Abstract
Many cellular processes are carried out by molecular ‘machines’— assemblies of multiple differentiated proteins that physically interact to execute biological functions1–8. Despite much speculation, strong evidence of the mechanisms by which these assemblies evolved is lacking. Here we use ancestral gene resurrection9–11and manipulative genetic experiments to determine how the complexity of an essential molecular machine—the hexameric transmembrane ring of the eukaryotic V-ATPase proton pump—increased hundreds of millions of years ago. We show that the ring of Fungi, which is composed of three paralogous proteins, evolved from a more ancient two-paralogue complex because of a gene duplication that was followed by loss in each daughter copy of specific interfaces by which it interacts with other ring proteins. These losses were complementary, so both copies became obligate components with restricted spatial roles in the complex. Reintroducing a single historical mutation from each paralogue lineage into the resurrected ancestral proteins is sufficient to recapitulate their asymmetric degeneration and trigger the requirement for the more elaborate three-component ring. Our experiments show that increased complexity in an essential molecular machine evolved because of simple, high-probability evolutionary processes, without the apparent evolution of novel functions. They point to a plausible mechanism for the evolution of complexity in other multi-paralogue protein complexes.
Comparative genomic approaches suggest that the components of many molecular machines have appeared sequentially during evolution and that complexity increased gradually by incorporating new parts into simpler assemblies2–8. Such horizontal analyses of extant systems, however, cannot decisively test these hypotheses or reveal the mechanisms by which additional parts became obligate components of larger complexes. In contrast, vertical approaches that combine computational phylogenetic analysis with gene synthesis and molecular assays allow changes in the sequence, structure and function of reconstructed ancestral proteins to be experimentally traced through time.9–11Here we apply this approach to characterize the evolution of a small molecular machine and dissect the mechanisms that caused it to increase in complexity.
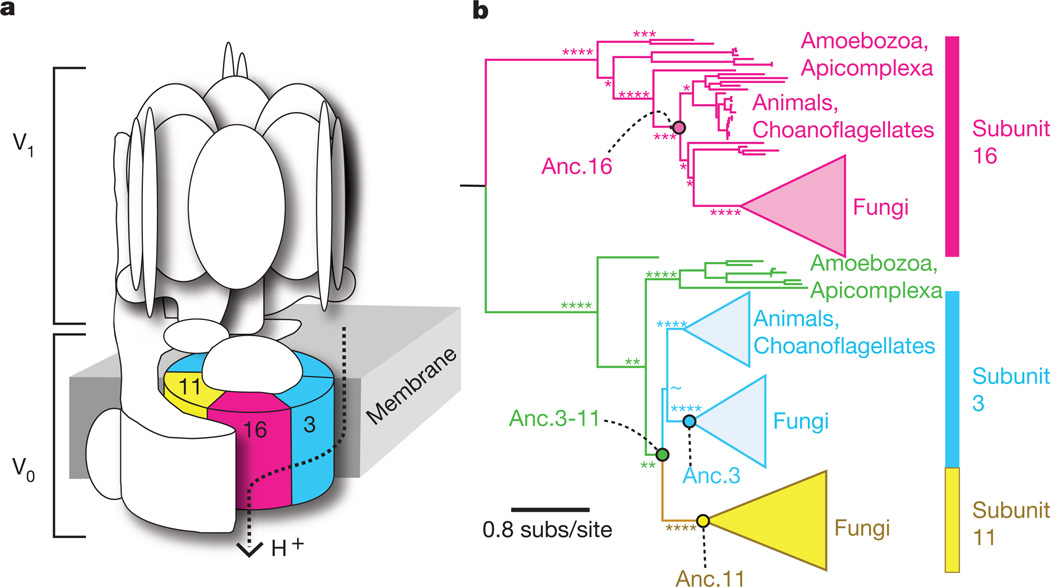
The vacuolar H+-ATPase (V-ATPase) is a multisubunit protein complex that pumps protons across membranes to acidify subcellular compartments; this function is required for intracellular protein trafficking, coupled transport of small molecules and receptor-mediated endocytosis1. V-ATPase dysfunction has been implicated in human osteoporosis, in acquired drug resistance in human tumours, and in pathogen virulence12–14. A key subcomplex of the V-ATPase is the V0protein ring, a hexameric assembly that uses a rotary mechanism to move protons across organelle membranes (Fig. 1a)15,16. Although the V-ATPase is found in all eukaryotes, the V0 ring varies in subunit composition among lineages. In animals and most other eukaryotes, the ring consists of one subunit of Vma16 protein and five copies of its paralogue, Vma3 (Fig. 1b)1. In Fungi, the ring consists of one Vma16 subunit, four copies of Vma3 and one Vma11 subunit, arranged in a specific orientation17. All three proteins are required for V-ATPase function in Fungi18,19, but the mechanisms are unknown by which both Vma3 and Vma11 became obligate components with specific positional roles in the complex.
