Abstract
The central role of RNA polymerase (RNAP) is to catalyze the processive synthesis of a growing RNA transcript. Recent structural and biophysical data have lead to a deeper understanding of the nucleotide addition cycle and insight into the structure-function relationships that govern transcription elongation. In this review, we discuss kinetic data on nucleotide incorporation in the context of crystal structures, which show RNAP in multiple conformations. We present a facilitated Brownian ratchet model of nucleotide incorporation, in which templated NTP binding to a non-catalytic site in the main channel promotes the conformational changes that lead to opening of the catalytic site and translocation.
Transcription, the DNA-directed synthesis of RNA, is the first step in the cascade of events that leads to gene expression. All cells utilize multi-subunit RNA polymerases (RNAPs) to synthesize the nascent RNA chain, using a well-conserved mechanism for nucleotide incorporation. For the addition of each nucleotide, RNAP must bind a nucleoside triphosphate (NTP) into the catalytic site, catalyze the phosphotransfer reaction, release pyrophosphate, and translocate along the DNA. In the past 10 years, there has been an explosion of structural data on elongation complexes (ECs), as well as many detailed kinetic studies on the mechanism of nucleotide incorporation. These data have led to a host of models of the mechanism and regulation of substrate loading, catalysis, and translocation. Despite all these data, the mechanism of nucleotide incorporation remains hotly debated. In this review, we focus on the structural elements that appear to be intimately involved in the nucleotide addition cycle and how these elements relate to the current kinetic models for nucleotide incorporation.
Structural Elements of RNA Polymerase
Crystal structures of both prokaryotic and eukaryotic RNAPs reveal an enzyme that has a basic overall structure resembling a crab claw [1,2]. The two largest subunits constitute the pincer of the enzyme and form a deep, ∼27Å wide channel, referred to as the main channel. In the EC crystal structures (Fig. 1), the downstream DNA enters the main channel making extensive contacts with the pincer. Fork loop 2 and the bridge helix separate the main channel from the catalytic site and delineate the front edge of the transcription bubble, where the two stands of DNA are separated. The non-template DNA passes on one side of fork loop 2 and exits to the outer surface of the enzyme, while the template DNA passes on the other side and continues across the bridge helix and into the catalytic site. At this point, the template DNA makes an ∼90° turn into the active site and forms a RNA-DNA hybrid with the nascent RNA chain. The RNA-DNA hybrid continues for 8-9 nucleotides before it is separated by the lid, which mark the upstream edge of the transcription bubble. The RNA passes the lid, and is extruded through the RNA exit channel. The template DNA continues on and re-anneals with the non-template DNA at the zipper [3,4••,5]. Mutations in these RNAP structural elements result in aberrant RNAP function [6].
Figure 1.
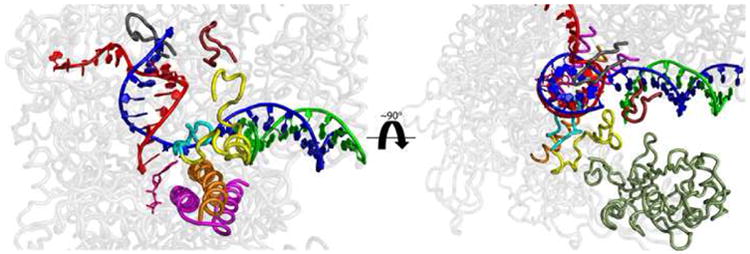
Important structural elements involved in translocation and the nucleotide addition cycle. Side (left) and top (right) views of the β and β′-subunits (transparent grey) of the T. aquaticus transcription elongation complex (PDB: 2O5J) are shown. Several key structural elements are highlighted. These structures have been shown to either adopt multiple conformations in crystal structures or to be involved in translocation and/or the nucleotide addition cycle, based on mutational analysis [6]. The template and non-template DNA strands are highlighted in green and blue, respectively. These strands are separated at fork loop 2 (β413-441; yellow). The βD-loopII (β442-454), which extends from fork loop 2, is cyan. The nascent RNA chain is red. The DNA and RNA hybrid are separated at the lid (β′525-538; grey) and rudder (β′580-600; dark red). The catalytically essential trigger loop (β′1225-1265; magenta) is located below the bridge helix (β′1066-1095; orange) and is shown interacting with a nucleotide (pink) in the catalytic site. The pincer, (β140-332) which closes down on the downstream DNA, is shown in dark green and is omitted on the left for clarity. All residue numbers correspond to T. aquaticus RNAP.
