is the number of observations in cluster k. Then the sparse clustering algorithm seeks to identify weights w1, w2, …, wp for each feature to maximize
| (5) |
subject to the constraints that , and wj ≥ 0 for all j. The variable s is a tuning parameter. As s increases, the number of nonzero wj’s will increase. Thus, by choosing an appropriate value of s, the clustering will be performed using only a subset of the features (the features for which wj > 0). Note that sparse clustering imposes an L1 penalty on the feature weights, which is similar to the L1 penalty imposed on the regression coefficients in lasso regression 68 (which also causes an increasing number of coefficients to be equal to 0 as the value of the tuning parameter changes). See Witten and Tibshirani 67 for a more detailed description of the sparse clustering algorithm, including a method for choosing the tuning parameter s. In particular, Witten and Tibshirani 67 show that this sparse clustering method tends to produce better results than several previously published methods for reducing the dimension of a data set prior to clustering, such as clustering on PCA scores69,70.
Witten and Tibshirani 67 maximize (5) by using an algorithm that sets at the beginning of the procedure and then updates the wj’s iteratively. The supervised sparse clustering method of Gaynor and Bair 17 is similar to sparse clustering but chooses the initial feature weights as follows:
For each feature in the data set, calculate a test statistic Tj for testing the null hypothesis of no association between the jth feature and the outcome variable.
-
Choose a threshold M, and define the initial weights w1, w2, …, wp as follows:
where m is the number of features such that |Tj| > M.
In other words, rather than giving equal initial weights to all the features in the data set, supervised sparse clustering gives equal initial weights to the features most strongly associated with the outcome variable and an initial weight of 0 to all other features. Gaynor and Bair 17 show that this modification of sparse clustering is more likely to identify clusters that are associated with an outcome variable.
Note that supervised sparse clustering is similar to several other semi-supervised clustering methods. The method for choosing the initial cluster weights is analogous to the method for choosing the features in the supervised clustering algorithm of Bair and Tibshirani 62. Indeed, the first step of the supervised sparse clustering algorithm applies k-means clustering to the features most strongly associated with the outcome variable, which is identical to the supervised clustering method. The difference is that supervised sparse clustering updates the feature weights after identifying the initial set of clusters and iterates the procedure until convergence. Gaynor and Bair 17 show that this procedure can produce better results than supervised clustering in some situations, particularly when the outcome variable is only weakly associated with the clusters. The supervised sparse clustering procedure is also similar to the seeded k-means clustering algorithm of Basu et al. 16 since it uses the known outcome data to “seed” the initial step of the sparse clustering method and then iterates the remainder of the sparse clustering algorithm without further consideration of the outcome variable.
Conclusion
There has been considerable methodological research activity in the area of semi-supervised clustering (particularly constrained clustering) in the past decade. There now exists numerous methods for performing constrained clustering (including the special case of partially labeled data) that can be applied to a wide variety of different data sets. In particular, several methods have been developed for the special case of clustering genes in DNA microarray data, where biological information often exists about the relationships between some subset of the genes.
Nevertheless, there are several important unanswered questions in the area of semi-supervised clustering. Although many algorithms exist for performing constrained clustering, there does not appear to be extensive research comparing the performance of the various algorithms (either in terms of running time or in terms of their ability to identify clusters correctly). Thus, users of these methods may be uncertain about which method should be applied to a given data set given the large number of options. Also, in the important special case of genetic data, most existing research has focused on clustering data from DNA microarrays. One might also wish to identify gene clusters based on other types of modern high-throughput genetic data, including data from genome-wide association studies, RNA-Seq, or next-generation DNA sequencing. There is a need for semi-supervised clustering methods that can be applied to these other types of genetic data sets. Finally, as noted earlier, the problem of identifying clusters associated with an outcome variable has not been studied extensively in the literature. Only a handful of methods currently exist. Development of new methods for this problem is another potential area for future research.
Acknowledgments
This work was partially supported by NIEHS grant P30ES010126 and NCATS grant UL1RR025747. We thank the two anonymous reviewers for their helpful suggestions.