Figure 1. Structure and evolution of the V-ATPase complex.
a, In S. cerevisiae, the V-ATPase contains two subcomplexes: the octameric V1 domain is on the cytosolic side of the organelle membrane, and the hexameric V0 ring is membrane bound. Protein subunits Vma3, Vma11 and Vma16 are labelled and coloured. b, Maximum likelihood phylogeny of V-ATPase subunits Vma3, Vma11 and Vma16. All eukaryotes contain subunits 3 and 16, but Fungi also contain subunit 11. Circles show ancestral proteins reconstructed in this study. Colours correspond to those of subunits in panel a; unduplicated orthologues of Vma3 and Vma11 are green. Asterisks show approximate likelihood ratios for major nodes: ****, >103; ***, >102; **, >10; *,<10; ~, <2. The complete phylogeny is presented in Supplementary Information, section 2.
To understand how the three-component ring evolved, we reconstructed ancestral V0 proteins from just before and after the increase in complexity, synthesized and functionally characterized them in a yeast genetic system, and used manipulative methods to identify the genetic and molecular mechanisms by which their functions changed. We first inferred the phylogeny and best-fit evolutionary model of the protein family of which Vma3, Vma11 and Vma16 are members, using the sequences of all 139 extant family members available in GenBank (Supplementary Table 1). The maximum likelihood phylogeny (Fig. 1b and Supplementary Information, section 2) indicates that Vma3 and Vma11 are sister proteins that were produced by duplication of an ancestral gene (Anc.3–11) before the last common ancestor of all Fungi (~800 million years ago20). Whether this duplication occurred before or after the divergence of Fungi from other eukaryotes (~1 billion years ago20) is not clearly resolved, although the latter scenario is more parsimonious. TheVma3/Vma11 andVma16 lineages, in turn, descend from an older gene duplication deep in the eukaryotic lineage (Fig. 1b). We used a maximum likelihood algorithm21to infer the ancestral amino acid sequences with the highest probability of producing all the extant sequence data, given the best-fit phylogeny and model. We reconstructed the ancestral proteins (Anc.3–11 and Anc.16) that made up the ancient two-paralogue eukaryotic ring, as well as the duplicated subunits Anc.3 and Anc.11 from the three-component ring in the common ancestor of all Fungi (Supplementary Information, sections 3 and 4).
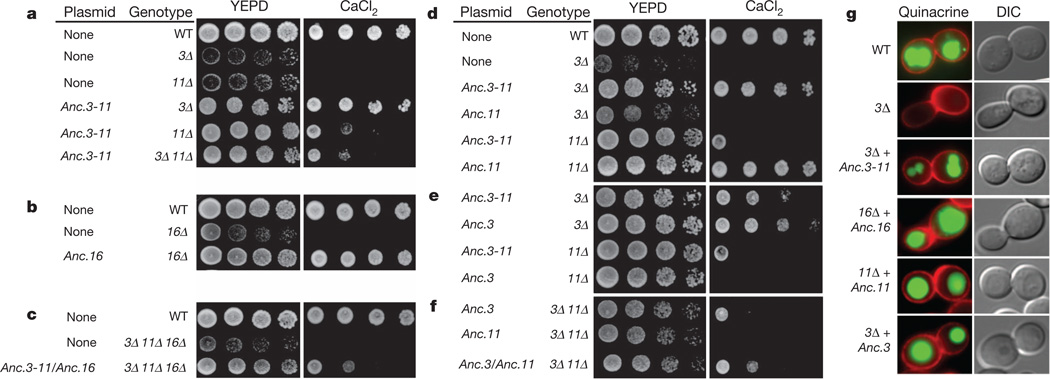
To characterize the functions of these reconstructed proteins, we synthesized coding sequences and transformed them into Saccharomyces cerevisiae deficient for various ring components and therefore incapable of growth in the presence of elevated CaCl2 (ref. 22).We found that the ancestral two-subunit ring can functionally replace the three-subunit ring of extant yeast. When the resurrected Anc.3–11 was transformed into yeast deficient for Vma3 (vma3Δ) or Vma11(vma11Δ), growth in the presence of elevated CaCl2 was rescued, indicating that the functions of the present-day Vma3 and Vma11 proteins were already present before the duplication that generated them (Fig. 2a). Furthermore, Anc.3–11—unlike either of its present-day descendants—can partially rescue growth in yeast that are doubly deficient for both Vma3 and Vma11 (vma3Δvma11Δ). The reconstructed Anc.16 also rescued growth in Vma16-deficient S. cerevisiae (vma16Δ) (Fig. 2b), and co-expression of Anc.3–11 and Anc.16 together rescued cell growth in vma3Δvma11Δvma16Δ yeast, which lack all three ring subunits (Fig. 2c). The ancestral genes specifically restore proper V-ATPase function in acidification of the vacuolar lumen (Fig. 2g). In addition, mutation of the ancestral subunits to remove glutamic acid residues known to be essential for V-ATPase enzyme function17,23abolished their ability to rescue growth on CaCl2 (Supplementary Information, section 7). These inferences about the functions ofAnc.3–11 andAnc.16 are robust to uncertainty about ancestral amino acid states. We reconstructed alternative versions of Anc.3–11 and Anc.16 by introducing amino acid states with posterior probability >0.2, but none of these abolished the ability of the ancestral genes to substitute functionally for the extant subunits (Supplementary Information, section 8). These results establish that during the increase in complexity, neither the V0 complex nor its component proteins evolved new functions required for growth under the conditions in which the ring is known to be important.