Another major structural element seen in all multi-subunit RNAP structures is the secondary channel, which is ∼12 Å wide and ∼45 Å deep and leads directly into the catalytic site. The 3′-end of a transcript in a backtracked EC is extruded into this channel [7-10], and several transcription factors, such as GreA, GreB, and DksA in prokaryotes [11-13] and TFIIS in eukaryotes [5], bind in this channel and interact with the RNA and/or the catalytic site. In addition, this channel is generally believed to be the main route for NTP entry into the catalytic site [1,3,14-16].
Nucleotide Incorporation and the Trigger Loop and Bridge Helix
The catalytically essential trigger loop interacts with the bridge helix and spans the main channel and secondary channel and can adopt “open” and “closed” conformations (Figs. 1 & 2) [4•4•,17••,18•-20]. In the open conformation, the trigger loop lies primarily in the main channel, leaving free access to the catalytic site via the secondary channel; whereas, in the closed conformation, it makes extensive interactions with the NTP in the catalytic site and closes off entry and exit of NTPs or pyrophosphate to/from the catalytic site. These observations led to the suggestions that the closed conformation represents the catalytic complex and that the trigger loop needs to open and close for every round of nucleotide incorporation [17••,18•,20]. Changes in the conformation of the trigger loop also appear to coincide with bending and straightening of the bridge helix (Fig. 2). Cramer and coworkers recently published a review and a movie of nucleotide incorporation, based on several RNAP crystal structures, that shows the conformational changes in the bridge helix and trigger loop that are thought to be important for catalysis and translocation [21]. One paradox regarding the role of the trigger loop in nucleotide incorporation is that TFIIS binds in the secondary channel in such a way that it would appear to block the interaction of the trigger loop with the NTP in the catalytic site [5,14], however it does not affect the rate of nucleotide incorporation [22].
Figure 2.
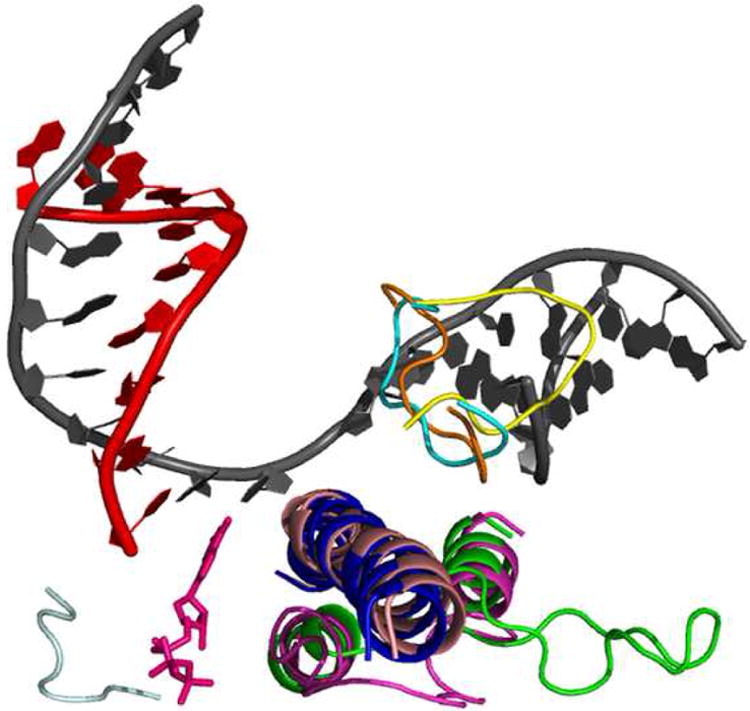
Conformational changes in the S. cerevisiae EC thought to be involved in translocation. Shown are the different conformations of the trigger loop, bridge helix, and fork loop 2. The trigger loop can adopt an open (green, PDB:1Y1V) and closed (magenta, PDB:2E2H) conformation. When closed, the trigger loop contributes several residues that are essential for catalysis. The bridge helix can adopt a bent (light pink, PDB:2VUM) and unbent (blue, PDB:1Y77) state. Fork loop 2 has been seen in three distinct conformations: open (yellow, PDB:2E2I), partially closed (orange, PDB:1Y1V), and closed (cyan, PDB:1Y77). The DNA strands are in grey and the RNA is red. The catalytic nucleotide (pink) is shown bound to the catalytic residues (light grey).