References
- 1.Hastie T, Tibshirani R, Friedman JH. Springer Series in Statistics. 2 Springer; New York, NY: 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- 2.Gordon AD. Monographs on Statistics and Applied Probability. 2 Chapman & Hall; 1999. Classification. [Google Scholar]
- 3.Blum A, Mitchell T. Combining labeled and unlabeled data with co-training. Proceedings of the 11th Annual Conference on Computational Learning Theory; 1998. pp. 92–100. [Google Scholar]
- 4.Joachims T. Transductive inference for text classification using support vector machines. Proceedings of the 16th International Conference on Machine Learning (ICML-1999); 1999. pp. 200–209. [Google Scholar]
- 5.Nigam K, Mccallum AK, Thrun S, Mitchell T. Text classification from labeled and unlabeled documents using EM. Machine Learning. 2000;39:103–134. [Google Scholar]
- 6.Basu S, Bilenko M, Mooney R. A probabilistic framework for semi-supervised clustering. Proceedings of the 10th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM. 2004. pp. 59–68. [Google Scholar]
- 7.Forgy EW. Cluster analysis of multivariate data: efficiency versus interpretability of classifications. Biometrics. 1965;21:768–769. [Google Scholar]
- 8.MacQueen J. Some methods for classification and analysis of multivariate observations. In: Le Cam LM, Neyman J, editors. Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability; Berkeley, CA: University of Cali-fornia Press; 1967. pp. 281–297. [Google Scholar]
- 9.Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. Journal of the Royal Statistical Society Series C (Applied Statistics) 1979;28(1):100–108. [Google Scholar]
- 10.Lloyd S. Least squares quantization in PCM. IEEE Transactions on Information Theory. 1982;28(2):129–137. [Google Scholar]
- 11.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2001;63(2):411–423. doi: 10.1111/1467-9868.00293. [DOI] [Google Scholar]
- 12.Milligan G, Cooper M. An examination of procedures for determining the number of clusters in a data set. Psychometrika. 1985;50(2):159–179. doi: 10.1007/BF02294245. [DOI] [Google Scholar]
- 13.Sugar CA, James GM. Finding the number of clusters in a dataset. Journal of the American Statistical Association. 2003;98(463):750–763. doi: 10.1198/016214503000000666. [DOI] [Google Scholar]
- 14.Tibshirani R, Walther G. Cluster validation by prediction strength. Journal of Computational and Graphical Statistics. 2005;14(3):511–528. doi: 10.1198/106186005X59243. [DOI] [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu S, Banerjee A, Mooney R. Semi-supervised clustering by seeding. Proceedings of the 19th International Conference on Machine Learning (ICML-2002); 2002. pp. 19–26. [Google Scholar]
- 17.Gaynor S, Bair E. Identification of biologically relevant subtypes via preweighted sparse clustering. ArXiv e-prints. 2013 doi: 10.1016/j.csda.2017.06.003. arXiv:1304.3760. http://arxiv.org/abs/1304.3760. [DOI] [PMC free article] [PubMed]
- 18.Brown MPS, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proceedings of the National Academy of Sciences. 2000;97(1):262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateos A, Dopazo J, Jansen R, Tu Y, Gerstein M, Stolovitzky G. Systematic learning of gene functional classes from dna array expression data by using multilayer perceptrons. Genome Research. 2002;12(11):1703–1715. doi: 10.1101/gr.192502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J, Cline M, Martin J, Finkelstein D, Awad T, Kulp D, Siani-Rose MA. A knowledge-based clustering algorithm driven by gene ontology. Journal of Biopharmaceutical Statistics. 2004;14(3):687–700. doi: 10.1081/BIP-200025659. [DOI] [PubMed] [Google Scholar]
