Abstract
Oncometabolites are defined as small-molecule components (or enantiomers) of normal metabolism whose accumulation causes signaling dysregulation to establish a milieu that initiates carcinogenesis. In a similar manner, we propose the term “gerometabolites” to refer to small-molecule components of normal metabolism whose depletion causes signaling dysregulation to establish a milieu that drives aging. In an investigation of the pathogenic activities of the currently recognized oncometabolites R(-)-2-hydroxyglutarate (2-HG), fumarate, and succinate, which accumulate due to mutations in isocitrate dehydrogenases (IDH), fumarate hydratase (FH), and succinate dehydrogenase (SDH), respectively, we illustrate the fact that metabolic pseudohypoxia, the accumulation of hypoxia-inducible factor (HIFα) under normoxic conditions, and the subsequent Warburg-like reprogramming that shifts glucose metabolism from the oxidative pathway to aerobic glycolysis are the same mechanisms through which the decline of the “gerometabolite” nicotinamide adenine dinucleotide (NAD)+ reversibly disrupts nuclear–mitochondrial communication and contributes to the decline in mitochondrial function with age. From an evolutionary perspective, it is reasonable to view NAD+-driven mitochondrial homeostasis as a conserved response to changes in energy supplies and oxygen levels. Similarly, the natural ability of 2-HG to significantly alter epigenetics might reflect an evolutionarily ancient role of certain metabolites to signal for elevated glutamine/glutamate metabolism and/or oxygen deficiency. However, when chronically altered, these responses become conserved causes of aging and cancer. Because HIFα-driven pseudohypoxia might drive the overproduction of 2-HG, the intriguing possibility exists that the decline of gerometabolites such as NAD+ could promote the chronic accumulation of oncometabolites in normal cells during aging. If the sole activation of a Warburg-like metabolic reprogramming in normal tissues might be able to significantly increase the endogenous production of bona fide etiological determinants in cancer, such as oncometabolites, this undesirable trade-off between mitochondrial dysfunction and activation of oncometabolites production might then pave the way for the epigenetic initiation of carcinogenesis in a strictly metabolic-dependent manner. Perhaps it is time to definitely adopt the view that aging and aging diseases including cancer are governed by a pivotal regulatory role of metabolic reprogramming in cell fate decisions.
Keywords: cancer, aging, metabolism, oncometabolites, HIF, pseudohypoxia, Warburg effect, Myc, geroncogenesis
Over the past decade, Otto Warburg’s initially discarded hypothesis from the 1920s, that mitochondrial dysfunction and the acquisition of a glycolytic phenotype were both metabolic features at the root of cancer, has finally attained the status of a core cancer hallmark.1-7 The metabolic signatures of cancer cells, which have been frequently perceived by traditional biochemists as indirect, secondary phenomena merely required to support oncogene-directed anabolic proliferation and survival,8 are not passive responses to damaged mitochondria as originally proposed by Otto Warburg; we now know that the metabolism and metabolites of cancer cells can be oncogenic themselves. Thus, proto-oncogenes and tumor suppressors possibly originated through evolution as components of metabolic regulation and, more importantly, so-called oncometabolites can directly impair the normal epigenetic regulation of cell differentiation and alter cell signaling to drive cellular transformation and oncogenesis.9-22
If Otto Warburg were alive today, he would surely be surprised not only by the new convention of metabolic reprogramming as a bona fide cancer attribute, but also by the latest discovery that dysfunctional mitochondria and Warburg-like metabolic reprogramming are crucial contributors to aging triggered by the reversible decline of nuclear nicotinamide adenine dinucleotide (NAD+) metabolite levels.23,24 We here propose that, given that the term oncometabolite refers to a small-molecule component (or enantiomer) of normal metabolism whose accumulation causes signaling dysregulation to establish a milieu that initiates carcinogenesis,20,22 it might be reasonable to coin the term gerometabolite to denote a small-molecule component of normal metabolism whose depletion causes metabolic and nonmetabolic dysregulation to establish a milieu that drives aging.
Oncometabolites: Reprogramming the Epigenetic Landscape in Cancer
The term oncometabolite was first coined to describe (R)-2-hydroxyglutarate [(R)-2HG], the reduced form of 2-oxoglutarate (2OG).9-11,13,19,20,22 (R)-2HG is a by-product of gain-of-function mutations in the genes encoding the isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2), which normally catalyze the reversible NADP+-dependent oxidative-decarboxylation of isocitrate to produce 2OG in the cytoplasm or mitochondria, respectively. There has been increasing interest in (R)-2HG due to its apparent novelty as a rare metabolite found only in trace amounts in non-diseased mammalian cells under normal conditions; in addition, (R)-2HG is truly pathogenic and not just an indolent byproduct of a loss-of-function mutation in human malignancies. The pathological accumulation of small organic acids such as (R)-2HG due to the acquisition of neomorphic enzymatic activity by cancer-associated mutations in cytosolic IDH1 and mitochondrial IDH2 is apparently sufficient to impair cellular differentiation by competing with the normal functioning of α-ketoglutarate, which is produced in part by wild-type IDH and is used as a substrate for dioxygenase enzymes that modify nuclear epigenetic marks.4,20,22 For example, (R)-2HG can inhibit certain members of the ten-eleven translocation (TET) family of dioxygenase enzymes, including TET2 DNA hydroxylase, which converts 5′-methylcytosine (5mC) to 5-hydroxymethylcytosine, an intermediate in either passive or active DNA demethylation. In addition, (R)-2HG can inhibit specific Jumonji C domain-containing histone lysine demethylase (KDM) enzymes, which demethylate the lysine residues on histone tails (e.g., histone H3 lysine 9 [H3K9]). The ability of (R)-2HG to competitively inhibit chromatin-modifying α-ketoglutarate-dependent dioxygenase enzymes alters histone and DNA methylation in a synergistic manner, thus drastically impairing normal epigenetic regulation by promoting hypermethylation at CpG islands in some cases. Indeed, the oncogenic activity of the (R)-2HG oncometabolite likely relies on its ability to epigenetically block the acquisition of differentiation markers while inducing the expression of stem cell-maintenance genes.4,20,22,25,26
Loss-of-function mutations in tumor suppressor genes encoding the Krebs cycle enzymes fumarate hydratase (FH) and succinate dehydrogenase (SDH) lead to an accumulation of the oncometabolites fumarate and succinate, respectively.12,14,16-18,21 Abnormal accumulation of fumarate and succinate has potential tumorigenic effects via several mechanisms including the following: overproduction of reactive oxygen species (ROS), which may participate in oncogenic signaling and tumor progression by the irreversible modification of DNA and oxidation of proteins; irreversible modification of cysteine residues in proteins by succination, which may result in the constitutive activation of the NRF2-mediated antioxidant defense pathway that can promote tumorigenesis not only by enhancing ROS detoxification, but also by producing a reductive milieu that can promote cell survival and proliferation; and dysregulation of the enzymatic reactions involved in the biosynthesis of arginine and purine. As observed for (R)-2HG, the aberrant accumulation of fumarate and succinate can also inhibit the activities of α-ketoglutarate-dependent TET and the KDM family of 5mC hydroxylases, thus remodeling the cancer epigenome toward undifferentiated and aggressive phenotypes (e.g., neuroendocrine differentiation and epithelial-to-mesenchymal transition). Accordingly, there is an overlap in the hypermethylation patterns of tumors containing IDH and SDH mutations,16 strongly suggesting a shared role for (R)-2HG, succinate, and fumarate oncometabolites in the reprogramming of the epigenetic cancer landscape.