Is RNA Polymerase Governed by a Brownian Ratchet?
Several models for nucleotide incorporation propose that RNAP functions as a Brownian ratchet, with thermal energy driving an equilibrium between the pre- and post-translocated states (and even backtracked states) at each nucleotide position [14,21,23,24]. Although studies have shown that complexes stalled at a given template position can reside in the pre- and post-translocated states and that cognate NTPs can shift the complexes to the post-translocated state [20,23,25•,26], there is no data pertaining to the rate of interconversion between these states. RNAP undoubtedly reaches an equilibrium between the pre- and post-translocated states if given enough time (such as in a stalled EC due to nucleotide deprivation); however, it remains unclear if RNAP achieves an equilibrium between these states on the time scale faster than nucleotide incorporation (∼30 nt/s), and conversion between the pre-translocated and backtracked states appears to be slow [27]. It is clear that Brownian ratchet models, in which NTP binding to the catalytic site acts as a pawl and the rate of incorporation is governed by the NTP concentration and the percentage of complexes in the post-translocated state [23], cannot explain much of the kinetic data on nucleotide incorporation and pyrophosphate release. First, a recent study of a fidelity mutant of yeast RNAP II (rpb1-E1103A) demonstrated that this mutant increases the rate of nucleotide incorporation (both correct and incorrect), even though it is shifted to the pre-translocated state relative to wild-type RNAP [25•]. This study suggests either that the translocation state does not govern the rate of nucleotide incorporation or that NTPs bind preferentially to the pre-translocated state. Second, kinetic studies of misincorporation reveal that RNAP can exist in two states which are not in rapid equilibrium: an “activated” state which catalyzes misincorporation and an “unactivated” state which does not [28,29]. These data suggest that if there is an equilibrium between translocation states, it is either slow relative to the rate of nucleotide incorporation, or rapid, but does not govern the rate-limiting step of nucleotide incorporation. Finally, as discussed below, several kinetic studies indicate that RNAP contains a non-catalytic binding site for templated NTPs that regulates nucleotide incorporation and pyrophosphate release.
NTP-Assisted Nucleotide Incorporation
Early presteady-state kinetic studies of E. coli RNAP demonstrated that the addition of a templated, but non-incorporatable, NTP analog increases the rate of incorporation of the templated NTP [30••]. This result, coupled with the observation that the rate of nucleotide incorporation shows a sigmoidal dependence on NTP concentration, indicates the presence of an additional NTP binding site on RNAP. If there were only a single binding site for templated NTPs, the presence of a non-incorporatable NTP analog would inhibit the rate of nucleotide incorporation because it will compete for binding to the catalytic site [30••]. It was proposed that binding of a templated NTP to an allosteric site transitions RNAP from a slow, high fidelity “unactivated” state into a fast but lower fidelity “activated” state. Subsequent studies, which coupled sequence and structural analyses of RNAP with examination of the effects of the downstream DNA sequence on nucleotide incorporation, led to the proposal that a templated NTP binds to fork loop 2 and, upon pairing with the template DNA, increases the rate of nucleotide incorporation by altering the active site geometry and by acting as a ratchet that induces and/or maintains the post-translocated state prior to binding of a NTP in the catalytic site [28,30••,31•].