Figure 2. Two reconstructed ancestral V0 subunits functionally replace the three-paralogue ring in extant yeast.
S. cerevisiae were plated in decreasing concentrations on permissive medium (YEPD) buffered with elevated CaCl2. a, Expression of Anc.3–11 rescues growth in yeast that are deficient for endogenous subunit Vma3 (3Δ), subunit Vma11 (11Δ) or both (3Δ11Δ). Growth of wild-type (WT) yeast is shown for comparison. b, Anc.16 rescues growth in yeast that are deficient for subunit Vma16 (16Δ). c, Expression of Anc.3–11 and Anc.16 together rescues growth in yeast that are deficient for Vma3, Vma11 and Vma16. d, Anc.11 rescues growth in vma11Δ but not in vma3Δ yeast. e, Anc.3 rescues growth in vma3Δ but not vma11Δ yeast. f, Anc.3 and Anc.11 together rescue growth in vma3Δvma11Δ mutants. g, Yeast expressing reconstructed ancestral subunits properly acidified the vacuolar lumen. Red signal shows yeast cell walls; green signal (quinacrine) shows acidified compartments. Yeast were visualized by differential interference contrast microscopy.
Similar experiments with the components of the ancestral three-component ring show that after the duplication of Anc.3–11, its descendants Anc.3 and Anc.11 both became necessary for a functional complex because of complementary losses of ancestral functions. Unlike Anc.3–11, expression of Anc.3 can rescue growth and vacuole acidification in vma3Δ but not vma11Δ yeast, and Anc.11 can rescue growth in vma11Δ but not vma3Δ yeast (Fig. 2d, e, g). Furthermore, both Anc.3 and Anc.11 are required to rescue growth fully in vma3Δvma11Δ yeast (Fig. 2f). These data indicate that after its origin by gene duplication, Anc.11 lost the ancestral protein’s ability to carry out at least some functions of Vma3, and Anc.3 lost the ancestral capacity to carry out those of Vma11.
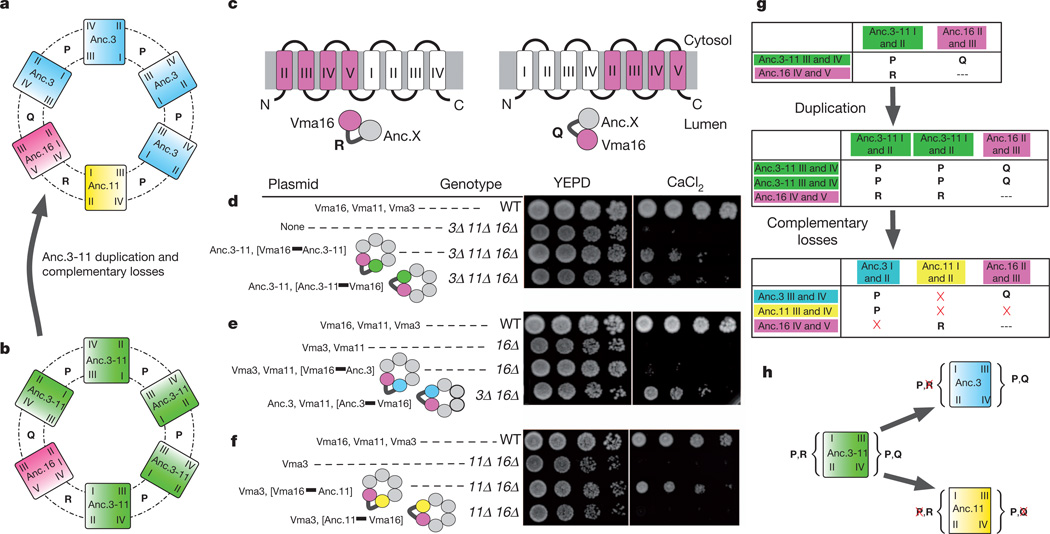
We conjectured that Vma3 and Vma11 evolved their specialized roles because they lost specific interfaces present in their ancestor that are required for ring assembly. Previous experiments with fusions of extant yeast proteins have shown that the arrangement of subunits in the ring is constrained by the capacity of each subunit to form specific interfaces (which we labelled P, Q and R) with the other subunits24. Specifically, Vma11 is restricted to a single position between Vma16 and Vma3, because its clockwise interface can participate only in interfaceRwithVma16, and its anticlockswise interface can participate only in interface P with the clockwise side ofVma3 (Fig. 3). By contrast, copies of Vma3 occupy several positions in the ring, because they form interface P with other copies of Vma3 or Vma11, as well as interface Q withVma16.However,Vma3 cannot form interface R withVma16. As a result, both Vma3 and Vma11 are required in extant yeast to form a complete ring with Vma16.