Oncometabolites and Pseudohypoxia: A Metabolic Link to Gerometabolites?
As mentioned above, a common feature of the mutations in IDH, FH, and SDH enzymes is the reduced activity of α-ketoglutarate-dependent dioxygenases such as TET and KDMs, which leads to an inhibition of histone and DNA demethylation. The (R)-2HG, succinate, and fumarate oncometabolites also target hypoxia-inducible factor α (HIFα) prolyl hydroxylases (PHDs). Early studies showed that (R)-2HG cannot only reduce levels of α-ketoglutarate, but also inhibit HIFα-PHDs, leading to decreased HIFα degradation and an enhanced HIFα-orchestrated “pseudohypoxic” response (Fig. 1); accordingly, HIFα has been shown to be upregulated in cells treated with exogenous (R)-2HG and in cells that overexpress mutant IDH1. Conversely, later studies suggested that in contrast to (S)-2HG, which acts as an inhibitor of HIFα-PHD, (R)-2HG stimulates the activity of this enzyme;27,28 accordingly, the expression of mutant IDH1 has been shown to enhance HIFα degradation and diminish HIFα response levels, whereas the loss of HIFα-PHD activity can block the transformation ability of mutant IDH. In certain cellular contexts, therefore, HIFα or other specific targets of hydroxylation by HIFα-PHD appear to suppress the oncogenic potential of (R)-2HG. Thus, it remains somewhat unclear whether (R)-2HG has an agonistic or antagonistic effect on HIFα-PHD at tumor-relevant concentrations and whether HIFα, which has traditionally been viewed as oncogenic, could act as a tumor suppressor in some IDH-mutated tumors. Nevertheless, these data were generated in vitro and highlight contradictory (and difficult to integrate with analyses of tumors) but not mutually exclusive results, obtained from different models, in which (R)-2HG-induced stabilization of HIFα as a consequence of competitive inhibition of PHDs, the 2OG-dependent dioxygenases that regulate HIFα, is a potential mechanism for oncogenesis closely related to HIFα-driven glycolytic response.
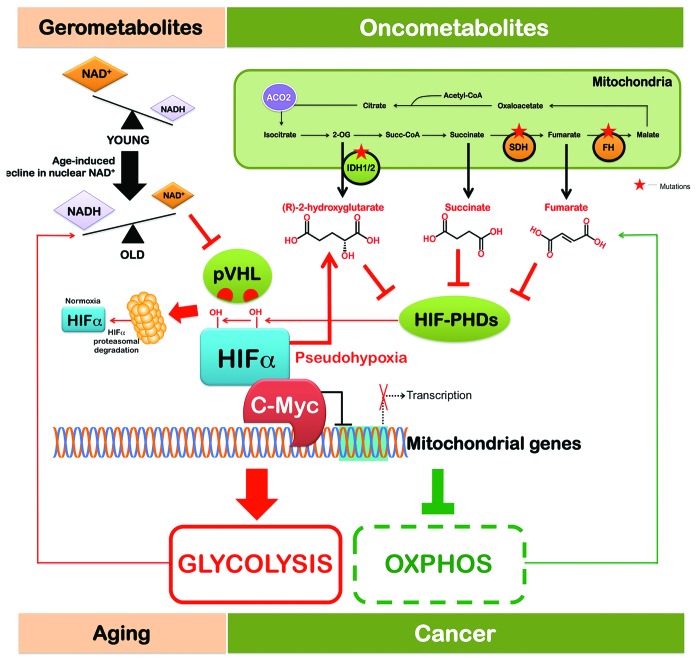
Figure 1. The pseudohypoxia-switching hub: A unifying link between gerometabolites and oncometabolites. Transition from oxidative metabolism into Warburg-like aerobic glycolysis sets a cell metabotype commonly shared by aging and cancer. The intriguing convergence of gerometabolites and oncometabolites on Myc, the so-called “oncogene from hell”, provides not only evidence for a key molecular “funnel factor” linking metabolism with aging–cancer signatures, but also provocatively implicates Myc as a distinctive mechanistic target to decelerate aging and postpone age-related diseases such as cancer without the emergence of resistance phenomena.
SDH- and FH-deficient cells and tumors have been reported to exhibit the activation of an HIFα-orchestrated “pseudohypoxic” response, which could, at least in part, be attributed to the allosteric inhibition of HIFα-PHD by elevated levels of succinate or fumarate (Fig. 1).29-36 Succinate was initially found to impair PHD2, the α-ketoglutarate-dependent enzyme regulating HIFα stability, through product inhibition, and subsequent work confirmed that succinate could also inhibit the related enzyme PHD3. Succinate impedes PHD activity by product inhibition and prevents the decarboxylation of α-ketoglutarate to succinate, an essential co-reaction in the hydroxylation of targets by PHDs. Inhibited PHDs can no longer hydroxylate HIFα, resulting in its stabilization. Because fumarate can similarly inhibit PHD2, these observations link the elevated levels of HIFα-dependent oncogenic pathways observed in SDH- and FH-deficient tumors to the anti-HIFα-PHD activity induced by elevated levels of the oncometabolites succinate and fumarate.