- 21.Qu Y, Xu S. Supervised cluster analysis for microarray data based on multivariate gaussian mixture. Bioinformatics. 2004;20(12):1905–1913. doi: 10.1093/bioinformatics/bth177. [DOI] [PubMed] [Google Scholar]
- 22.Fang Z, Yang J, Li Y, Luo Q, Liu L, et al. Knowledge guided analysis of microar-ray data. Journal of Biomedical Informatics. 2006;39(4):401–411. doi: 10.1016/j.jbi.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Huang D, Pan W. Incorporating biological knowledge into distance-based clustering analysis of microarray gene expression data. Bioinformatics. 2006;22(10):1259–1268. doi: 10.1093/bioinformatics/btl065. [DOI] [PubMed] [Google Scholar]
- 24.Brameier M, Wiuf C. Co-clustering and visualization of gene expression data and gene ontology terms for saccharomyces cerevisiae using self-organizing maps. Journal of Biomedical Informatics. 2007;40(2):160–173. doi: 10.1016/j.jbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Chopra P, Kang J, Yang J, Cho H, Kim H, Lee MG. Microarray data mining using landmark gene-guided clustering. BMC Bioinformatics. 2008;9(1):92. doi: 10.1186/1471-2105-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tari L, Baral C, Kim S. Fuzzy c-means clustering with prior biological knowledge. Journal of Biomedical Informatics. 2009;42(1):74–81. doi: 10.1016/j.jbi.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu S, Davidson I, Wagstaff K. Chapman & Hall/CRC Data Mining and Knowledge Discovery Series. CRC Press; Boca Raton, FL: 2009. Constrained Clustering: Advances in Algorithms, Theory, and Applications. [Google Scholar]
- 28.Wagstaff K, Cardie C, Rogers S, Schrödl S. Constrained k-means clustering with background knowledge. Proceedings of the 18th International Conference on Machine Learning (ICML-2001); 2001. pp. 577–584. [Google Scholar]
- 29.Basu S, Banerjee A, Mooney R. Active semi-supervision for pairwise constrained clustering. Proceedings of the 4th SIAM International Conference on Data Mining (SDM-2004); 2004. pp. 333–344. [Google Scholar]
- 30.Bilenko M, Basu S, Mooney R. Integrating constraints and metric learning in semi-supervised clustering. Proceedings of the 21st International Conference on Machine learning (ICML-2004); 2004. pp. 81–88. [Google Scholar]
- 31.Klein D, Kamvar S, Manning C. From instance-level constraints to space-level constraints: Making the most of prior knowledge in data clustering. Proceedings of the 19th International Conference on Machine Learning (ICML-2002); 2002. pp. 307–314. [Google Scholar]
- 32.Bilenko M, Mooney R. Adaptive duplicate detection using learnable string similarity measures. Proceedings of the 9th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2003. pp. 39–48. [Google Scholar]
- 33.Xing E, Ng A, Jordan M, Russell S. Distance metric learning, with application to clustering with side-information. Advances in Neural Information Processing Systems. 2003;15:505–512. [Google Scholar]
- 34.Bar-Hillel A, Hertz T, Shental N, Weinshall D. Learning distance functions using equivalence relations. Proceedings of the 20th International Conference on Machine learning (ICML-2003); 2003. pp. 11–18. [Google Scholar]
- 35.Kamvar S, Klein D, Manning C. Spectral learning. Proceedings of the 17th International Joint Conference of Artificial Intelligence; 2003. pp. 561–566. [Google Scholar]
- 36.Chang H, Yeung DY. Locally linear metric adaptation for semi-supervised clustering. Proceedings of the 21st International Conference on Machine learning (ICML-2004); 2004. pp. 153–160. [Google Scholar]
- 37.Lange T, Law M, Jain A, Buhmann J. Learning with constrained and unlabelled data. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2005); 2005. pp. 731–738. [Google Scholar]