Figure 3. Increasing complexity by complementary loss of interactions in the fungal V0 ring.
a, Model of the ancestral three-paralogue ring, arranged as in extant yeast24.Unique intersubunit interfaces are labelled P, Q and R. Subunits are colour-coded as in Fig. 1. b, Model of the ancestral two-paralogue ring, before duplication of Anc.3–11. c, To constrain the location of specific subunits, gene fusions were constructed by tethering an ancestral subunit to either the aminoor carboxy-terminal side of yeast Vma16. Roman numerals indicate the locations of transmembrane helices (I, II, III, IV and V)24. d–f, Growth assays of yeast with fused V0 subunits identify the interfaces that ancestral subunits can form. For each experiment, expressed V0 subunits are listed. Tethered subunits are in brackets and connected by a thick line. Cartoons show the constrained location of the tethered subunit relative to Vma16. Anc.3–11 can function on either side of Vma16 (d). Anc.3 can function only on the clockwise side of Vma16 (e). Anc.11 can function only on the anticlockwise side of Sc.16 (f). g, Interfaces that are formed byV0 subunits before and after duplication and complementary loss of interfaces, based on the data in panels d–f. Red crosses indicate lost interfaces. h, Schematic of interfaces formed by Anc.3–11 that were lost in Anc.3 and Anc.11, based on data in panels d–f.
To determine whether interaction interfaces were lost during evolution, we engineered fusions of ancestral ring proteins to assess the capacity of each to form the specific interfaces with other subunits that are required for a functional complex. Because Anc.3–11 can complement the loss of both Vma3 and Vma11, we proposed that the Anc.3–11 subunit could participate in all three specific interaction interfaces, and that these capacities were then partitioned between Anc3 and Anc11 after the duplication of Anc.3–11 (Fig. 3a, b). To test this hypothesis, we created six reciprocal gene fusions between yeast subunit Vma16 and ancestral subunits Anc.3–11, Anc.3 and Anc.11 (Fig. 3c and Supplementary Information, section 9). Each fusion constrains the structural position of subunits relative to subunit Vma16, making it possible to determine which arrangements yield a functional ring. As predicted, Anc.3–11 functioned on either side of Vma16 (Fig. 3d), indicating that it could form all three interfaces P, Q and R. By contrast, Anc.3 functioned when constrained to participate in interface Q with Vma16 and interface P with Vma3; however, ring function was lost when Anc.3 was constrained to form interface R with Vma16 (Fig. 3e). Anc.11 functioned when constrained to participate in interface R with Vma16 and interface P with Vma3, but ring function was lost when Anc.11 was constrained to participate in interface Q with Vma16 and interface P with Vma3. This result indicates that Anc.11 lost the capacity to form one or both of these interfaces during its post-duplication divergence from Anc.3–11 (Fig. 3f).
Taken together, these data indicate that the specificity of the ring arrangement and the obligate roles of Vma3 and Vma11 evolved by complementary loss of asymmetric interactions with other members of the ring (Fig. 3g, h). Before Anc.3–11 duplicated, the protein ring contained copies of only undifferentiated subunit Vma3/Vma11 and subunit 16. Immediately after Anc.3–11 duplicated, the two descendant subunits must have been functionally identical, so the protein ring could have assembled with many possible combinations of the two descendants, including copies of only one of the descendant proteins. This flexibility disappeared when Anc.3 lost the ancestral interface that allowed it to interact with the anticlockwise side of Vma16, and Anc.11 lost the ability to interact with the clockwise side of Vma16 and/or the anticlockwise side of Vma3. These complementary losses are sufficient to explain the specific arrangement of contemporary subunits in reconstructed and present-day fungal transmembrane rings.