A landmark study by David Sinclair’s group at Harvard revealed that a decrease in a small-molecule component of normal metabolism (i.e., NAD+) causally triggers an HIFα-driven metabolic reprogramming that disrupts mitochondrial homeostasis in normal tissues during aging.23 The biogenesis of complex I, the largest enzyme of the mammalian mitochondrial oxidative phosphorylation (OXPHOS) respiratory chain, is a very complex process due to the large size and number of subunits (45 in humans). The situation is further complicated, because the complex I subunits have a double genomic origin; some of the genes encoding complex I subunits have been retained in the genome of the mitochondrion, the ancient symbiont of eukaryotes, thus complicating the transcriptional regulation of OXPHOS complex I. The functional communication between the nucleus and mitochondria necessary for the formation of stoichiometric OXPHOS complexes, which is largely driven by the peroxisome proliferator-activated receptor-γ coactivators α and β (PGC-1α and PGC-1β),37 is lost during the aging process, causing a specific loss of mitochondrial, but not nuclear, encoded OXPHOS subunits. Gomes et al.23 described the existence of a previously unrecognized PGC-1α/β-independent pathway of nuclear–mitochondrial communication that is induced by a decline in nuclear NAD+, a central metabolic cofactor, by virtue of its redox capacity. NAD+ is consumed as a co-substrate by the so-called sirtuins to deacetylate proteins in different subcellular compartments for a variety of functions, such as optimizing mitochondrial function and biogenesis and stabilizing HIF-1α under normoxic conditions,38-46 thus causing a pseudohypoxia-driven imbalance between nuclear- and mitochondrial-encoded OXPHOS subunits (Fig. 1). This process is accelerated by deleting the NAD+-dependent histone deacetylase sirtuin 1 (SIRT1), whereas treatment with a compound that boosts NAD+ levels is sufficient to restore the mitochondrial homeostasis in old mice to a state similar to that of young mice in a SIRT1-dependent manner.23
In the aging scenario, therefore, the decline in nuclear NAD+ is the causal inducer of the accumulation of HIF-1α under normoxic conditions, which promotes a pseudohypoxic state that disrupts nuclear–mitochondrial communication and contributes to the decline in mitochondrial function with age. Mechanistically, the decline in nuclear NAD+ levels reduces the activity of SIRT1 in the nucleus, causing the levels of the von Hippel–Lindau (VHL) gene product (pVHL), an ubiquitin ligase that recognizes and ubiquitylates HIF to promote its degradation by the proteasome,47-50 to decline and HIF-1α to be stabilized (Fig. 1). Although Gomes et al.23 failed to detect SIRT1-related changes in the hydroxylation of HIF-1α, it was clear that SIRT1 regulated the pVHL status at the posttranscriptional level, suggesting that SIRT1 is constantly required to maintain mitochondrial homeostasis by inducing pVHL and by ensuring that HIF-1α is degraded efficiently. Because pseudohypoxia will undoubtedly shift glucose metabolism from the oxidative pathway to aerobic glycolysis, the reprogramming toward an HIFα-directed, Warburg-like, pseudohypoxic state appears to be a metabolic phenomenon shared by the most common aging diseases, including cancer,51,52 type 2 diabetes and metabolic syndrome,53-55 high fat diet-induced liver steatosis,56 white adipose tissue-related dietary obesity,57 and likely also affects other key organs such as the heart and brain during aging.23
Oncometabolites, Gerometabolites, Gerogenes, and Gerosuppressors: An HIFα-Driven Pseudohypoxic Metabolic Signature
The fact that the accumulation of oncometabolites and the decline of gerometabolites similarly activate the same metabolic, pseudohypoxic response to elicit oncogenic and aging effects is, intriguingly, consistent with the paradoxical ability of the best-characterized gerogene (i.e., the mechanistic target of rapamycin [mTOR]) to contribute, when stimulated, to the transformation and growth of cancer cells (oncogenesis) while leading to stem cell depletion through the activation of senescence programs (aging).58,59 The prolonged stimulation of mTOR in normal cells can lead to stem cell depletion and reduced organismal health and life span, and mTOR activation is a common feature in most human malignancies. Although the ultimate molecular mechanism(s) underlying the aimless continuation of developmental growth driven by nutrient-sensing, growth-promoting signaling pathways such as the mTOR gerogene are not completely understood, it should be noted that the hyperfunctions (aging), loss of homeostasis, and age-related disease (cancer) triggered by the continuous post-developmental activity of such gerogenic pathways60-71 can be easily linked to the pseudo-hypoxic scenario triggered by the accumulation of oncometabolites and the decline of gerometabolites.
HIFα homeostasis is controlled by its rate of translation and degradation (Fig. 2). Under physiological conditions in young tissues, HIFα is constantly degraded in a process dependent on oxygen, prolyl hydroxylation by HIFα-PHDs, and ubiquitylation mediated by pVHL. In cancer, HIFα levels are elevated owing to hypoxia (lack in oxygen) or pseudo-hypoxia (PHD inhibition or pVHL mutation), which inhibits HIFα degradation. We now know that HIFα-PHDs respond to stimuli other than oxygen, including the oncometabolites (R)-2HG, succinate, or fumarate, as illustrated by the pseudo-hypoxic response in SDH- and FH-deficient tumors. Declining NAD+ gerometabolite levels also induce a pseudohypoxic state during aging by blocking pVHL-dependent HIFα stability in a SIRT1-dependent manner. Levels of HIFα in cancer can also be elevated due to accelerated translation. In this regard, a pseudohypoxic response in tumors can be achieved by accelerated mTOR-dependent translation of HIFα.72-78 The mTOR-dependent translation of HIFα is induced by the loss of the TSC1, TSC2, or LKB1 tumor-suppressor/gerosuppressor genes and possibly by the activation via the AKT pathway. Indeed, mTOR inhibition has been shown to lead to a profound attenuation of HIFα protein levels in the majority of primary and cancer cells that have been studied. Under severe hypoxia, however, no influence of mTOR inhibitors on HIFα expression status has been observed; thus, stimulation of HIFα by gerogenic mTOR signaling may only be relevant under mild hypoxia or normoxia. The pseudo-hypoxic hypothesis of the gerogenic activity of mTOR is further supported by the fact that the function of the PHD–HIF feedback loop (hypoxia inactivates PHDs, causing accumulation of HIF-1α; in turn, HIF-1α further transactivates PHDs) has been suggested to limit the induction of HIF-1α by geropromoters (i.e., mTOR activators) such as insulin, growth factors, hormones, cytokines, and nutrients under normoxia.79-82 The failure to limit mTOR-dependent induction of HIF-1α may therefore contribute to age-related diseases (Fig. 1). Intriguingly, given that 2OG is a limiting co-substrate for PHD activity during normoxia, and that 2OG levels depend on amino acid availability, it is plausible that PHD activity depends not only on oxygen or oncometabolites, but also on amino acid availability, suggesting a global metabolic sensor function for PHDs as a signal for both HIFα and mTOR.83
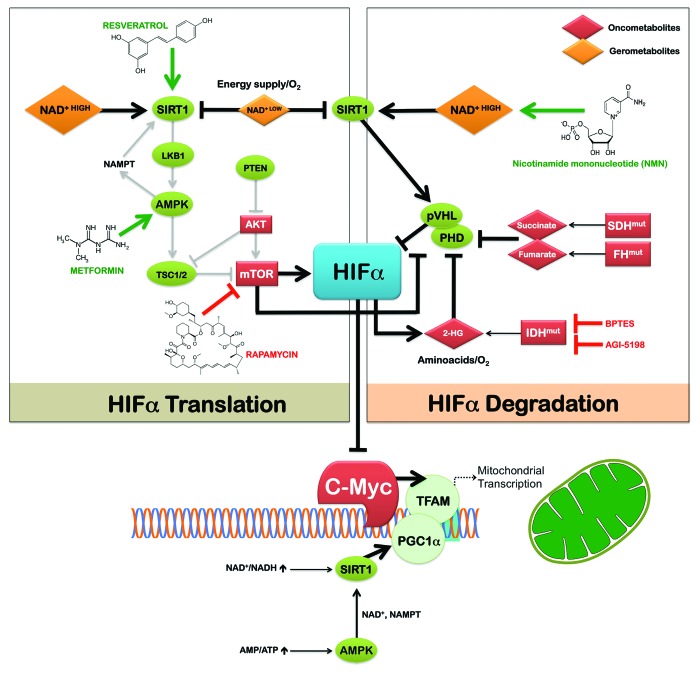
Figure 2. Gerometabolites and oncometabolites could control HIFα homeostasis by its rate of translation (left) and degradation (right). Proteins colored in green represent HIFα downregulators (e.g., gerosuppressors) whereas those in red represent HIFα inducers (e.g., gerogenes, oncometabolites). While it is obvious that the antifungal antibiotic rapamycin, the polyphenol resveratrol, and the biguanide metformin already belong to the family of gerosuppressor-targeting drugs, other SIRT1 activators such as NMN and oncometabolite inhibitors (e.g., BPTES, AGI-5198) should be viewed as drug candidates that could reverse or postpone the pseudohypoxia-switching hub driving aging and cancer.
