Abstract
Adipogenesis is regulated by a complex interplay between transcription factors, in concert with—among others—transcriptional cofactors, signaling cascades and miRNAs. Several studies have implicated the transcriptional cofactor and acetyltransferase Tip60 in PPARγ signaling and adipocyte differentiation. Since Tip60 protein levels, but not mRNA levels, are upregulated during adipogenesis, and since Tip60 can be degraded by the proteasome, we hypothesized that Tip60 protein may be stabilized through deubiquitination during adipogenesis. Indeed, Tip60 is protected from proteasomal degeradation by the deubiquitinase USP7, which is particularly important for mitotic clonal expansion (MCE), an early step in adipogenesis. Besides this novel role in early differentiation, earlier studies indicated that Tip60 is also important during the later stages of differentiation, indicating a dual role for this protein in adipogenesis. Our recent study sheds new light on the role of Tip60 in cellular differentiation and provide new insights into the importance of a regulatory process that has not been studied intensively in adipogenesis: protein (de)ubiquitination.
Keywords: adipogenesis, (de)ubiquitination, Tip60, USP7, mitotic clonal expansion (MCE)
Since adipose tissue is increasingly being recognized as a key regulator of whole-body energy homeostasis and consequently as a prime therapeutic target for metabolic syndrome, adipocyte differentiation and biology are under intensive study.
Adipocyte differentiation is regulated by a complex network of transcription factors, the activity and expression of which is regulated—among others—by transcriptional cofactors, signaling cascades, and miRNAs.1 Increasing evidence shows that adipogenesis is a hierarchical sequence of molecular events: different factors are regulated at different time points, ultimately leading to increased expression and activity of the master regulator of adipogenesis, PPARγ. A detailed understanding of the chronological steps in adipogenesis is therefore essential to understand the role of adipocytes in energy metabolism and obesity-related human health problems like type 2 diabetes.
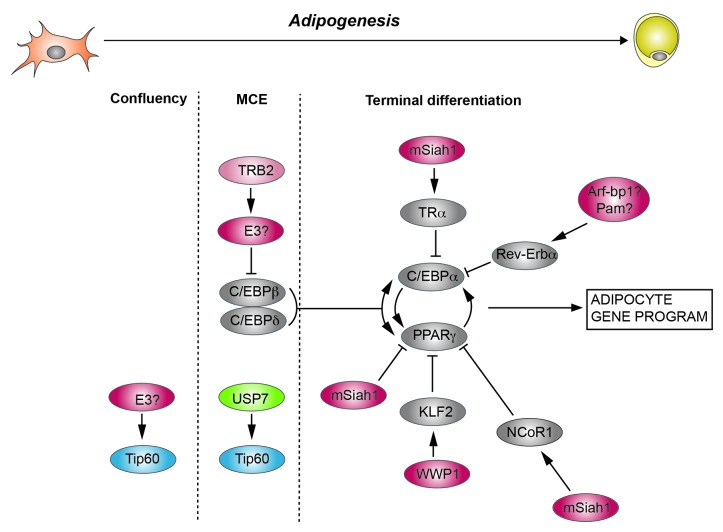
Many of the molecular mechanisms underlying adipocyte differentiation have been discovered using various preadipocyte cell culture systems, the 3T3-L1 preadipocyte cell line being the best known.2 3T3-L1 is a clonal cell line derived from mouse 3T3 cells, selected on basis of its ability to differentiate into mature adipocytes upon appropriate stimulation.3 This differentiation process can be divided into three phases: pre-adipocytes are first cultured in normal medium till reaching confluence, and second, the confluent adipocytes are cultured for 2–3 d in the presence of a hormonal cocktail containing dexamethasone, glucocorticoid, and high dosage of insulin (Fig. 1). This period is also called mitotic clonal expansion (MCE) because the preadipocytes re-enter the cell cycle and undergo another two rounds of cell division. Third, cells undergo terminal differentiation to become mature white adipocytes when cultured in the presence of media containing only insulin for another 3–10 d. During this period many genes involved in lipid uptake (e.g., LPL, CD36, FATP), lipid droplet formation (e.g., Tip47, PLIN), lipogenesis (e.g., FAS), glycerol uptake (e.g., AQP7), and glucose uptake (e.g., Glut4) are upregulated and lipid droplets appear.