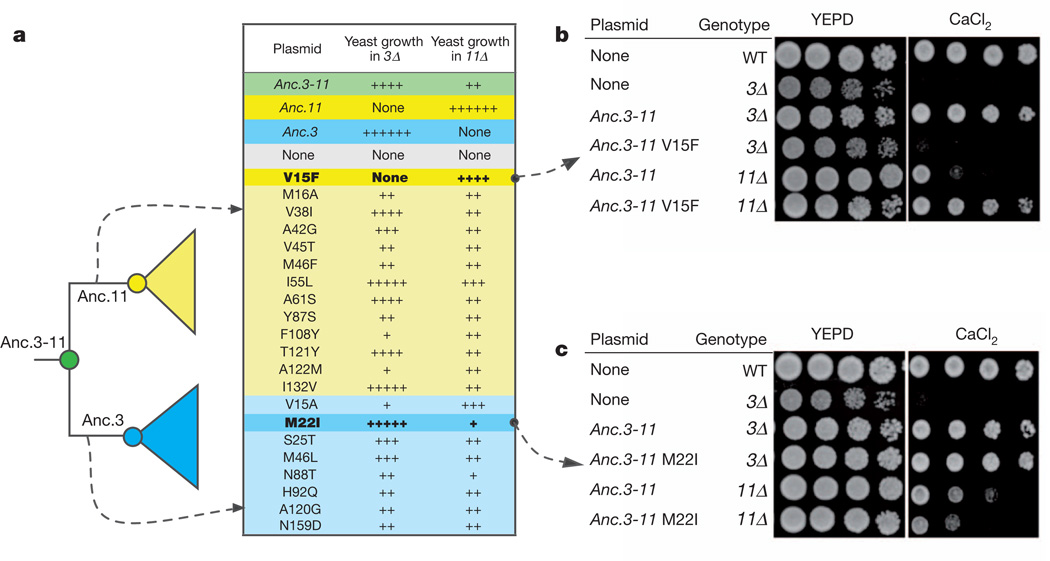
To establish the genetic basis for the partitioning of the functions of Anc.3–11 between Vma3 and Vma11, we introduced historical mutations into Anc3.11 by directed mutagenesis and determined whether they recapitulated the shifts in function that occurred during the evolution of Anc.3 and Anc.11. The two phylogenetic branches leading from Anc.3–11 to Anc.3 and to Anc.11 contain 25 and 31 amino acid substitutions, respectively, but only a subset of these are strongly conserved in subunits Vma3 or Vma11 from extant Fungi (Fig. 4a). We introduced each of these ‘diagnostic’ substitutions into Anc.3–11 and experimentally evaluated whether they recapitulated the loss by Anc.3 or Anc.11 of the capacity to complement Vma gene deletions. We found that a single amino-acid replacement that occurred on the branch leading to Anc.11 (V15F) abolished the capacity of Anc.3–11 to function as subunit 3; it also enhanced the ability of Anc.3–11 to function as subunit 11 (Fig. 4b). V15F is located in transmembrane helix I, which participates in the P interface that our experiments indicate may have been lost on the same branch (Fig. 3 and Supplementary Information, section 4). Conversely, a single historical replacement (M22I) on the branch leading to Anc.3 radically reduced the capacity of Anc.3–11 to function as subunit 11 (Fig. 4c). M22I is also in transmembrane helix I, which participates in formation of the R interface that was lost on this branch (Fig. 3 and Supplementary Information, section 4). The Anc.3–11 M22I mutant retains some of the capacity of the ancestral protein to rescue growth in the Vma11-deficient background, suggesting that other mutations also contributed to the functional evolution of Vma3. One other historical mutation (N88T) on this branch also impaired the capacity of Anc.3–11 to function as subunit 11, but it reduced the capacity of the protein to function asVma3as well, suggesting that epistatic interactions with other residues allow this mutation to be tolerated in Anc.3 and its descendants. Several of the replacements on the branch leading to Anc.11 show a similar pattern, reducing the capacity of the protein to replace Vma3, indicating that these historical replacements function better together than in isolation.
Figure 4. Genetic basis for functional differentiation of Anc.3 and Anc.11.
a, Experimental analysis of historical amino acid replacements. The table lists replacements that occurred on the branches leading from Anc.3–11 to Anc.11 (yellow) or to Anc.3 (blue) and that were subsequently conserved. Each derived residue was introduced singly into Anc.3–11; the variant genes were transformed into S. cerevisiae, and growth was assayed on elevated CaCl2. The table shows growth semiquantitatively from zero (none) to wild type (++++++). Bold mutations entirely or partly recapitulate the functional evolution of Anc.11 and Anc.3. b, Replacement V15F abolishes the capacity of Anc.3–11 to function as subunit 3 and enhances the capacity of Anc.3–11 to function as subunit 11. c, Replacement M22I impairs the capacity of Anc.3–11 to function as subunit 11 without affecting its capacity to function as subunit 3.