It is readily apparent that by protecting adult cells from initiating premature cell senescence using pharmacological agents that simultaneously prevent oncogenesis, we might delay aging without the potential increase in cancer risk; this scenario is well-recognized for pharmacological inhibitors of mTOR activity, such as rapamycin. By similarly affecting the mTOR gerogenic pathway, calorie restriction (CR) is likely the most-recognized anti-aging intervention that contradicts the paradigm that increasing adult cell function to levels found in the young must be counterbalanced against the risk of higher incidences of cancer. New findings reported by David Sinclair’s group strongly suggest that not only long-term CR, but also acute increases in NAD+ levels and/or small compounds that prevent HIF-1α stabilization (e.g., NMN, a precursor to NAD+ that increases NAD+ levels in vivo) can rapidly prevent the decline in pVHL and accumulation of HIF-1α in old mice to restore mitochondrial function and notably reverse many biochemical aspects of aging to that of a young mouse in an SIRT1-dependent manner.23 Based on the proposed model, which integrates the interconnected signaling of an HIF-1α-driven pseudohypoxic aging/cancer signature by oncometabolites, gerometabolites, gerogenes, and gerosuppressors (Fig. 2), we can make several predictions:
First, the activation of metabolic gerosuppressors (e.g., the evolutionarily conserved energy-sensor AMP-activated protein kinase), which critically antagonize the mTOR gerogenic activity,60-71,84 might exert anti-aging effects by restoring nuclear–mitochondrial communication during aging via the gerometabolite NAD+. Indeed, AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. To date, we have evidence that AMPK activation enhances SIRT1 activity by increasing cellular NAD+ levels, resulting in the deacetylation and modulation of the activity of downstream SIRT1 targets that include the PGC-1α transcription factor.85-87 We know that, in response to metformin, SIRT1 can be activated through an AMPK-mediated increase in gene expression in nicotinamide phosphoribosyltransferase, the rate-limiting enzyme of the salvage pathway for NAD+.85,88,89 Moreover, metformin has been shown to inhibit the ability of insulin and IGF-1 to induce HIF-1α expression, and, accordingly, HIF-1α levels appear to decrease significantly in diabetic patients treated with metformin.90-92 Because NAD+-dependent SIRT1 can regulate mitochondria homeostasis independent of PGC-1α/β, and AMPK activity dictates the dominant process in response to the energetic state of the cell,23 further work is necessary to unambiguously elucidate whether gerosuppressant agents with AMPK agonistic activity (e.g., metformin) can elicit their anti-aging (and anti-cancer) effects by restoring nuclear–mitochondrial communication via the suppression of HIF-1α-driven, pseudo-hypoxic cellular states in a PGC-1α/β-independent manner (Fig. 2).
Second, if the pseudohypoxic state that disrupts PGC-1α/β-independent nuclear–mitochondrial communication contributes to the decline in mitochondrial function with age, a process that is apparently reversible, it would be relevant to study whether changes in the NAD+-SIRT1-HIFα-OXPHOS pathway similarly disrupt mitochondrial homeostasis in pre-malignant and cancer tissues. In addition, it would be important to determine whether increasing NAD+ (e.g., with NMT) is beneficial for cancer prevention and treatment, especially in tumors characterized by the aberrant accumulation of pathogenic oncometabolites (Fig. 2).
Third, the direct targeting of (R)-2HG production by inhibiting the conversion of glutamine to α-ketoglutarate using small-molecule inhibitors (e.g., bis-2-[5-phenyl-acetamido-1,2,4-thiadiazol-2-yl]ethyl sulfide)93 or selective inhibitors directed against the neomorphic forms of mutated IDH (e.g., AGI-5198)94,95 can significantly slow the growth rate of IDH mutant cancer cells (Fig. 2). This finding suggests that the “starvation” of IDH cells of α-ketoglutarate or direct blocking of the ability of the mutant enzyme to produce (R)-2HG may have therapeutic benefits. However, it is also possible that treatment with exogenous α-ketoglutarate may be beneficial due to a reduction in the competitive inhibitory effect of 2-HG on α-ketoglutarate-dependent enzymes. For example, the introduction of cell-permeating α-ketoglutarate derivatives has been shown to restore normal PHD activity, which targets HIF1α for ubiquitylation and proteasomal degradation.96 This restores normal, low levels of HIF1α in SDH-deficient cells, thus indicating that pharmacological elevation of intracellular α-ketoglutarate alleviates pseudohypoxia in tumor cells with mitochondrial dysfunction of OXPHOS enzymes. Moreover, because increased levels of intracellular α-ketoglutarate have a marked effect on the basal levels of HIF-1α protein (i.e., α-ketoglutarate may be a limiting factor for HIF-PHD activity under normoxia), it is possible that the mTOR-mediated increased translation of HIF-1α can be overcome if HIF-1α degradation can be accelerated by α-ketoglutarate. This scenario raises several new and challenging questions. Can the decline of gerometabolites such as NAD+ promote the chronic accumulation of oncometabolites in normal cells during aging? Is the sole activation of a Warburg-like metabolic reprogramming in normal tissues able to significantly increase the endogenous production of oncometabolites, even in the absence of mutations in IDHs or OXPHOS enzymes? If so, oncometabolites, as bona fide etiological determinants in cancer, might then pave the way for the initiation of carcinogenesis in a strictly metabolic-dependent manner (Fig. 3). Moreover, it is possible that therapeutic strategies that target oncometabolites might significantly impact the aging process.