A similar model was proposed based on studies of eukaryotic RNAPII that showed that NTPs complementary to the template DNA one and two bases downstream from the nucleotide to be incorporated increase the rate of nucleotide incorporation [32••,33,34•,35]. In this model, a downstream templated NTP (or NTPs) binds to a site in the main channel and regulates opening of the downstream transcription bubble. This nucleotide is then shuttled along with the template DNA into the catalytic site. Further support for an additional templated NTP binding site comes from recent studies showing that the presence of the downstream templated NTP enhances pyrophosphate release from the previous incorporation event by ∼ 200-fold (from ∼3s-1 to ∼600s-1) [36••]. Presumably, pyrophosphate is occupying the active site, with the trigger loop closed (which also blocks the pre-insertion site); therefore, the templated NTP must bind somewhere else, and the downstream template DNA lies in the main channel.
It has been suggested that there cannot be a templated NTP binding site in the main channel because in crystal structures of T. thermophilus RNAP ECs, the downstream DNA in the main channel is completely paired [4••,17••]. These structures, however, lack a non-template strand upstream of the insertion site, which may affect downstream DNA opening [4••]. Furthermore, DNA footprinting studies of ECs stalled at multiple positions along the DNA template revealed that in some complexes, the downstream DNA is fully paired, while in others, one or two base pairs of the downstream DNA are open [28,37]. Finally, in the structure of RNAP II with α-amanitin bound, the base entering the active site is unpaired and lies above the bridge helix pointing into the main channel [20]. It is likely that the state of the DNA (paired or unpaired) immediately downstream of the insertion site depends on the details of the sequence and possibly on the presence of downstream NTPs. It is certainly difficult to explain the data discussed in this section without allowing for templated NTP binding in the main channel.
Fork Loop 2 and the β-pincer May Comprise a Non-catalytic NTP Binding Site
Fork loop 2 lies across from the template DNA at the front of the transcription bubble. It interacts with the β-pincer (β140-332), which makes direct contact with the double-stranded downstream DNA (Fig. 1). The pincer has been observed in “open” and “closed” conformations, and opening of the pincers is thought to be necessary to allow translocation [4••,17••]. Fork loop 2 also makes several interactions with the bridge helix as it crosses over and forms βD-loopII, which, in turn, interacts with the closed trigger loop, RNA-DNA hybrid, and the frayed 3′-end of a backtracked complex [14,17••,38]. It has been suggested that conformational changes of fork loop 2 (Fig. 2) are coupled with opening and closing of the pincers [4••,17••], unwinding of the downstream DNA [4••,5,17••,18•,20,39,40], and changes in βD-loopII, the trigger loop, and the bridge helix [17••,38].
Sequence and structural analyses revealed that fork loop 2, the β-pincer, and several surrounding elements resemble NTP binding domains seen in ATPases and GTPases [31•]. Fork loop 2 contains several glycines in both prokaryotes and eukaryotes and has sequences similar to “P-loops” of NTP binding domains. In addition, a conserved Walker B motif (DxxG) is located at the rear of fork loop 2. Finally, fork loop 2 makes extensive hydrophobic interactions with the β-pincer, which is also characteristic of NTP binding domains. Given that fork loop 2 has all the elements of a NTP binding domain and is connected to the structural elements that appear to be directly involved in translocation and nucleotide incorporation, it seems probable that templated NTPs may bind to this site to regulate transcription. Consistent with this suggestion, mutations in fork loop 2 result in defects in nucleotide incorporation that cannot be explained by fork loop 2 functioning only to open the downstream DNA (Unpublished results) [41].
Is the Secondary Channel the Only Route for Nucleotide Entry?