Figure 1. Protein (de)ubiquitination in adipogenesis. Indicated are the 3 stages of 3T3-L1 differentiation: growing to confluency, mitotic clonal expansion (MCE), and terminal differentiation. Also depicted are selected transcription factors and co-regulators that are subject to (de)ubiquitination. Ubiquitin E3 ligases are depicted in red, deubiquitinases in green. Please note that TRB2 itself is not an E3 ligase, but stimulates C/EBPβ ubiquitination through an unidentified E3 ligase.
Tip60: An Essential Transcriptional Cofactor in Adipogenesis
A number of transcriptional cofactors, non-DNA binding proteins that can activate or repress transcription (coactivators and corepressors, respectively), have been implicated in adipogenesis, by regulating the activity of PPARγ and/or other adipogenic transcription factors.4,5 In the absence of ligand, nuclear receptor corepressors like nuclear receptor corepressor protein (NCoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) can bind to PPARγ and recruit histone deacetylases (HDACs) to repress transcription.6,7 Ligand binding alters PPARγ’s affinity for a number of coactivators, which are involved in chromatin remodeling by histone modification and nucleosome mobilization, leading to the recruitment of the basal transcription machinery to PPAR target genes.8 Among others, PPARγ coactivators include SRC family members, the SWI/SNF chromatin remodeling complex, and the mediator complex (also referred to as thyroid receptor associated protein [TRAP]/vitamin D receptor interacting proteins [DRIP] complex). SRC belongs to the p160 family. These proteins have weak histone acetyltransferase (HAT) activities while their main function probably lies in providing scaffolds upon which coactivator complexes are assembled.9 The coactivators they recruit include cAMP responsive element binding protein (CREB) binding protein (CBP)/p300. CBP/p300 complex possesses HAT activity and aids in remodelling chromatin to allow transcriptional activation.10 The SWI/SNF complex is thought to be targeted to nuclear receptors by interaction with receptors, coactivators or general transcription machinery.10 This complex also functions in PPARγ-mediated transcription.11-13 The Mediator complex, a multi-protein co-activator complex essential for most RNA polymerase II transcription, also contributes to PPARγ-mediated transcription by bridging this transcription factor to the basal transcription machinery.14-16 More recently, we and others identified the acetyltransferase HIV-1 Tat interacting protein 60 (Tip60), also referred to as K(lysine) acetyltransferase 5 (KAT5), as an essential cofactor in adipogenesis.17,18
Tip60 is a member of a small family of MYST acetyltransferases, named after its founding members MOZ, Ybf2/Sas3 (yeast), Sas2 (yeast), and Tip60, which share a highly conserved MYST acetyltransferase domain, but display limited homology outside this region.19 Tip60 can acetylate both histones and non-histone proteins.19 Tip60 is part of a large multi-protein complex named NuA4,20 implicated in many fundamental cellular processes like transcription, DNA damage repair, cell cycle control, and apoptosis.21 In agreement with this, homozygous Tip60 knockout mice display embryonic lethality.22
We have previously identified Tip60 as a PPARγ interacting protein.17 Chromatin immunoprecipitation experiments showed that the endogenous Tip60 protein is recruited to PPARγ target genes Fabp4 and perilipin in mature 3T3-L1 adipocytes, but not in pre-adipocytes, indicating that Tip60 recruitment critically depends on PPARγ. Importantly, we showed that in common with disruption of PPARγ function, siRNA-mediated reduction of Tip60 protein impairs differentiation of 3T3-L1 pre-adipocytes. Taken together, these findings qualify the acetyltransferase Tip60 as a adipogenic transcriptional co-factor.17 An interesting observation made at that time was that expression of the Tip60 protein, but not mRNA, increases during the first stages of 3T3-L1 differentiation, suggesting that regulation of Tip60 protein levels may play an important role in early adipogenesis. Data obtained in other biological settings indicate that Tip60 can be degraded by the proteasome.23,24 We therefore wished to investigate the molecular mechanisms regulating Tip60 protein stability in adipogenesis.