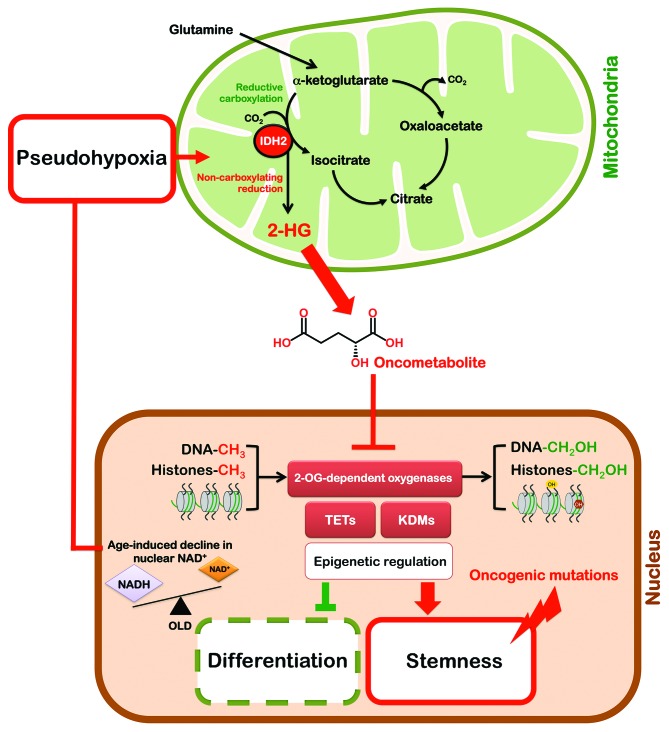
Figure 3. Gero-oncometabolite mitochondrial retrograde-like signaling: A new candidate signaling pathway linking aging with cancer. Mitochondrial retrograde signaling is a pathway of communication from mitochondria to the nucleus under normal and pathophysiological conditions. It involves multiple factors that sense and transmit mitochondrial signals to effect changes in nuclear gene expression; these changes lead to a reconfiguration of metabolism to accommodate cells to defects in mitochondria. Although during aging there is a specific loss of mitochondrial, but not nuclear, encoded OXPHOS subunits, the pseudohypoxic state that disrupts the PGC1-α/β-independent nuclear–mitochondrial communication can contributes not only to the decline in mitochondrial function with age, but can also mediate an undesirable trade-off between Warburg reprogramming and oncometabolites production in normal cells. Glutamine-derived α-ketoglutarate is reductively carboxylated by the NADPH-linked mitochondrial IDH2 to form isocitrate, which can then be isomerized to citrate. However, the increased IDH2-dependent carboxylation of glutamine-derived α-ketoglutarate in hypoxia has been associated with a concomitant increased synthesis of 2-hydroxyglutarate (2HG) in cells with wild-type IDH1 and IDH2. Moreover, reprogramming of metabolism by HIF1α in the absence of hypoxia is sufficient to induce reductive activity on α-ketoglutarate that is uncoupled from carboxylation, thus producing the oncometabolite 2HG. This hypothetical scenario offers a plausible connection between gerometabolites-induced pseudohypoxia and oncometabolites-induced block in differentiation to promote tumorigenesis, likely through epigenetic mechanisms.
Gerometabolites and Oncometabolites Share a Communication Link to the “Oncogene From Hell” (c-Myc)
The extremely powerful gene transcription activator c-Myc has long been thought to be a key, if not “the” key, protein target for the development of cell proliferation-inhibiting drugs.97 The “oncogene from hell”,98 Myc, is a critical link between altered cellular metabolism and the genesis of many human cancers.99-104 c-Myc regulates genes involved in the biogenesis of ribosomes and mitochondria as well as the regulation of glucose and glutamine metabolism but can also induce DNA damage, increase reactive oxygen species, and mitigate p53 function to consequently induce genome instability. Moreover, recent publications have revealed that the “evil arsenal” of Myc includes the coordination of the crosstalk between tumor and microenvironment as it engages a complex inflammatory response in the tumor stroma and induces angiogenesis.105,106 This c-Myc-dependent connection between a Warburg-like OXPHOS-to-glycolysis shift in cellular bioenergetics and tumor-initiating capacity is well illustrated in the metabolo–genetic processes accompanying the generation of induced pluripotent stem cells (iPSCs).107-109 The inclusion of the c-Myc oncogene in the cocktail of nuclear reprogramming factors OSKM (i.e., Oct4, Sox2, Klf4, and c-Myc) potentiates the pluripotent glycolytic behavior and the tumorigenic incidence of derived iPSCs; conversely, c-Myc removal decreases the tumorigenicity of iPSCs and facilitates OXPHOS-dependent lineage commitment and terminal differentiation. In cancer, metabolic reprogramming can be mediated by crosstalk between HIF-1α and c-Myc. Moreover, SIRT1 is known to directly regulate c-Myc transcriptional activity in cancer cells, either via the deacetylation of c-Myc or by binding c-Myc and promoting its association with Max.110-113 In this regard, it is intriguing that David Sinclair’s group now reveals for the first time that the nuclear ability of SIRT1/HIF-1α to inhibit mitochondrial OXPHOS genes functions by decreasing c-Myc-regulated transcription of the key mitochondrial transcription factor A (TFAM).23 TFAM is required for mitochondrial DNA replication, transcription, and maintenance, and low levels of TFAM appear to lead to first-phase OXPHOS dysfunction triggered by the aging-associated decline of NAD+.114,115 Although the transition to an irreversible phase 2 OXPHOS dysfunction remains to be characterized, this process might be related to increased ROS production. It is important to note that mitochondrial dysfunction due to TFAM downregulation been shown to enhance tumorigenesis via mitochondrial genome instability and the production of ROS.116
Despite the pervasive role of Myc in human cancer, its requirement for proliferation and maintenance of adult stem cell compartments has raised considerable skepticism regarding the therapeutic value of Myc, given the expected toxicity of Myc inhibition for healthy tissues. However, the development of the Myc-interfering molecule termed Omomyc, which binds c- and N-Myc, Max, and Miz-1 but does not bind Mad or certain HLH proteins, has demonstrated that it might cause edge-specific perturbations in the Myc interactome that destroy specific protein interactions of the Myc node while leaving others intact.117-123 This results in a “rewriting” of the Myc transcriptome in a manner that produces opposing effects on the 2 arms of Myc activity, namely transactivation and transrepression of gene transcription. Specifically, Omomyc prevents Myc binding to promoter E-boxes and the transactivation of target genes while allowing Myc to retain Miz-1-dependent binding to promoters and transrepression function. Remarkably, Omomyc has a significant effect on the expression of genes encoding metabolic enzymes, including those involved in glycolysis (e.g., phosphoglycerate kinase 1 and lactate dehydrogenase B) and those involved in the expression of oncometabolites such as IDH2. In this scenario, it would be of interest to determine how Omomyc not only impacts the ability of declining NAD+ to disrupt nuclear–mitochondrial communication with age, but also affects the malignant behavior of tumors with metabolic enzyme mutations that produce oncometabolites. Critically, Omomyc might be utilized in malignant scenarios in which the accumulation of the oncometabolite 2-HG appears in the absence of metabolic enzyme mutations. A recent, pioneering study by Terunuma et al.124 revealed that the oncometabolite 2-HG accumulates at high levels in a subset of tumors and human breast cancer cell lines.124 Importantly, the activation of the Myc pathway mechanistically phenocopied IDH mutations, which were absent in 2-HG-overexpressing breast carcinomas. Myc/2-HG-high tumors exhibited a distinct, increased DNA methylation pattern that was associated with a poor prognosis, a stem cell-like transcriptional signature, and a tendency to overexpress glutaminase, thus suggesting a functional relationship between glutamine and 2-HG metabolism in breast cancer.124 Because these findings clearly implicate 2-HG as a candidate breast cancer oncometabolite associated with Myc activation and poor prognosis, Myc/2-HG-high tumors become an ideal scenario in which to explore whether Myc-disrupting strategies (e.g., Omomyc) can impede the signaling between oncometabolites and pathways resulting in malignancy. Importantly, as is the case for anti-aging interventions, this unique therapeutic strategy may prevent selection for resistance.