Consistent with nucleotides entering the catalytic site via the secondary channel, NTPs have been observed bound in the secondary channel in crystal structures of RNAP II, although most of the binding sites partially overlap the catalytic site [5,21,42]. In addition, the antibiotic microcin J25 binds in the secondary channel and appears to compete with NTP binding, leading to the suggestion that it acts by blocking the entry of NTPs into the catalytic site via the secondary channel [43]. A major criticism of the secondary channel model, however, is that the channel is too small to accommodate more than one nucleotide at a time, which may lead to an NTP trafficking problem [31•,44]. Furthermore, the secondary channel has a negative electrostatic potential which would be expected to disfavor nucleotide entry, although computer simulations suggest that an elongation rate of ∼20 nt/s is possible with NTP binding via the secondary channel [15]. In addition, several transcription factors and small molecules (ppGpp) that bind in the secondary channel would appear to block the entry of NTPs [11-13,45].
As outlined above, the evidence for a nucleotide-binding site in the main channel is overwhelming; however, it remains unclear if NTPs can be shuttled from the main channel into the catalytic site. The primary argument against this idea is that there does not appear to be sufficient room for an NTP to pass over the bridge helix [1,44]. The large number of conformational rearrangements that seem to accompany nucleotide incorporation, coupled with the observation that the antibiotic streptolydigin lies along the top of the bridge helix while simultaneously interacting with fork loop 2 and the trigger loop, at least raise the specter of its possibility [17••]. Alternatively, a NTP could be transferred to the trigger loop and shuttled underneath the bridge helix along with closing of the trigger loop, although this path would result in the NTP losing contact with the template DNA. Currently, there is no direct evidence for NTPs entering the catalytic site via the main channel or the secondary channel.
Facilitated Brownian Ratchet Incorporation Model
During processive synthesis, RNAP may function as a “facilitated” Brownian ratchet with two pawls: one pawl being NTP binding into the catalytic site and the second pawl being NTP binding to a non-catalytic binding site (fork loop 2). We call it a facilitated Brownian ratchet because binding of a templated NTP to fork loop 2 may induce the conformational changes that lead to opening of the catalytic site and/or translocation. In other words, binding of a templated NTP to fork loop 2 may act as a “pry bar” to help move the ratchet (i.e. RNAP) forward and binding of a NTP into the catalytic site may act as a pawl that maintains the post-translocated state immediately prior to incorporation.
Figure 3 presents a model for NTP-assisted nucleotide incorporation that encompasses all of the results discussed above. After a nucleotide is incorporated, RNAP is in the pre-translocated state, with the trigger loop closed and pyrophosphate bound. The next NTP binds to fork loop 2 and melts the downstream DNA to pair with the next template base (Fig. 3, step 1), which, in turn, facilitates opening of the pincer and destabilizes the closed trigger loop, promoting pyrophosphate release (Fig. 3, step 2). These conformational changes allow the enzyme to rapidly translocate (Fig. 3, step 3). The NTP bound in fork loop 2 may be shuttled directly into the catalytic site along with the template base (Fig. 3, step 4a) [31•,32••,33,34•], or upon translocation, a NTP from bulk solution may bind to the newly opened catalytic site via the secondary channel (Fig. 3, step 4b) [31•]. After the NTP is bound to the catalytic site, the trigger loop closes down on the catalytic site (Fig. 3, step 5) and induces bond formation (Fig. 3, step 6), and the cycle begins again.
Figure 3.
