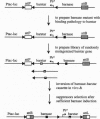
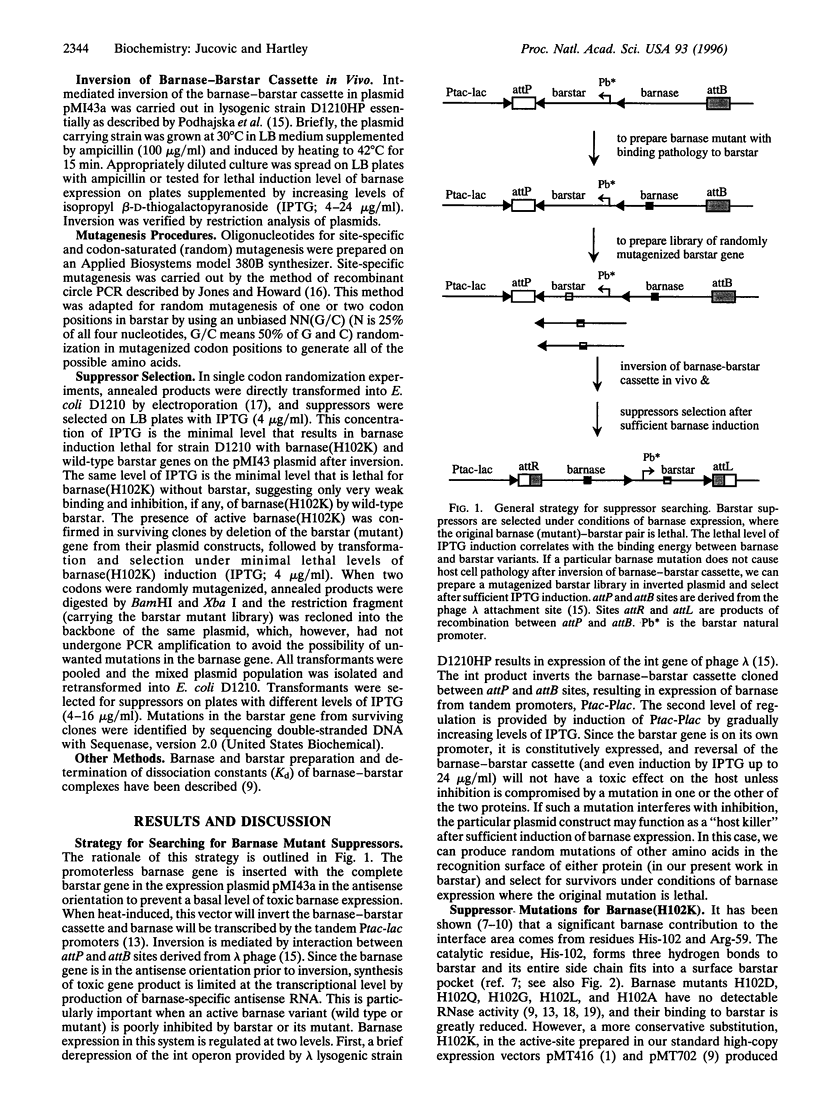
Abstract
Barnase and barstar are trivial names of the extracellular RNase and its intracellular inhibitor produced by Bacillus amyloliquefaciens. Inhibition involves the formation of a very tight one-to-one complex of the two proteins. With the crystallographic solution of the structure of the barnase-barstar complex and the development of methods for measuring the free energy of binding, the pair can be used to study protein-protein recognition in detail. In this report, we describe the isolation of suppressor mutations in barstar that compensate for the loss in interaction energy caused by a mutation in barnase. Our suppressor search is based on in vivo selection for barstar variants that are able to protect host cells against the RNAse activity of those barnase mutants not properly inhibited by wild-type barstar. This approach utilizes a plasmid system in which barnase expression is tightly controlled to keep the mutant barnase gene silent. When expression of barnase is turned on, failure to form a complex between the mutant barnase and barstar has a lethal effect on host cells unless overcome by substitution of the wild-type barstar by a functional suppressor derivative. A set of barstar suppressors has been identified for barnase mutants with substitutions in two amino acid positions (residues 102 and 59), which are critically involved in both RNase activity and barstar binding. The mutations selected as suppressors could not have been predicted on the basis of the known protein structures. The single barstar mutation with the highest information content for inhibition of barnase (H102K) has the substitution Y30W. The reduction in binding caused by the R59E mutation in barnase can be partly reversed by changing Glu-76 of barstar, which forms a salt bridge with the Arg-59 in the wild-type complex, to arginine, thus completing an interchange of the two charges.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Buckle A. M., Schreiber G., Fersht A. R. Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. Biochemistry. 1994 Aug 2;33(30):8878–8889. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- Guillet V., Lapthorn A., Hartley R. W., Mauguen Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure. 1993 Nov 15;1(3):165–176. doi: 10.1016/0969-2126(93)90018-c. [DOI] [PubMed] [Google Scholar]
- Hartley R. W. Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol. 1988 Aug 20;202(4):913–915. doi: 10.1016/0022-2836(88)90568-2. [DOI] [PubMed] [Google Scholar]
- Hartley R. W. Directed mutagenesis and barnase-barstar recognition. Biochemistry. 1993 Jun 15;32(23):5978–5984. doi: 10.1021/bi00074a008. [DOI] [PubMed] [Google Scholar]
- Hasan N., Szybalski W. Control of cloned gene expression by promoter inversion in vivo: construction of improved vectors with a multiple cloning site and the Ptac promoter. Gene. 1987;56(1):145–151. doi: 10.1016/0378-1119(87)90167-3. [DOI] [PubMed] [Google Scholar]
- Jones D. H., Howard B. H. A rapid method for site-specific mutagenesis and directional subcloning by using the polymerase chain reaction to generate recombinant circles. Biotechniques. 1990 Feb;8(2):178–183. [PubMed] [Google Scholar]