Corollary
From an evolutionary perspective, it is reasonable to view the alterations in mitochondrial homeostasis driven by the gerometabolite NAD+ as a conserved response to changes in energy supplies and oxygen levels. Similarly, the natural ability of 2-HG in the absence of IDH mutations to significantly alter epigenetics might reflect an evolutionarily ancient status of certain metabolites to signal for elevated glutamine/glutamate metabolism and/or oxygen deficiency. For example, in human cells proliferating under hypoxia, α-ketoglutarate can accumulate and be metabolized through an enhanced reductive activity of wild-type IDH2 in the mitochondria, leading to 2-HG accumulation.125 Crucially, the constitutive activation of HIF-1α is sufficient to recapitulate IDH2-dependent carboxylation of glutamine-derived α-ketoglutarate and the associated, concomitant increased synthesis of 2-HG in cells with wild-type IDH1 and IDH2 even in normoxic conditions, thus offering a plausible connection between gerometabolites-induced pseudohypoxia and oncometabolites-induced epigenetic deregulation (Fig. 3). As mentioned above, a functional relationship between glutamine metabolism and 2-HG accumulation has also been identified in biologically aggressive, Myc-overexpressing breast carcinomas in the absence of IDH mutations.
We are now beginning to understand how the chronic alteration of cellular responses to gerometabolites and oncometabolites became conserved causes of aging and cancer, at least in part by aberrantly connecting a Warburg-like reprogramming of cellular bioenergetics with pseudohypoxia-related (epi)transcriptional circuitries. It appears that a yet-to-be-defined complex, interacting with steps of inhibition, disruption, or activation in metabolic pathways, is likely to be involved in the causal connection between gerometabolites (e.g., those having gerosuppressant activities similar to that of NAD+ or yet-to-be discovered metabolites with geropromoting activities) and aging as well as between oncometabolites and an enhanced potential for cell transformation to malignancy. This paradigmatic shift in the discussion of the links between cellular metabolism and aging diseases can be further fueled by the identification of well-characterized, cancer-associated mutations in genes encoding enzymes with significant roles in cellular metabolism and by adopting the view that aging and aging diseases such as cancer are governed not only by genetic and epigenetic controllers, but also by a pivotal regulatory role of metabolic reprogramming in cell fate decisions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was financially supported by the Ministerio de Ciencia e Innovación (SAF2012-38914), Plan Nacional de I+D+I, MICINN, Spain.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/28079
References
- 1.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–48. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–98. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–46. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 7.Menendez JA, Joven J, Cufí S, Corominas-Faja B, Oliveras-Ferraros C, Cuyàs E, Martin-Castillo B, López-Bonet E, Alarcón T, Vazquez-Martin A. The Warburg effect version 2.0: metabolic reprogramming of cancer stem cells. Cell Cycle. 2013;12:1166–79. doi: 10.4161/cc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frezza C, Pollard PJ, Gottlieb E. Inborn and acquired metabolic defects in cancer. J Mol Med (Berl) 2011;89:213–20. doi: 10.1007/s00109-011-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 2012;31:2491–8. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Soga T, Pollard PJ, Adam J. The emerging role of fumarate as an oncometabolite. Front Oncol. 2012;2:85. doi: 10.3389/fonc.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–6. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 16.Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–52. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Yang M, Pollard PJ. Succinate: a new epigenetic hacker. Cancer Cell. 2013;23:709–11. doi: 10.1016/j.ccr.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, Licht JD, Deberardinis RJ, Chandel NS. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell. 2013;51:236–48. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3:730–41. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 20.Krell D, Mulholland P, Frampton AE, Krell J, Stebbing J, Bardella C. IDH mutations in tumorigenesis and their potential role as novel therapeutic targets. Future Oncol. 2013;9:1923–35. doi: 10.2217/fon.13.143. [DOI] [PubMed] [Google Scholar]
- 21.Adam J, Yang M, Soga T, Pollard PJ. Rare insights into cancer biology. Oncogene. 2013;••• doi: 10.1038/onc.2013.222. [DOI] [PubMed] [Google Scholar]
- 22.Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. J Clin Invest. 2013;123:3652–8. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) Induces a Pseudohypoxic State Disrupting Nuclear–mitochondrial communication during Aging. Cell. 2013;155:1624–38. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu LE, Gomes AP, Sinclair DA. Geroncogenesis: Metabolic Changes during Aging as a Driver of Tumorigenesis. Cancer Cell. 2014;25:12–9. doi: 10.1016/j.ccr.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borodovsky A, Salmasi V, Turcan S, Fabius AW, Baia GS, Eberhart CG, Weingart JD, Gallia GL, Baylin SB, Chan TA, et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget. 2013;4:1737–47. doi: 10.18632/oncotarget.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4:1729–36. doi: 10.18632/oncotarget.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–8. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gimenez-Roqueplo AP, Favier J, Rustin P, Mourad JJ, Plouin PF, Corvol P, Rötig A, Jeunemaitre X. The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am J Hum Genet. 2001;69:1186–97. doi: 10.1086/324413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brière JJ, Favier J, Bénit P, El Ghouzzi V, Lorenzato A, Rabier D, Di Renzo MF, Gimenez-Roqueplo AP, Rustin P. Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum Mol Genet. 2005;14:3263–9. doi: 10.1093/hmg/ddi359. [DOI] [PubMed] [Google Scholar]
- 31.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, Kung AL, Sanso G, Powers JF, Tischler AS, et al. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:72–80. doi: 10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brière JJ, Favier J, Gimenez-Roqueplo AP, Rustin P. Tricarboxylic acid cycle dysfunction as a cause of human diseases and tumor formation. Am J Physiol Cell Physiol. 2006;291:C1114–20. doi: 10.1152/ajpcell.00216.2006. [DOI] [PubMed] [Google Scholar]
- 34.Dahia PL, Familial Pheochromocytoma Consortium Transcription association of VHL and SDH mutations link hypoxia and oxidoreductase signals in pheochromocytomas. Ann N Y Acad Sci. 2006;1073:208–20. doi: 10.1196/annals.1353.023. [DOI] [PubMed] [Google Scholar]
- 35.Hewitson KS, Liénard BM, McDonough MA, Clifton IJ, Butler D, Soares AS, Oldham NJ, McNeill LA, Schofield CJ. Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates. J Biol Chem. 2007;282:3293–301. doi: 10.1074/jbc.M608337200. [DOI] [PubMed] [Google Scholar]