Protein Ubiquitination and Deubiquitination in Adipogenesis
The regulation of protein stability, i.e., the balance between protein ubiquitination and deubiquitination, in adipogenesis has not been studied intensively so far. The ubiquitin-proteasome pathway is an important mechanism to regulate protein stability, in which proteins are first ubiquitinated through the subsequent action of E1 (activating), E2 (conjugating), and E3 (ligating) enzymes and then degraded by the 26S proteasome.25 Substrate specificity of the ubiquitin-conjugation system is mainly mediated by the E3 ligases, which can belong to the RING finger, the HECT domain, and the U box family.25 A general decline of proteasome activity is observed in adipogenesis in 3T3-L1 cellular models.26 This is consistent with the observation that during the early stages of differentiation in human adipose-derived stem cells proteasome activity has been shown to be highest and it decreases as the stem cells become differentiated.27 At present only a limited number of E3 ubiquitin ligases and substrate proteins have been identified in adipogenesis (Fig. 1). Involvement of the ubiquitin-proteasome pathway in adipogenesis can be either negative, through degradation of pro-adipogenic players, or positive, through degradation of anti-adipogenic factors. Pro-adipogenic players that can be targeted by the ubiquitin-proteasome pathway in adipogenesis include C/EBPβ, PPARγ, C/EBPα, and Tip60. Overexpression of TRB2, a non-enzymatic signaling intermediate that is downregulated in early adipogenesis, inhibits adipogenesis by reducing the level of C/EBPβ through a proteasome-dependent way, but the E3 ubiquitin ligase has not been identified.28 C/EBPα is targeted for degradation by the E3 ligase F-box family member F-box- and WD repeat domain-containing 7 (Fbxw7).29 Together with ring-box 1 (Rbx1), cullin 1 (Cul1) and S-phase kinase-associated protein 1 (Skp1), F-box proteins like Fbxw7 form SCF type E3 ubiquitin ligase complexes.30 Fbxw7-mediated degradation of C/EBPα inhibits adipogenesis.29 Interestingly, Fbxw7 has also been implicated in lipid metabolism and cell fate decision in mouse liver.31 Finally, PPARγ has been identified as a protein that is ubiquitinated and degraded by the proteasome.32 Very recently, the E3 ubiquitin ligase mSiah1, which also ubiquitinates NCoR1 (see below), was shown to ubiquitinate PPARγ.33 While Tip60 is polyubiquitinated on multiple residues and degraded by the ubiquitin-proteasome pathway in pre-adipocytes,34 the E3 ligase responsible remains to be identified.
Anti-adipogenic players that can be targeted by the ubiquitin-proteasome pathway include KLF2, NCoR1, and Rev-erbAα. KLF2 plays a negative role in adipogenesis by directly inhibiting PPARγ expression.35 A HECT-domain E3-ubiquitin ligase, WWP1, interacts with KLF2 in vivo and mediates both poly-ubiquitination and proteasomal degradation of KLF2. Thus WWP1 has a positive role in regulation of adipogenesis.36 The E3 ubiquitin ligase mSiah1 targets NCoR1, a corepressor for PPARγ and other transcription factors,6 for proteasomal degradation.37 Through regulating the interaction of TRα (thyroid hormone receptor α) and NCoR1, which has an inhibiting effect on the promoter of C/EBPα, mSiah1 plays a positive role in adipogenesis. The third anti-adipogenic factor that is targeted by the proteasome is Rev-erbAα, an orphan nuclear receptor.38,39 The Rev-erbAα protein is necessary for the early mitotic events that are required for adipogenesis. The subsequent reduction in Rev-erbAα protein, due to increased degradation via the 26S proteasome, is also required for adipocyte differentiation because Rev-erbAα represses the expression of PPARγ2, the master transcriptional regulator of adipogenesis.40 The E3 ligases Arf-bp1 and Pam (Myc-bp2) can ubiquitinate Rev-erbAα in hepatocytes, but it is unknown if they play a similar role in adipocytes.41