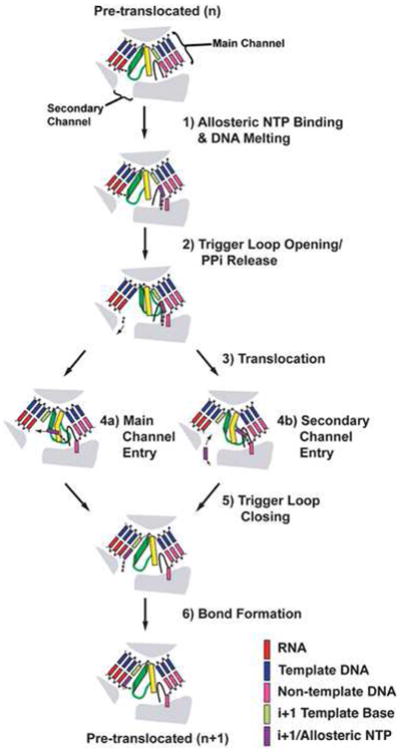
Model of the nucleotide addition cycle that utilizes a secondary NTP binding site. Fork loop 2 is black, the bridge helix is yellow, and the trigger loop is green. This model begins with the EC in the pre-translocated state with pyrophosphate still bound from the previous incorporation. See main text for details.
Although binding of a NTP to fork loop 2 increases the rate of nucleotide incorporation, it is not essential. In the proposed model, if fork loop 2 is not utilized, then the opening of the transcription bubble, trigger loop, and pincers (required for translocation) may be slow, leading to a slow entry into the post-translocated state. Furthermore, the trigger loop would spend more time in the closed conformation and sterically block NTP entry, thereby lowering the rate of synthesis. This path is equivalent to that proposed by Cramer and co-workers [21] and may come into play at certain sequences or with stalled complexes in which all the NTPs have been removed. All in all, there appear to be three possible “paths” to nucleotide incorporation: i) direct entry of a NTP into the catalytic site via the secondary channel when RNAP is in the post-translocated state, which does not involve fork loop 2 (non-allosteric path); ii) entry of a second templated NTP into the catalytic site via the secondary channel after a templated NTP binds into fork loop 2 (allosteric path); iii) shuttling of the NTP bound in fork loop 2 into the catalytic site (shuttle path). Which path is followed will likely depend on both the environmental conditions, NTP concentrations, and DNA sequence context.
A key point of this NTP-assisted incorporation model is the idea that during rapid processive elongation, the trigger loop remains closed and pyrophosphate stays bound until the next NTP binds fork loop 2 (or an equivalent templated site), which, in turn, allows rapid translocation. As long as the NTP concentration is sufficiently high or if the NTP is shuttled into the catalytic site from fork loop 2, the catalytic site is rapidly filled and the trigger loop closes, restarting the cycle. Such a regulatory mechanism would serve to keep the active site protected from the action of factors that bind in the secondary channel, because the closed trigger loop may block their binding sites. If RNAP encounters a disruption in synthesis due to either sequence context or NTP pools, the trigger loop would slowly open, allowing regulatory factors to bind via the secondary channel. These suggestions are consistent with misincorporation studies which demonstrated that GreA induces RNA cleavage in ECs only after they transition from the activated (fast) to the unactivated (slow) state [28]. Finally this idea could resolve the apparent paradox that TFIIS does not inhibit nucleotide incorporation even though it binds between the catalytic site and the trigger loop.