- 38.Handl J, Knowles J. On semi-supervised clustering via multiobjective optimization. Proceedings of the 8th Annual Conference on Genetic and Evolutionary Computation (GECCO 2006); 2006. pp. 1465–1472. [Google Scholar]
- 39.Li T, Ding C, Jordan M. Solving consensus and semi-supervised clustering problems using nonnegative matrix factorization. Proceedings of the 7th IEEE International Conference on Data Mining (ICDM 2007); 2007. pp. 577–582. [Google Scholar]
- 40.Xiang S, Nie F, Zhang C. Learning a Mahalanobis distance metric for data clustering and classification. Pattern Recognition. 2008;41(12):3600–3612. doi: 10.1016/j.patcog.2008.05.018. [DOI] [Google Scholar]
- 41.Wang F, Li T, Zhang C. Semi-supervised clustering via matrix factorization. Proceedings of the 8th SIAM International Conference on Data Mining (SDM-2008); 2008. pp. 1–12. [Google Scholar]
- 42.Cohn D, Caruana R, McCallum A. Semi-supervised clustering with user feedback. In: Basu S, Davidson I, Wagstaff K, editors. Constrained Clustering: Advances in Algorithms, Theory, and Applications. chapter 2. CRC Press; Boca Raton, FL: 2009. pp. 17–31. Chapman & Hall/CRC Data Mining and Knowledge Discovery Series. [Google Scholar]
- 43.Yin X, Chen S, Hu E, Zhang D. Semi-supervised clustering with metric learning: An adaptive kernel method. Pattern Recognition. 2010;43(4):1320–1333. doi: 10.1016/j.patcog.2009.11.005. [DOI] [Google Scholar]
- 44.Davidson I, Ravi S. Clustering with constraints: Feasibility issues and the k-means algorithm. Proceedings of the 5th SIAM International Conference on Data Mining (SDM-2005); 2005. pp. 138–149. [Google Scholar]
- 45.Davidson I, Ravi S. Agglomerative hierarchical clustering with constraints: Theoretical and empirical results. Knowledge Discovery in Databases (KDD 2005); 2005. pp. 59–70. [Google Scholar]
- 46.Law M, Topchy A, Jain A. Model-based clustering with probabilistic constraints. Proceedings of the 5th SIAM International Conference on Data Mining (SDM-2005); 2005. pp. 641–645. [Google Scholar]
- 47.Lu Z, Leen T. Semi-supervised learning with penalized probabilistic clustering. Advances in Neural Information Processing Systems. 2005;17:849–856. [Google Scholar]
- 48.Tang W, Xiong H, Zhong S, Wu J. Enhancing semi-supervised clustering: a feature projection perspective. Proceedings of the 13th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2007. pp. 707–716. [Google Scholar]
- 49.Kulis B, Basu S, Dhillon I, Mooney R. Semi-supervised graph clustering: a kernel approach. Machine Learning. 2009;74:1–22. doi: 10.1007/s10994-008-5084-4. [DOI] [Google Scholar]
- 50.Yoshida T, Okatani K. A graph-based projection approach for semi-supervised clustering. In: Kang BH, Richards D, editors. Knowledge Management and Acquisition for Smart Systems and Services. Springer-Verlag; Berlin, Germany: 2010. pp. 1–13. Lecture Notes in Computer Science. [Google Scholar]
- 51.Greene D, Cunningham P. Constraint selection by committee: an ensemble approach to identifying informative constraints for semi-supervised clustering. Proceedings of the 18th European Conf. on Machine Learning (ECML 2007); 2007. pp. 140–151. [Google Scholar]
- 52.Mallapragada P, Jin R, Jain A. Active query selection for semi-supervised clustering. 19th International Conference on Pattern Recognition (ICPR 2008); IEEE. 2008. pp. 1–4. [Google Scholar]
- 53.Zheng L, Li T. Semi-supervised hierarchical clustering. Proceedings of the 11th IEEE International Conference on Data Mining (ICDM 2011); 2011. pp. 982–991. [DOI] [Google Scholar]
- 54.Miyamoto S, Terami A. Semi-supervised agglomerative hierarchical clustering algorithms with pairwise constraints. Proceedings of the 2010 IEEE International Conference on Fuzzy Systems (FUZZ 2010); 2010. pp. 1–6. [DOI] [Google Scholar]