- Jucovic M., Hartley R. W. In vivo system for the detection of low level activity barnase mutants. Protein Eng. 1995 May;8(5):497–499. doi: 10.1093/protein/8.5.497. [DOI] [PubMed] [Google Scholar]
- Meiering E. M., Serrano L., Fersht A. R. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992 Jun 5;225(3):585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V., Erickson R. E. Urea denaturation of barnase: pH dependence and characterization of the unfolded state. Biochemistry. 1992 Mar 17;31(10):2728–2734. doi: 10.1021/bi00125a013. [DOI] [PubMed] [Google Scholar]
- Paddon C. J., Hartley R. W. Expression of Bacillus amyloliquefaciens extracellular ribonuclease (barnase) in Escherichia coli following an inactivating mutation. Gene. 1987;53(1):11–19. doi: 10.1016/0378-1119(87)90088-6. [DOI] [PubMed] [Google Scholar]
- Palzkill T., Botstein D. Probing beta-lactamase structure and function using random replacement mutagenesis. Proteins. 1992 Sep;14(1):29–44. doi: 10.1002/prot.340140106. [DOI] [PubMed] [Google Scholar]
- Podhajska A. J., Hasan N., Szybalski W. Control of cloned gene expression by promoter inversion in vivo: construction of the heat-pulse-activated att-nutL-p-att-N module. Gene. 1985;40(1):163–168. doi: 10.1016/0378-1119(85)90038-1. [DOI] [PubMed] [Google Scholar]
- Prevost M., Wodak S. J., Tidor B., Karplus M. Contribution of the hydrophobic effect to protein stability: analysis based on simulations of the Ile-96----Ala mutation in barnase. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10880–10884. doi: 10.1073/pnas.88.23.10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUSHIZKY G. W., GRECO A. E., HARTLEY R. W., Jr, SOBER H. A. STUDIES ON B. SUBTILIS RIBONUCLEASE. I. CHARACTERIZATION OF ENZYMATIC SPECIFICITY. Biochemistry. 1963 Jul-Aug;2:787–793. doi: 10.1021/bi00904a028. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Tecklenburg M., Betz J. L. Plasmids containing many tandem copies of a synthetic lactose operator. Gene. 1980 Feb;8(3):279–300. doi: 10.1016/0378-1119(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Fersht A. R. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993 May 18;32(19):5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Fersht A. R. The refolding of cis- and trans-peptidylprolyl isomers of barstar. Biochemistry. 1993 Oct 19;32(41):11195–11203. doi: 10.1021/bi00092a032. [DOI] [PubMed] [Google Scholar]
- Serrano L., Matouschek A., Fersht A. R. The folding of an enzyme. VI. The folding pathway of barnase: comparison with theoretical models. J Mol Biol. 1992 Apr 5;224(3):847–859. doi: 10.1016/0022-2836(92)90566-3. [DOI] [PubMed] [Google Scholar]
- Shastry M. C., Agashe V. R., Udgaonkar J. B. Quantitative analysis of the kinetics of denaturation and renaturation of barstar in the folding transition zone. Protein Sci. 1994 Sep;3(9):1409–1417. doi: 10.1002/pro.5560030907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speyer J. F. A simple and effective electroporation apparatus. Biotechniques. 1990 Jan;8(1):28–30. [PubMed] [Google Scholar]