- 36.Favier J, Brière JJ, Burnichon N, Rivière J, Vescovo L, Benit P, Giscos-Douriez I, De Reyniès A, Bertherat J, Badoual C, et al. The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4:e7094. doi: 10.1371/journal.pone.0007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 38.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–90. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 39.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 40.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–90. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merksamer PI, Liu Y, He W, Hirschey MD, Chen D, Verdin E. The sirtuins, oxidative stress and aging: an emerging link. Aging (Albany NY) 2013;5:144–50. doi: 10.18632/aging.100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012;8:287–96. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 45.Webster BR, Lu Z, Sack MN, Scott I. The role of sirtuins in modulating redox stressors. Free Radic Biol Med. 2012;52:281–90. doi: 10.1016/j.freeradbiomed.2011.10.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong L, Mostoslavsky R. Fine tuning our cellular factories: sirtuins in mitochondrial biology. Cell Metab. 2011;13:621–6. doi: 10.1016/j.cmet.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–71. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haase VH. The VHL tumor suppressor: master regulator of HIF. Curr Pharm Des. 2009;15:3895–903. doi: 10.2174/138161209789649394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khacho M, Lee S. Subcellular dynamics of the VHL tumor suppressor: on the move for HIF degradation. Future Oncol. 2009;5:85–95. doi: 10.2217/14796694.5.1.85. [DOI] [PubMed] [Google Scholar]
- 50.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–60. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raimundo N, Baysal BE, Shadel GS. Revisiting the TCA cycle: signaling to tumor formation. Trends Mol Med. 2011;17:641–9. doi: 10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullen AR, DeBerardinis RJ. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol Metab. 2012;23:552–9. doi: 10.1016/j.tem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williamson JR, Chang K, Frangos M, Hasan KS, Ido Y, Kawamura T, Nyengaard JR, van den Enden M, Kilo C, Tilton RG. Hyperglycemic pseudohypoxia and diabetic complications. Diabetes. 1993;42:801–13. doi: 10.2337/diab.42.6.801. [DOI] [PubMed] [Google Scholar]
- 54.Nyengaard JR, Ido Y, Kilo C, Williamson JR. Interactions between hyperglycemia and hypoxia: implications for diabetic retinopathy. Diabetes. 2004;53:2931–8. doi: 10.2337/diabetes.53.11.2931. [DOI] [PubMed] [Google Scholar]
- 55.Ptitsyn A, Hulver M, Cefalu W, York D, Smith SR. Unsupervised clustering of gene expression data points at hypoxia as possible trigger for metabolic syndrome. BMC Genomics. 2006;7:318. doi: 10.1186/1471-2164-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carabelli J, Burgueño AL, Rosselli MS, Gianotti TF, Lago NR, Pirola CJ, Sookoian S. High fat diet-induced liver steatosis promotes an increase in liver mitochondrial biogenesis in response to hypoxia. J Cell Mol Med. 2011;15:1329–38. doi: 10.1111/j.1582-4934.2010.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan J, Danzer C, Simka T, Ukropec J, Walter KM, Kumpf S, Mirtschink P, Ukropcova B, Gasperikova D, Pedrazzini T, et al. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–70. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iglesias-Bartolome R, Gutkind SJ. Exploiting the mTOR paradox for disease prevention. Oncotarget. 2012;3:1061–3. doi: 10.18632/oncotarget.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blagosklonny MV. Big mice die young but large animals live longer. Aging (Albany NY) 2013;5:227–33. doi: 10.18632/aging.100551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blagosklonny MV. Hormesis does not make sense except in the light of TOR-driven aging. Aging (Albany NY) 2011;3:1051–62. doi: 10.18632/aging.100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blagosklonny MV. Molecular damage in cancer: an argument for mTOR-driven aging. Aging (Albany NY) 2011;3:1130–41. doi: 10.18632/aging.100422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blagosklonny MV. NCI’s provocative questions on cancer: some answers to ignite discussion. Oncotarget. 2011;2:1352–67. doi: 10.18632/oncotarget.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blagosklonny MV. Cell cycle arrest is not yet senescence, which is not just cell cycle arrest: terminology for TOR-driven aging. Aging (Albany NY) 2012;4:159–65. doi: 10.18632/aging.100443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blagosklonny MV. Once again on rapamycin-induced insulin resistance and longevity: despite of or owing to. Aging (Albany NY) 2012;4:350–8. doi: 10.18632/aging.100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blagosklonny MV. Prospective treatment of age-related diseases by slowing down aging. Am J Pathol. 2012;181:1142–6. doi: 10.1016/j.ajpath.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 66.Blagosklonny MV. Rapalogs in cancer prevention: anti-aging or anticancer? Cancer Biol Ther. 2012;13:1349–54. doi: 10.4161/cbt.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leontieva OV, Paszkiewicz GM, Blagosklonny MV. Mechanistic or mammalian target of rapamycin (mTOR) may determine robustness in young male mice at the cost of accelerated aging. Aging (Albany NY) 2012;4:899–916. doi: 10.18632/aging.100528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blagosklonny MV. Common drugs and treatments for cancer and age-related diseases: revitalizing answers to NCI’s provocative questions. Oncotarget. 2012;3:1711–24. doi: 10.18632/oncotarget.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blagosklonny MV. MTOR-driven quasi-programmed aging as a disposable soma theory: blind watchmaker vs. intelligent designer. Cell Cycle. 2013;12:1842–7. doi: 10.4161/cc.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blagosklonny MV. Rapamycin extends life- and health span because it slows aging. Aging (Albany NY) 2013;5:592–8. doi: 10.18632/aging.100591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blagosklonny MV. Aging is not programmed: genetic pseudo-program is a shadow of developmental growth. Cell Cycle. 2013;12:3736–42. doi: 10.4161/cc.27188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Treins C, Giorgetti-Peraldi S, Murdaca J, Monthouël-Kartmann MN, Van Obberghen E. Regulation of hypoxia-inducible factor (HIF)-1 activity and expression of HIF hydroxylases in response to insulin-like growth factor I. Mol Endocrinol. 2005;19:1304–17. doi: 10.1210/me.2004-0239. [DOI] [PubMed] [Google Scholar]
- 73.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–31. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A. Akt1 activation can augment hypoxia-inducible factor-1alpha expression by increasing protein translation through a mammalian target of rapamycin-independent pathway. Mol Cancer Res. 2006;4:471–9. doi: 10.1158/1541-7786.MCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 75.Yuan G, Nanduri J, Khan S, Semenza GL, Prabhakar NR. Induction of HIF-1alpha expression by intermittent hypoxia: involvement of NADPH oxidase, Ca2+ signaling, prolyl hydroxylases, and mTOR. J Cell Physiol. 2008;217:674–85. doi: 10.1002/jcp.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harada H, Itasaka S, Kizaka-Kondoh S, Shibuya K, Morinibu A, Shinomiya K, Hiraoka M. The Akt/mTOR pathway assures the synthesis of HIF-1alpha protein in a glucose- and reoxygenation-dependent manner in irradiated tumors. J Biol Chem. 2009;284:5332–42. doi: 10.1074/jbc.M806653200. [DOI] [PubMed] [Google Scholar]
- 77.Sudhagar S, Sathya S, Lakshmi BS. Rapid non-genomic signalling by 17β-oestradiol through c-Src involves mTOR-dependent expression of HIF-1α in breast cancer cells. Br J Cancer. 2011;105:953–60. doi: 10.1038/bjc.2011.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agani F, Jiang BH. Oxygen-independent regulation of HIF-1: novel involvement of PI3K/AKT/mTOR pathway in cancer. Curr Cancer Drug Targets. 2013;13:245–51. doi: 10.2174/1568009611313030003. [DOI] [PubMed] [Google Scholar]