- 55.Davidson I, Ravi S. Using instance-level constraints in agglomerative hierarchical clustering: theoretical and empirical results. Data Mining and Knowledge Discovery. 2009;18(2):257–282. doi: 10.1007/s10618-008-0103-4. [DOI] [Google Scholar]
- 56.Miyamoto S, Terami A. Constrained agglomerative hierarchical clustering algorithms with penalties. Proceedings of the 2011 IEEE International Conference on Fuzzy Systems (FUZZ 2011); 2011. pp. 422–427. doi:10.1109/FUZZY. 2011.6007351. [Google Scholar]
- 57.Bade K, Nurnberger A. Personalized hierarchical clustering. Proceedings of the 2006 IEEE/WIC/ACM International Conference on Web Intelligence (WI 2006); 2006. pp. 181–187. [DOI] [Google Scholar]
- 58.Zhao H, Qi Z. Hierarchical agglomerative clustering with ordering constraints. Proceedings of the 3rd International Conference on Knowledge Discovery and Data Mining (WKDD 2010); 2010. pp. 195–199. [DOI] [Google Scholar]
- 59.Hamasuna Y, Endo Y, Miyamoto S. Semi-supervised agglomerative hierarchical clustering using clusterwise tolerance based pairwise constraints. Proceedings of the 7th International Conference on Modeling Decision for Artificial Intelligence (MDAI 2010); 2010. pp. 152–162. [Google Scholar]
- 60.Hamasuna Y, Endo Y, Miyamoto S. Semi-supervised agglomerative hierarchical clustering with ward method using clusterwise tolerance. Proceedings of the 8th International Conference on Modeling Decision for Artificial Intelligence (MDAI 2011); 2011. pp. 103–113. [Google Scholar]
- 61.Hamasuna Y, Endo Y, Miyamoto S. On agglomerative hierarchical clustering using clusterwise tolerance based pairwise constraints. Journal of Advanced Computational Intelligence and Intelligent Informatics. 2012;16(1):174–179. [Google Scholar]
- 62.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2(4):e108. doi: 10.1371/journal.pbio.0020108. doi:10.1371/journal.pbio. 0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowak G, Tibshirani R. Complementary hierarchical clustering. Biostatistics. 2008;9(3):467–483. doi: 10.1093/biostatistics/kxm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bullinger L, Döhner K, Bair E, Fröhling S, Schlenk R, Tibshirani R, Döhner H, Pollack JR. Gene expression profiling identifies new subclasses and improves outcome prediction in adult myeloid leukemia. The New England Journal of Medicine. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 65.Koestler DC, Marsit CJ, Christensen BC, Karagas MR, Bueno R, Sugarbaker DJ, Kelsey KT, Houseman EA. Semi-supervised recursively partitioned mixture models for identifying cancer subtypes. Bioinformatics. 2010;26(20):2578–2585. doi: 10.1093/bioinformatics/btq470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Houseman EA, Christensen B, Yeh RF, Marsit C, Karagas M, Wrensch M, Nelson H, Wiemels J, Zheng S, Wiencke J, et al. Model-based clustering of dna methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9(1):365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Witten DM, Tibshirani R. A framework for feature selection in clustering. Journal of the American Statistical Association. 2010;105(490):713–726. doi: 10.1198/jasa.2010.tm09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996;58(1):267–288. [Google Scholar]
- 69.Ghosh D, Chinnaiyan AM. Mixture modelling of gene expression data from microarray experiments. Bioinformatics. 2002;18(2):275–286. doi: 10.1093/bioinformatics/18.2.275. [DOI] [PubMed] [Google Scholar]
- 70.Liu JS, Zhang JL, Palumbo MJ, Lawrence CE. Bayesian clustering with variable and transformation selections. Bayesian Statistics 7: Proceedings of the Seventh Valencia International Meeting; 2003. pp. 249–275. [Google Scholar]