While the stabilization of protein expression through deubiquitination is under intense study in other research fields (e.g., p53 signaling in cancer42), very little information was available on such processes in adipogenesis until recently. The human genome contains 63 deubiquitinases,43 but we identified only two major enzymatic activities in adipocytes: the cytoplasmatic UCHL3 enzyme and the nuclear USP7 protein.34,44 While the cytoplasmatic deubiquitinase UCHL3 has been implicated in adipogenesis and insulin signaling both in vitro and in vivo,44-46 its substrate(s) are currently unknown. Recently, we reported the adipogenic transcriptional cofactor and acetyltransferase Tip60 to be a USP7 substrate.34
USP7 Deubiquitinates Tip60 in Early Adipogenesis
Using an HA-tagged probe that covalently binds active DUBs47 we identified USP7 as a major DUB activity in 3T3-L1 adipocytes and in WAT and BAT from mice.34 USP7 plays an important role in adipogenesis, as siRNA-mediated inhibition of USP7 expression or treatment of cells with a broad range inhibitor of DUB activity through a small-molecule inhibitor decreased adipogenesis. One of the substrates of USP7 is Tip60: USP7 interacts with Tip60 and deubiquitinates this protein, resulting in increased Tip60 protein levels. Importantly, USP7-mediated deubiquitination of Tip60 was also observed in vitro with purified proteins, indicating that Tip60 is indeed a direct substrate. To identify which genes are regulated by this newly identified USP7-Tip60 pathway, knockdown of either factor was performed, followed by microarray analysis. Interestingly, a subset of Tip60 target genes that were also controlled by USP7 were involved in cell cycle regulation. Knockdown of either factor resulted in impaired MCE (i.e., reduced cell division in the early stages of differentiation).34 Besides this novel role in early differentiation, earlier studies described above indicated that Tip60 is also important during the later stages of differentiation, indicating a dual role for this protein in adipogenesis (Fig. 2). Interestingly, Dar et al. also recently reported that USP7 can interact with and deubiquitinate Tip60, and they show stabilization of Tip60 to be required for an effective p53-dependent apoptotic pathway.48 Together with our current findings in adipogenesis, these studies underscore the relevance of USP7-mediated Tip60 stabilization in multiple independent biological pathways.
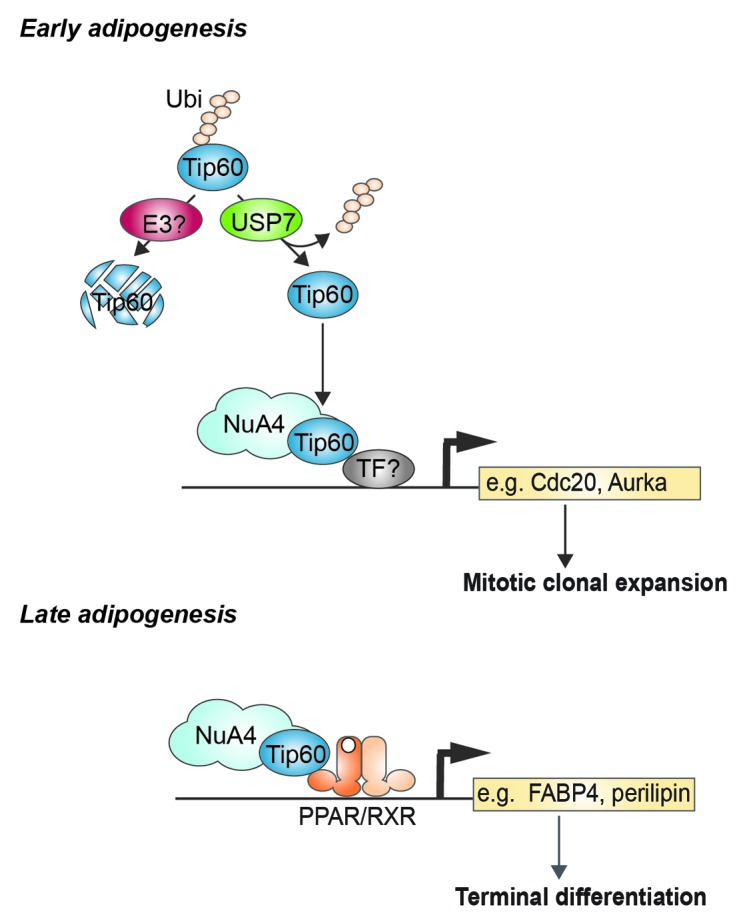
Figure 2. A dual role for Tip60 in adipogenesis. Tip60 is protected from proteasomal degradation by the deubiquitinase USP7 in early adipogenesis and plays a role in regulating genes involved in MCE, probably as part of the Tip60-containing NuA4 complex. Whether Tip60/NuA4 is recruited to cell cycle genes (e.g., Cdc20, Aurka) through a specific transcription factor (TF) during MCE is currently unknown. During late adipogenesis Tip60 is recruited to lipid handling genes (e.g., FABP4, perilipin) by PPARγ-RXR.