- 79.Demidenko ZN, Blagosklonny MV. The purpose of the HIF-1/PHD feedback loop: to limit mTOR-induced HIF-1α. Cell Cycle. 2011;10:1557–62. doi: 10.4161/cc.10.10.15789. [DOI] [PubMed] [Google Scholar]
- 80.Leontieva OV, Natarajan V, Demidenko ZN, Burdelya LG, Gudkov AV, Blagosklonny MV. Hypoxia suppresses conversion from proliferative arrest to cellular senescence. Proc Natl Acad Sci U S A. 2012;109:13314–8. doi: 10.1073/pnas.1205690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leontieva OV, Blagosklonny MV. Hypoxia and gerosuppression: the mTOR saga continues. Cell Cycle. 2012;11:3926–31. doi: 10.4161/cc.21908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blagosklonny MV. Hypoxia, MTOR and autophagy: converging on senescence or quiescence. Autophagy. 2013;9:260–2. doi: 10.4161/auto.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boulahbel H, Durán RV, Gottlieb E. Prolyl hydroxylases as regulators of cell metabolism. Biochem Soc Trans. 2009;37:291–4. doi: 10.1042/BST0370291. [DOI] [PubMed] [Google Scholar]
- 84.Menendez JA, Joven J, Aragonès G, Barrajón-Catalán E, Beltrán-Debón R, Borrás-Linares I, Camps J, Corominas-Faja B, Cufí S, Fernández-Arroyo S, et al. Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: a new family of gerosuppressant agents. Cell Cycle. 2013;12:555–78. doi: 10.4161/cc.23756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cantó C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–9. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Liang Y, Vanhoutte PM. SIRT1 and AMPK in regulating mammalian senescence: a critical review and a working model. FEBS Lett. 2011;585:986–94. doi: 10.1016/j.febslet.2010.11.047. [DOI] [PubMed] [Google Scholar]
- 88.Caton PW, Kieswich J, Yaqoob MM, Holness MJ, Sugden MC. Metformin opposes impaired AMPK and SIRT1 function and deleterious changes in core clock protein expression in white adipose tissue of genetically-obese db/db mice. Diabetes Obes Metab. 2011;13:1097–104. doi: 10.1111/j.1463-1326.2011.01466.x. [DOI] [PubMed] [Google Scholar]
- 89.Zheng Z, Chen H, Li J, Li T, Zheng B, Zheng Y, Jin H, He Y, Gu Q, Xu X. Sirtuin 1-mediated cellular metabolic memory of high glucose via the LKB1/AMPK/ROS pathway and therapeutic effects of metformin. Diabetes. 2012;61:217–28. doi: 10.2337/db11-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Treins C, Murdaca J, Van Obberghen E, Giorgetti-Peraldi S. AMPK activation inhibits the expression of HIF-1alpha induced by insulin and IGF-1. Biochem Biophys Res Commun. 2006;342:1197–202. doi: 10.1016/j.bbrc.2006.02.088. [DOI] [PubMed] [Google Scholar]
- 91.Takiyama Y, Harumi T, Watanabe J, Fujita Y, Honjo J, Shimizu N, Makino Y, Haneda M. Tubular injury in a rat model of type 2 diabetes is prevented by metformin: a possible role of HIF-1α expression and oxygen metabolism. Diabetes. 2011;60:981–92. doi: 10.2337/db10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ece H, Cigdem E, Yuksel K, Ahmet D, Hakan E, Oktay TM. Use of oral antidiabetic drugs (metformin and pioglitazone) in diabetic patients with breast cancer: how does it effect serum Hif-1 alpha and 8Ohdg levels? Asian Pac J Cancer Prev. 2012;13:5143–8. doi: 10.7314/APJCP.2012.13.10.5143. [DOI] [PubMed] [Google Scholar]
- 93.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–7. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim J, DeBerardinis RJ. Cancer. Silencing a metabolic oncogene. Science. 2013;340:558–9. doi: 10.1126/science.1238523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, Watson DG, Gottlieb E. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27:3282–9. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Watson J. Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol. 2013;3:120144. doi: 10.1098/rsob.120144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soucek L, Evan G. Myc-Is this the oncogene from Hell? Cancer Cell. 2002;1:406–8. doi: 10.1016/S1535-6108(02)00077-6. [DOI] [PubMed] [Google Scholar]
- 99.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 100.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–8. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 101.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 102.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 103.Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–4. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- 104.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–9. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 105.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25:907–16. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whitfield JR, Soucek L. Tumor microenvironment: becoming sick of Myc. Cell Mol Life Sci. 2012;69:931–4. doi: 10.1007/s00018-011-0860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Folmes CD, Martinez-Fernandez A, Faustino RS, Yamada S, Perez-Terzic C, Nelson TJ, Terzic A. Nuclear reprogramming with c-Myc potentiates glycolytic capacity of derived induced pluripotent stem cells. J Cardiovasc Transl Res. 2013;6:10–21. doi: 10.1007/s12265-012-9431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fang IM, Yang CM, Yang CH, Chiou SH, Chen MS. Transplantation of induced pluripotent stem cells without C-Myc attenuates retinal ischemia and reperfusion injury in rats. Exp Eye Res. 2013;113:49–59. doi: 10.1016/j.exer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 109.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–11. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mao B, Zhao G, Lv X, Chen HZ, Xue Z, Yang B, Liu DP, Liang CC. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol. 2011;43:1573–81. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 112.Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, Larsson LG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A. 2012;109:E187–96. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Menssen A, Hermeking H. c-MYC and SIRT1 locked in a vicious cycle. Oncotarget. 2012;3:112–3. doi: 10.18632/oncotarget.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mendelsohn AR, Larrick J. Partial reversal of skeletal muscle aging by restoration of normal NAD+ levels. Rejuvenation Res. 2014 doi: 10.1089/rej.2014.1546. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 116.Woo DK, Green PD, Santos JH, D’Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am J Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Soucek L, Jucker R, Panacchia L, Ricordy R, Tatò F, Nasi S. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res. 2002;62:3507–10. [PubMed] [Google Scholar]
- 118.Soucek L, Nasi S, Evan GI. Omomyc expression in skin prevents Myc-induced papillomatosis. Cell Death Differ. 2004;11:1038–45. doi: 10.1038/sj.cdd.4401443. [DOI] [PubMed] [Google Scholar]
- 119.Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006;66:4591–601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- 120.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–5. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS One. 2011;6:e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Soucek L, Whitfield JR, Sodir NM, Massó-Vallés D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–13. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Terunuma A, Putluri N, Mishra P, Mathé EA, Dorsey TH, Yi M, Wallace TA, Issaq HJ, Zhou M, Killian JK, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]