How complexity and novel functions evolve has been a longstanding question in evolutionary biology25–27, because mutations that compromise existing functions are far more frequent than those that generate new ones28. Our results indicate that the architectural complexity of molecular assemblies can evolve because of a few simple, relatively high-probability mutations that degrade ancestral interfaces but leave other functions intact. The specific roles of subunits Vma3 and Vma11 seem to have been acquired when duplicated genes lost some, but not all, of the capacity of the ancestral protein to participate in interactions with copies of itself and another protein required for proper ring assembly. Because complementary losses occurred in both lineages, the two descendant subunits became obligate components, and the complexity of the ring increased. It is possible that specialization of the duplicated subunits allowed increases in fitness, but genome-wide interaction screens and the phenotype of vma11Δ yeast provide no evidence that Vma11 evolved novel functions in addition to those that it inherited from Anc.3–11 in the V0 ring29.
We are aware of no other mechanistic analyses of a molecular machine’s evolutionary trajectory, so the generality of our observations is unknown. By definition, however, all molecular machines involve differentiated parts in specific spatial orientations, and many such complexes are entirely or partially composed of paralogous proteins2–8. In the evolution of any such assembly, additional paralogues could become obligate components because of gene duplication30and subsequent mutations that cause specific interaction interfaces among them to degenerate.
This view of the evolution of molecular machines is related to recent models that explain other biological phenomena—such as the retention of large numbers of duplicate genes and mobile genetic elements within genomes—as the product of degenerative processes acting on modular biological systems27. Although mutations that enhanced the functions of individual ring components may have occurred during evolution, our data indicate that simple degenerative mutations are sufficient to explain the historical increase in complexity of a crucial molecular machine. There is no need to invoke the acquisition of ‘novel’ functions caused by low-probability mutational combinations.
METHODS
In silico reconstruction of ancestral protein sequences
V0 complex subunits Vma3,Vma11 and Vma16 are sometimes referred to as subunits c, c′ and c” in the literature. We searched GenBank for all eukaryote V-ATPase V0 ring sequences (Supplementary Information, section 1). Our query returned subunit 3, 11 and 16 protein sequences for 26 species in Fungi, and subunit 3 and 11 sequences for 35 species in Metazoa, Amoebozoa and Apicomplexa. We aligned the sequences using PRANK v0.081202 (refs 32, 33). We selected the best-fit model (WAG with gamma-distributed rate variation and a proportion of invariant sites) using the Akaike Information Criterion as implemented in PROTTEST34,35. With this model, we used PhyML v3.0 to infer the maximum likelihood topology, branch lengths and model parameters36. We optimized the topology using the best result from nearest-neighbour interchange and subtree pruning and regrafting; we optimized all other free parameters using the default hill-climbing algorithm in PhyML. Phylogenetic support was calculated as the approximate likelihood ratio (converted from the approximate likelihood ratio statistic (aLRS) for branches reported by PhyML, using the equation aLR=exp[aLRS/2]) and as the likelihood ratio-based SH-like branch supports37. Nematoda subunit 3 and 11 sequences were connected by a very long branch basal to the Chromalveolata lineages. This result is inconsistent with the expectation that Nematoda are animals38, so we excluded Nematoda data from further downstream analysis.
We inferred ML ancestral states and posterior probability distributions at each site for all ancestral nodes in the ML phylogeny using our own set of Python scripts, called Lazarus, which wraps PAML version 4.1 (ref. 39). Lazarus parsimoniously places ancestral gap characters according to Fitch’s algorithm40. We characterized the overall support for Anc.3–11, Anc.16, Anc.3 and Anc.11 by binning the posterior probability of the ML state at each site into 5%-sized bins and then counting the proportion of total sites within each bin (Supplementary Information, section 2).
Robustness to alignment uncertainty
To assess the robustness of ancestral reconstructions to alignment uncertainty, we performed alignment using four algorithms: CLUSTAL version 2.0.10 (ref. 41), MUSCLE v3.7 (ref. 42), AMAP v2.2 (ref. 43), and PRANK v0.081202 (refs 32, 33). We then inferred the ML phylogeny and branch lengths for each alignment, using the methods described above. The resultant alignments varied in length from347 sites (using CLUSTAL) to 683 sites (using PRANK), but all four alignments yielded the same ML topology with nearly identical ML branch lengths.
To determine which alignment algorithm yields the most accurate ancestral inferences under V-ATPase phylogenetic conditions, we simulated sequences across the V-ATPase ML phylogeny using insertion and deletion rates ranging from 0.0 to 0.1 indels per site. For each indel rate, we generated ten random unique indel-free ancestral sequences of 400 amino acids in length and then used INdelible44 to simulate the ancestral sequence evolving along the branches of our ML phylogeny under the conditions of the WAG model with a proportion of invariant sites (+I) and a discrete gamma distribution of evolutionary rates (+G) with indel events randomly injected according to the specified indel rate. The size of each indel event was drawn from a Zipfian distribution with coefficient equal to 1.1 and the maximum length limited to 10 amino acids. We aligned the descendant sequences of each replicate using AMAP, CLUSTAL, MUSCLE and PRANK. For each alignment, we inferred the ML topology, branch lengths and model parameters using the methods described above. We used Lazarus to reconstruct all of the ancestral states, and queried Lazarus for the most-recent shared ancestor for opisthokont subunit 3/11 and opisthokont subunit 16 sequences. We measured the error of ancestral reconstructions as the proportion of ancestral sites that incorrectly contained an indel character (see Supplementary Information, section 6).