Acknowledgments
This work was funded by NIH Grant R01GM79480 (DAE) and NIH Grant R01CA102029 (SRK). We thank Mikhail Kashlev, Steve Block, Zachary Burton, and Jeff Gelles for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal Structure of Thermus aquaticus Core RNA Polymerase at 3.3 Å Resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P, Bushnell DA, Kornberg RD. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science. 2001;292:1863–1876. [Google Scholar]
- 3.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science. 2001;292:1876–1882. [Google Scholar]
- ••4.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural Basis for Transcription Elongation by Bacterial RNA Polymerase. Nature. 2007;448:157. doi: 10.1038/nature05932. See annotation for [17••] [DOI] [PubMed] [Google Scholar]
- 5.Kettenberger H, Armache KJ, Cramer P. Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Trinh V, Langelier MF, Archambault J, Coulombe B. Structural Perspective on Mutations Affecting the Function of Multisubunit RNA Polymerases. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artsimovitch I, Landick R. Pausing by Bacterial RNA Polymerase is Mediated by Mechanistically Distinct Classes of Signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komissarova N, Kashlev M. Transcriptional Arrest: Escherichia coli RNA Polymerase Translocates Backward, Leaving the 3′-end of the RNA Intact and Extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA–DNA Hybrid Maintains the Register of Transcription by Preventing Backtracking of RNA Polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 10.Reeder TC, Hawley DK. Promoter Proximal Sequences Modulate RNA Polymerase II Elongation by a Novel Mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 11.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and Function of the Transcription Elongation Factor GreB Bound to Bacterial RNA Polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 12.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the Secondary Channel—Structural Framework for ppGpp-DksA Synergism During Transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 13.Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of Catalytic Residues to RNA Polymerase Active Center by Transcription Factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural Basis of Transcription: Backtracked RNA Polymerase II at 3.4 Angstrom Resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of Nucleoside Triphosphates and Role of the Entry Site to the RNA Polymerase II Active Center. Proc Natl Acad Sci USA. 2004;101:17361–17364. doi: 10.1073/pnas.0408168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landick R. NTP-entry routes in multi-subunit RNA polymerases. Trends Biochem Sci. 2005;30:651–654. doi: 10.1016/j.tibs.2005.10.001. [DOI] [PubMed] [Google Scholar]
- ••17.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural Basis for Substrate Loading in Bacterial RNA Polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. This paper presents the first structure of an elongation complex for a bacterial RNAP. The authors observed important conformational changes in the trigger loop, fork loop 2, and β-pincer. These conformational changes have lead to the suggestion that the synchronized movements of these structures are involved in translocation and nucleotide loading. [DOI] [PubMed] [Google Scholar]
- •18.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. This paper was the first to show that the trigger loop was able to adopt multiple conformations and provided the structural basis for the trigger loop centric models for nucleotide incorporation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by α-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brueckner F, Cramer P. Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 21.Brueckner F, Ortiz J, Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Curr Opin Struct Biol. 2009;19:294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Burton ZF. Transcription Factors IIF and IIS and Nucleoside Triphosphate Substrates as Dynamic Probes of the Human RNA Polymerase II Mechanism. J Mol Biol. 2004;342:1085–1099. doi: 10.1016/j.jmb.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 23.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A Ratchet Mechanism of Transcription Elongation and Its Control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat Struct Mol Biol. 2008;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- •25.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. Using a combination of footprinting and kinetics, this paper demonstrates that the rate of nucleotide incorporation does not correlate with the fraction of elongation complexes in the post-translocated state and that nucleotide misincorporation is governed by sequestration of the NTP into the catalytic site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komissarova N, Kashlev M. RNA Polymerase Switches between Inactivated and Activated States By Translocating Back and Forth along the DNA and the RNA. J Bio Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 27.Erie DA. The Many Conformational States Of RNA Polymerase Elongation Complexes And Their Roles In The Regulation Of Transcription. Biochim Biophys Acta. 2002;1577:224–239. doi: 10.1016/s0167-4781(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 28.Erie DA, Hajiseyedjavadi O, Young MC, Hippel PHv. Multiple RNA Polymerase Conformations and GreA: Control of the Fidelity of Transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 29.Holmes SF, Santangelo TJ, Cunningham CK, Roberts JW, Erie DA. Kinetic Investigation of Escherichia coli RNA Polymerase Mutants That Influence Nucleotide Discrimination and Transcription Fidelity. J Biol Chem. 2006;281:18677–18683. doi: 10.1074/jbc.M600543200. [DOI] [PubMed] [Google Scholar]
