Abstract
The dynamin-related Eps15-homology domain-containing protein 2 (EHD2) is a membrane remodeling ATPase that regulates the dynamics of caveolae. Here, we established an electron paramagnetic resonance (EPR) approach to characterize structural features of membrane-bound EHD2. We show that residues at the tip of the helical domain can insert into the membrane and may create membrane curvature by a wedging mechanism. Using EPR and X-ray crystallography, we found that the N-terminus is folded into a hydrophobic pocket of the GTPase domain in solution and can be released into the membrane. Cryo electron microscopy demonstrated that the N-terminus is not essential for oligomerization of EHD2 into a membrane-anchored scaffold. Instead, we found a function of the N-terminus in regulating targeting and stable association of EHD2 to caveolae. Our data uncover an unexpected, membrane-induced regulatory switch in EHD2 and demonstrate the versatility of EPR to study structure and function of dynamin superfamily proteins.
Keywords: EHD ATPases, dynamin superfamily GTPases, X-ray crystallography, electron paramagnetic resonance (EPR), cryo electron microscopy, protein conformation, liposomes, caveolae
Introduction
Eps15-homology domain-containing proteins (EHDs) comprise a highly conserved dynamin-related ATPase family in eukaryotes with four members in mammals (EHD1-4) and one in C. elegans (called Rme-1) and Drosophila (Past-1) (Naslavsky et al., 2011). Studies in C. elegans identified a function of Rme-1 in mediating the exit of cargo proteins from the endocytic recycling compartment (ERC) to the plasma membrane (Grant et al., 2001), and a similar function was demonstrated for mammalian EHD1 (Lin et al., 2001; Caplan et al., 2002). Subsequently, mammalian EHDs were shown to be involved in a diverse set of membrane trafficking pathways, both emanating from the plasma membrane and internal membrane systems (Shao et al., 2002; Naslavsky et al., 2006; Lasiecka et al., 2010). We and others recently demonstrated that EHD2 specifically localizes to the neck of caveolae (Stoeber et al., 2012; Morén et al., 2012; Ludwig et al., 2013), which are cup-shaped invaginations of the plasma membrane enriched in the protein caveolin (Parton et al., 2013). EHD2 is not required for their formation, but stably associates with surface-connected caveolae and slows down their mobility within the plasma membrane (Stoeber et al., 2012; Morén et al., 2012).
EHDs are composed of an N-terminal extended GTPase domain (G domain), followed by a helical domain and a C-terminal Eps15-homology (EH) domain. The G domains of EHDs bind to adenine rather than guanine nucleotides (Lee et al., 2005a; Daumke et al., 2007). X-ray structure analysis showed that the G domains of EHD2 and dynamin are structurally related (Daumke et al., 2007). Similar to other dynamin superfamily members, EHDs can tubulate negatively charged liposomes and oligomerize in ring-like structures around them (Daumke et al., 2007; Pant et al., 2009). In the case of EHD2, this leads to a 10-fold increase of its intrinsic ATPase activity. However, the rate of stimulated ATPase activity is still two orders of magnitude lower compared to that of dynamin under similar conditions (Faelber et al., 2011), pointing to a different function or regulation of nucleotide hydrolysis in these two proteins.
G domains of EHD2 stably dimerize via a nucleotide-independent interface, which is not conserved in other dynamin superfamily proteins. A second interface in the G domain promotes nucleotide-dependent assembly in dynamin and septin superfamily proteins (Schwefel et al., 2010). Dimer assembly via this nucleotide-dependent interface may mediate oligomerization of EHD2 into rings (Daumke et al., 2007). Two helical domains protrude in parallel from the G domain dimer. Based on mutagenesis, we suggested that the tips of two adjacent helical domains form a composite membrane-binding surface involving hydrophobic and positively charged residues. The C-terminal EH domains interact with linear peptide motifs containing an Asn-Pro-Phe (NPF) motif (de Beer et al., 1998). In the EHD2 dimer, EH domains bind on top of the opposing G domains and may block the nucleotide-dependent assembly interface of the G domain. Upon EHD2 assembly, the EH domains were suggested to switch to a KPFxxxNPF containing loop in the G domain of the adjacent EHD2 dimer. The KPFxxxNPF motif also mediates direct interactions with caveolae and specific caveolar targeting (Daumke et al., 2007; Morén et al., 2012).