Plasmids and yeast strains
Bacterial and yeast manipulations were performed using standard laboratory protocols for molecular biology45. Plasmids that were used are listed in Supplementary Information, section 5. Ancestral sequences (pGF140, pGF139, pGF506 and pGF508) were synthesized by GenScript with a yeast codon bias. Triple haemagglutinin epitope tags were included before each stop codon. The Anc.3–11, Anc.16, Anc.3 and Anc.11 genes were subcloned to single-copy, CEN-based yeast vectors. The ADH terminator sequence (247 base pairs (bp)) and Natr drug resistance marker46 were amplified using polymerase-chain-reaction (PCR) containing 40-bp tails homologous to the 3′ end of each coding region and vector sequence. Vectors were gapped, co-transformed into SF838-1Dα yeast with PCR fragments and cells were selected for Natr. A second round of in vivo ligation was used to place the ancestral genes under 500 bp of the VMA3 or VMA16 promoters to create pGF140 and pGF139, respectively. For vectors pGF240, pGF241, 1pGF252, pGF253, pGF503–pGF508, pGF510, pGF512–pGF515, pGF517–pGF519, pGF521, pGF523, pGF528, pGF529, pGF531, pGF534–pGF537 and pGF542, the relevant locus (Anc.3–11, Anc.16 or Anc.3) was PCR amplified with 5′ and 3′ untranslated flanking sequence and cloned into pCR4Blunt-TOPO (Invitrogen). When necessary, a modified Quikchange protocol47 was used to introduce point mutations before the gene was subcloned into a yeast vector (pRS316 or pRS415). To generate pGF502, sequence from codon 31 to the stop codon of Anc.16 was amplified with the ADH::Natr cassette from pGF139, cloned into TOPO, and in vivo ligated downstream of the VMA16 promoter (including a start codon) in pRS415.
A triple-fragment in vivo ligation was used to generate pGF646–pGF651. Gapped vector containing the VMA16 promoter was transformed into yeast with two PCR fragments of the ring genes to be fused. For pGF646, the coding region of (1) VMA16 (without codons 2–41) and (2) the coding region of Anc.11 (without codons 2–5 were amplified by PCR. The proteolipid on the C-terminal portion of the gene fusion also contained the ADH terminator and Natr cassette; the amplified products contained PCR tails with homology to link the genes to both the gapped vector and to each other. Gene fusions were modelled after the experimental design of Wang et al. (2007)24 in which the lumenal protein sequence linking the two proteolipids was designed to be exactly 14 amino acids. To meet these criteria, additional amino acids were inserted into the following vectors linking the two subunits: pGF646 (Thr-Arg-Val-Asp), pGF648, pGF650 (Thr-Arg), pGF649, pGF651 (Gly-Ser).
Yeast strains that were used are listed in Supplementary Information, section 2. Strains containing deletion cassettes other than KanR 45 were constructed by PCR amplifying the HygR or Natr cassette from pAG32 or pAG25, respectively, with primer tails with homology to flanking sequences to the VMA11 or VMA16 loci. 11Δ ::KanR and 16Δ ::KanR strains (SF838-1Dα) were transformed with the HygR and NatR PCR fragments, respectively, and selected for drug resistance. The 11Δ::HygR locus was amplified and transformed into LGY113 (to create LGY125) and LGY115 (to create LGY124). This was repeated with the 16Δ ::NatR locus to create LGY139 and LGY143.
Yeast Growth Assays
Yeast were grown in liquid culture, diluted fivefold and spotted onto YEPD media buffered to pH 5.0 or yeast extract peptone dextrose media containing 25mM (Figs 2, 3, 4) or 30mM CaCl2 (Fig. 2f).
Whole-cell extract preparation and immunoblotting
Yeast extracts and western blots were performed as previously described31. Antibodies that were used in this study included monoclonal primary anti-HA (Sigma-Aldrich), anti-Dpm1 (5C5; Invitrogen) and secondary horseradish-conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratory, West Grove, Pennsylvania, USA).
Fluorescence microscopy
Staining with quinacrine was performed as previously described31. The cell wall (shown in red) was visualized using concanavalin A tetramethylrhodamine (Invitrogen). Microscopy images were obtained using an Axioplan 2 fluorescence microscope (Carl Zeiss). A ×100 objective, AxioVision software (Carl Zeiss) and Adobe Photoshop Creative Suite (v. 8.0) were used.