Conclusions
Our recent study reveals deubiquitination of a transcriptional coregulator (Tip60) to be a key mechanism in the regulation of early adipogenesis.34 While giving a first glimpse on the role of a USP7-Tip60 pathway in adipocyte differentiation, it also gives rise to several new questions. First, our data suggest that the enzymatic activity of USP7 rather than its expression is upregulated during adipogenesis, but the underlying mechanism remains to be established.34 Second, as several different substrates have been identified for USP7 in other cellular models, it seem likely that also in adipocytes USP7 substrates other than Tip60 exist. Third, the critical Tip60 substrates in adipogenesis remain to be established: Tip60 may be partly responsible for the increased histone acetylation observed in specific key adipogenesis regulatory genes during adipogenesis,49 but Tip60 can also acetylate non-histone proteins. Finally, the in vivo relevance of the USP7-Tip60 pathway should be addressed, which is hampered by the embryonic lethality observed in USP7 and Tip60 knockout mice.22,50 Taken together, our findings shed new light on the role of Tip60 in cellular differentiation and provide new insights into the importance of a regulatory process that has not been studied intensively in adipogenesis: protein (de)ubiquitination. Future studies are needed to identify the critical enzymes and substrates, followed by assessment of their in vivo relevance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/adipocyte/article/28307
References
- 1.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–34. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulos SP, Dodson MV, Hausman GJ. Cell line models for differentiation: preadipocytes and adipocytes. Exp Biol Med (Maywood) 2010;235:1185–93. doi: 10.1258/ebm.2010.010063. [DOI] [PubMed] [Google Scholar]
- 3.Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 4.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Koppen A, Kalkhoven E. Brown vs white adipocytes: the PPARgamma coregulator story. FEBS Lett. 2010;584:3250–9. doi: 10.1016/j.febslet.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 6.Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–5. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 7.Stanley TB, Leesnitzer LM, Montana VG, Galardi CM, Lambert MH, Holt JA, Xu HE, Moore LB, Blanchard SG, Stimmel JB. Subtype specific effects of peroxisome proliferator-activated receptor ligands on corepressor affinity. Biochemistry. 2003;42:9278–87. doi: 10.1021/bi034472c. [DOI] [PubMed] [Google Scholar]
- 8.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/S1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 9.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/S0378-1119(00)00024-X. [DOI] [PubMed] [Google Scholar]
- 10.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- 11.Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol Cell Biol. 2004;24:4651–63. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–86. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 13.Caramel J, Medjkane S, Quignon F, Delattre O. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene. 2008;27:2035–44. doi: 10.1038/sj.onc.1210847. [DOI] [PubMed] [Google Scholar]
- 14.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–7. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 15.Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol Cell Biol. 2008;28:1081–91. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grøntved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol Cell Biol. 2010;30:2155–69. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Beekum O, Brenkman AB, Grontved L, Hamers N, van den Broek NJ, Berger R, Mandrup S, Kalkhoven E. The adipogenic acetyltransferase Tip60 targets activation function 1 of peroxisone proliferator-activated receptor gamma. Endocrinology. 2008;149:1840–9. doi: 10.1210/en.2007-0977. [DOI] [PubMed] [Google Scholar]
- 18.Grönniger E, Wessel S, Kühn SC, Söhle J, Wenck H, Stäb F, Winnefeld M. A new protocol for functional analysis of adipogenesis using reverse transfection technology and time-lapse video microscopy. Cell Biol Int. 2010;34:737–46. doi: 10.1042/CBI20090299. [DOI] [PubMed] [Google Scholar]