- ••30.Foster JE, Holmes SF, Erie DA. Allosteric Binding of Nucleoside Triphosphates to RNA Polymerase Regulates Transcription Elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. This paper provided the first evidence that RNAP contains a non-catalytic templated NTP binding site. The data in this paper suggested that NTP binding to this site allosterically enhances the rate of nucleotide incorporation. [DOI] [PubMed] [Google Scholar]
- •31.Holmes SF, Erie DA. Downstream DNA Sequence Effects on Transcription Elongation: Allosteric Binding of Nucleoside Triphosphates Facilitates Translocation Via a Ratchet Motion. J Bio Chem. 2003;278:35597–35608. doi: 10.1074/jbc.M304496200. This paper examines the kinetics of nucleotide incorporation, and the authors present a structural analysis showing that fork loop 2 and its surrounding structures contain all the components of a nucleotide binding site. The authors propose a model in which templated NTPs bind to fork loop 2 (in the main channel) and facilitate nucleotide incorporation. [DOI] [PubMed] [Google Scholar]
- ••32.Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic Error Correction and Regulation of Downstream Bubble Opening by Human RNA Polymerase II. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. This paper demonstrates that in RNAP II, the rate of sequestration of a substrate nucleotide in the active site is significantly faster than the rate of bond formation. Additional findings in this paper show that the presence of templated downstream nucleotides increases fidelity of RNAP. [DOI] [PubMed] [Google Scholar]
- 33.Burton ZF, Feig M, Gong XQ, Zhang C, Nedialkov YA, Xiong Y. NTP-Driven Translocation and Regulation of Downstream Template Opening by Multi-Subunit RNA Polymerases. Biochem Cell Biol. 2005;83:486–496. doi: 10.1139/o05-059. [DOI] [PubMed] [Google Scholar]
- •34.Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, Yan H, Burton ZF. NTP-driven Translocation by Human RNA Polymerase II. J Biol Chem. 2003;278:18303–18312. doi: 10.1074/jbc.M301103200. This paper presents the first examination of the kinetics of nucleotide incorporation by RNAP II. The authors suggest that RNAP utilizes a nucleotide-driven model of translocation. In addition, the authors proposed the idea that the main channel, and not the secondary channel, may be the main route by which NTPs enter the catalytic site. [DOI] [PubMed] [Google Scholar]
- 35.Xiong Y, Burton ZF. A Tunable Ratchet Driving Human RNA Polymerse II Translocation Adjusted by Accurately Templated Nucleoside Triphosphates Loaded at Downstream Sites and by Elongation Factors. J Biol Chem. 2007;282:36582–36592. doi: 10.1074/jbc.M707014200. [DOI] [PubMed] [Google Scholar]
- ••36.Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J Mol Biol. 2008;381:1106–1113. doi: 10.1016/j.jmb.2008.06.089. This paper demonstrates that the presence of a downstream templated NTP increases the rate of pyrophosphate release from the previous nucleotide incorporation event; providing the first indication of the functional role of the secondary binding site in the nucleotide addition cycle. [DOI] [PubMed] [Google Scholar]
- 37.Zaychikov E, Denissova L, Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20:“Jumping”of RNA polymerase and asymmetric expansion and contraction of the “transcription bubble”. Proc Natl Acad Sci USA. 1995;92:1739–43. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toulokhonov I, Zhang J, Palangat M, Landick R. A Central Role of the RNA Polymerase Trigger Loop in Active-Site Rearrangement during Transcriptional Pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Meyer PA, Suh MH, Mincheng Z, Fu J. Structure of the 12-Subunit RNA Polymerase II Refined with the Aid of Anomalous Diffraction Data. J Biol Chem. 2009;284:12933–12939. doi: 10.1074/jbc.M809199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P. Structural Basis of Transcription: Mismatch-Specific Fidelity Mechanisms and Paused RNA Polymerase II with Frayed RNA. Mol Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M. Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 2008;36:676–687. doi: 10.1093/nar/gkm1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westover KD, Bushnell DA, Kornberg RD. Structural Basis of Transcription: Nucleotide Selection by Rotation in the RNA Polymerase II Active Center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial Peptide Microcin J25 Inhibits Transcription by Binding within and Obstructing the RNA Polymerase Secondary Channel. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A Structural Model of Transcription Elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 45.Artsimovitch I, Patlan V, Sekine Si, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural Basis for Transcription Regulation by Alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]