Supplementary Material
Acknowledgements
This study was supported by National Institutes of Health (NIH) grants R01-GM081592 (to J.W.T.) and R01-GM38006 (to T.H.S.), National Science Foundation (NSF) grants IOB-0546906 (to J.W.T.) and DEB-0516530 (to J.W.T.), NIH Genetics Training grant T32-GM007257 (to G.C.F.), NSF IGERT grant DGE-9972830 (to V.H.-S.) and the Howard Hughes Medical Institute (J.W.T.). We thank L. Graham, G. Butler and B. Houser for generating yeast strains and other assistance. We thank members of the Stevens and Thornton laboratories for helpful comments.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions V.H.-S. performed the phylogenetic analysis and statistical reconstructions. G.C.F. performed functional experiments. All authors conceived the experiments, interpreted the results and wrote the paper.
The authors declare no competing financial interests.
References
- 1.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nature Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Pallen MJ, Matzke NJ. From the origin of species to the origin of bacterial flagella. Nature Rev. Microbiol. 2006;4:784–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, Ochman H. Stepwise formation of the bacterial flagellar system. Proc.Natl Acad. Sci. USA. 2007;104:7116–7121. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nature Rev. Microbiol. 2007;5:892–899. doi: 10.1038/nrmicro1767. [DOI] [PubMed] [Google Scholar]
- 5.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 6.Clements A, et al. The reducible complexity of a mitochondrial molecular machine. Proc. Natl Acad. Sci. USA. 2009;106:15791–15795. doi: 10.1073/pnas.0908264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archibald JM, Logsdon JM, Jr, Doolittle WF. Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol. Biol. Evol. 2000;17:1456–1466. doi: 10.1093/oxfordjournals.molbev.a026246. [DOI] [PubMed] [Google Scholar]
- 8.Gabaldón T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I) J. Mol. Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 9.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nature Rev. Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 10.Liberles D, editor. Ancestral Sequence Reconstruction. Oxford Univ. Press; 2007. [Google Scholar]
- 11.Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frattini A, et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nature Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Sayáns M, Somoza-Martìn JM, Barros-Angueira F, Rey JM, Garcìa- Garcìa A. V-ATPase inhibitors and implication in cancer treatment. Cancer Treat. Rev. 2009;35:707–713. doi: 10.1016/j.ctrv.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, et al. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirata T, et al. Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the g and c subunits. J. Biol. Chem. 2003;278:23714–23719. doi: 10.1074/jbc.M302756200. [DOI] [PubMed] [Google Scholar]
- 16.Imamura H, et al. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl Acad. Sci. USA. 2005;102:17929–17933. doi: 10.1073/pnas.0507764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell B, Graham LA, Stevens TH. Molecular characterization of the yeast vacuolar H+-ATPase proton pore. J. Biol. Chem. 2000;275:23654–23660. doi: 10.1074/jbc.M004440200. [DOI] [PubMed] [Google Scholar]
- 18.Umemoto N, Yoshihisa T, Hirata R, Anraku Y. Roles of the VMA3 gene product, subunit c of the vacuolar membrane H+-ATPase on vacuolar acidification and protein transport. A study with VMA3-disrupted mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:18447–18453. [PubMed] [Google Scholar]
- 19.Umemoto N, Ohya Y, Anraku Y. VMA11, a novel gene that encodes a putative proteolipid, is indispensable for expression of yeast vacuolar membrane H+-ATPase activity. J. Biol. Chem. 1991;266:24526–24532. [PubMed] [Google Scholar]
- 20.Taylor JW, Berbee ML. Dating divergences in the fungal tree of life: review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y. Vma11 and vma16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J. Biol. Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Cipriano DJ, Forgac M. Arrangement of subunits in the proteolipid ring of the V-ATPase. J. Biol. Chem. 2007;282:34058–34065. doi: 10.1074/jbc.M704331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno S. Evolution by Gene Duplication. Springer; 1970. [Google Scholar]
- 26.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 27.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl Acad. Sci. USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc. Natl Acad. Sci. USA. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong AHY, et al. Globalmapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 30.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan M, Graham LA, Stevens TH. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol. Biol. Cell. 2008;19:5131–5142. doi: 10.1091/mbc.E08-06-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 32.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 34.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 35.Abascal F, Zardoya R, Posada D. Prottest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 36.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 37.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 38.Aguinaldo AMA, et al. Evidence for a clade of nematodes, arthropods, and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z. PAML4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 40.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 1971;20:406–416. [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fletcher W, Yang Z. Indelible: a flexible simulator of biological sequence evolution. Mol. Biol. Evol. 2009;26:1879–1888. doi: 10.1093/molbev/msp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 46.Goldstein AL, McCuster JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 47.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.