- 19.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–76. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–54. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–42. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 23.Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 2002;21:1704–12. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, Yoshida M, Benkirane M, Trouche D, Khochbin S. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 2005;24:2634–45. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–76. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 26.Dasuri K, Zhang L, Ebenezer P, Fernandez-Kim SO, Bruce-Keller AJ, Szweda LI, Keller JN. Proteasome alterations during adipose differentiation and aging: links to impaired adipocyte differentiation and development of oxidative stress. Free Radic Biol Med. 2011;51:1727–35. doi: 10.1016/j.freeradbiomed.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto K, Sato Y, Sei M, Ewis AA, Nakahori Y. Proteasome activity correlates with male BMI and contributes to the differentiation of adipocyte in hADSC. Endocrine. 2010;37:274–9. doi: 10.1007/s12020-009-9298-4. [DOI] [PubMed] [Google Scholar]
- 28.Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J Biol Chem. 2007;282:24075–82. doi: 10.1074/jbc.M701409200. [DOI] [PubMed] [Google Scholar]
- 29.Bengoechea-Alonso MT, Ericsson J. The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBPalpha for degradation. Proc Natl Acad Sci U S A. 2010;107:11817–22. doi: 10.1073/pnas.0913367107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hindley CJ, McDowell GS, Wise H, Philpott A. Regulation of cell fate determination by Skp1-Cullin1-F-box (SCF) E3 ubiquitin ligases. Int J Dev Biol. 2011;55:249–60. doi: 10.1387/ijdb.103171ch. [DOI] [PubMed] [Google Scholar]
- 31.Onoyama I, Suzuki A, Matsumoto A, Tomita K, Katagiri H, Oike Y, Nakayama K, Nakayama KI. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J Clin Invest. 2011;121:342–54. doi: 10.1172/JCI40725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J Biol Chem. 2000;275:18527–33. doi: 10.1074/jbc.M001297200. [DOI] [PubMed] [Google Scholar]
- 33.Kilroy G, Kirk-Ballard H, Carter LE, Floyd ZE. The ubiquitin ligase Siah2 regulates PPARγ activity in adipocytes. Endocrinology. 2012;153:1206–18. doi: 10.1210/en.2011-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, Koppen A, Rakhshandehroo M, Tasdelen I, van de Graaf SF, van Loosdregt J, van Beekum O, Hamers N, van Leenen D, Berkers CR, et al. Early adipogenesis is regulated through USP7-mediated deubiquitination of the histone acetyltransferase TIP60. Nat Commun. 2013;4:2656. doi: 10.1038/ncomms3656. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Krüppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278:2581–4. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Krüppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316:139–48. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 37.Zhu XG, Kim DW, Goodson ML, Privalsky ML, Cheng SY. NCoR1 regulates thyroid hormone receptor isoform-dependent adipogenesis. J Mol Endocrinol. 2011;46:233–44. doi: 10.1530/JME-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–9. [PubMed] [Google Scholar]
- 39.Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart-Najib J, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–80. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28:2213–20. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin L, Joshi S, Wu N, Tong X, Lazar MA. E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erb alpha. Proc Natl Acad Sci U S A. 2010;107:11614–9. doi: 10.1073/pnas.1000438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood SA. Dubble or nothing? Is HAUSP deubiquitylating enzyme the final arbiter of p53 levels? Sci STKE. 2002;2002:pe34. doi: 10.1126/stke.2002.143.pe34. [DOI] [PubMed] [Google Scholar]
- 43.Nijman SMB, Luna-Vargas MPA, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 44.van Beekum O, Gao Y, Berger R, Koppen A, Kalkhoven E. A novel RNAi lethality rescue screen to identify regulators of adipogenesis. PLoS One. 2012;7:e37680. doi: 10.1371/journal.pone.0037680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Setsuie R, Suzuki M, Kabuta T, Fujita H, Miura S, Ichihara N, Yamada D, Wang YL, Ezaki O, Suzuki Y, et al. Ubiquitin C-terminal hydrolase-L3-knockout mice are resistant to diet-induced obesity and show increased activation of AMP-activated protein kinase in skeletal muscle. FASEB J. 2009;23:4148–57. doi: 10.1096/fj.09-132217. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki M, Setsuie R, Wada K. Ubiquitin carboxyl-terminal hydrolase l3 promotes insulin signaling and adipogenesis. Endocrinology. 2009;150:5230–9. doi: 10.1210/en.2009-0332. [DOI] [PubMed] [Google Scholar]
- 47.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–59. doi: 10.1016/S1074-5521(02)00248-X. [DOI] [PubMed] [Google Scholar]
- 48.Dar A, Shibata E, Dutta A. Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Mol Cell Biol. 2013;33:3309–20. doi: 10.1128/MCB.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Ramlee MK, Brunmeir R, Villanueva CJ, Halperin D, Xu F. Dynamic and distinct histone modifications modulate the expression of key adipogenesis regulatory genes. Cell Cycle. 2012;11:4310–22. doi: 10.4161/cc.22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–9. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]