Despite recent progress in structural studies, our previous structural analysis did not reveal the conformational changes associated with membrane-binding of EHD2. These transitions are difficult to address with conventional X-ray crystallography, since liposomes cannot be included in protein crystals. Also with nuclear magnetic resonance, structures of EHD2 oligomers, due to their large size, cannot easily be resolved. To circumvent these problems, we used a combination of site-directed spin labeling (SDSL), electron paramagnetic resonance (EPR), X-ray crystallography, cryo electron microscopy (cryoEM) and cell biology. We found that residues at the tip of the helical domain directly insert into membranes. Furthermore, we identified a membrane-dependent N-terminal switch that regulates cellular targeting of EHD2.
Results
The tip of the helical domain is a primary membrane-binding site
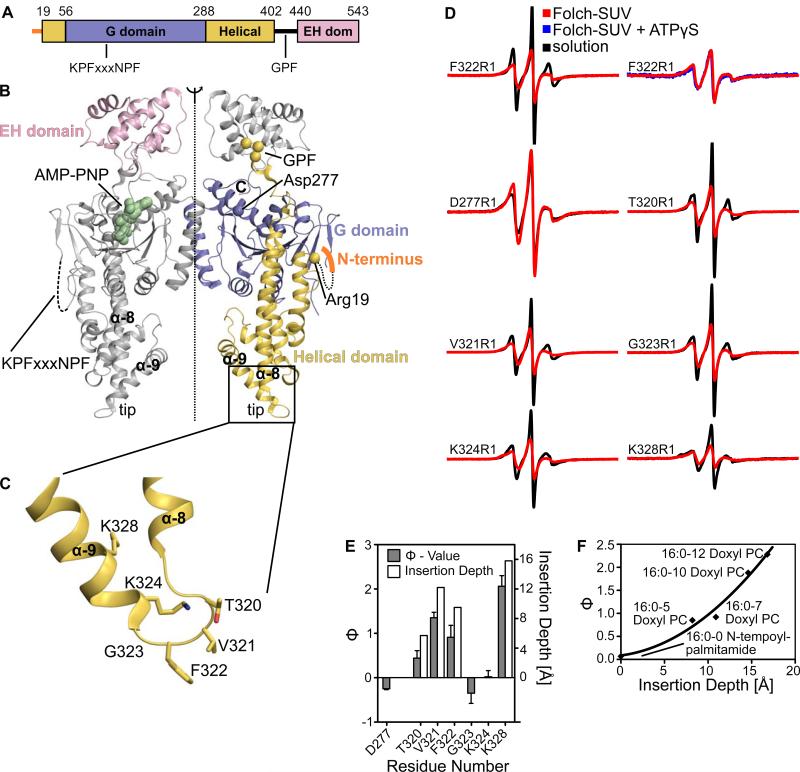
Mammalian EHDs share a sequence identity of 70 - 85% and display a common domain architecture (Figure 1A, B). Based on their location in the crystal structure and mutagenesis experiments, it has been inferred that residues at the tip of the helical domain (residues 320-340, Figure 1C) mediate membrane interaction (Daumke et al., 2007). Mutations in some of these residues reduce liposome binding and result in a cytoplasmic distribution of the protein when expressed in HeLa cells. To test whether this region recruits EHD2 to membranes via a direct membrane interaction, we established a site-directed spin labeling (SDSL) approach in combination with EPR spectroscopy. The three internal cysteines of EHD2 were mutated to serines. This cysteine-less EHD2 still bound and tubulated liposomes (data not shown). Subsequently, six residues in the helical domains were individually replaced with a single cysteine which was then coupled to the paramagnetic spin label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSL) to generate the new spin-labeled side chain R1.
Figure 1. Identification of the primary membrane-binding site by EPR.
A) Structure-based domain architecture of EHD2. Amino acid numbers at the domain boundaries and the location of two conserved peptide motifs are indicated.
B) Structure of the EHD2 dimer in the presence of a non-hydrolysable ATP analogue, AMP-PNP. The positions of the first visible amino acid in the helical domain (Arg19), a residue in the G domain (Asp277) and the KPFxxxNPF motif at the distal side of the G domain are indicated.
C) Magnification of the boxed area in Figure 1B showing details of the primary membrane-binding site of EHD2. Amino acids which have been modified by a spin-labelled side chain in this study are shown in stick representation.
D) Continuous wave EPR spectra of nucleotide-free EHD2 spin-labeled at residues 277, 320-324 and 328, in the absence (black) and presence (red) of Folch-SUVs. Spectra of residue 277 in the G domain of EHD2 showed no immobilization upon addition of Folch-SUVs, in contrast to the spectra of all other residues. Addition of ATPγS did not lead to further spectral changes of membrane-bound EHD2 F322R1 (blue).
E) The logarithmic ratio Φ of the accessibilities of spin-labels to the paramagnetic colliders O2 and NiEDDA was calculated for EHD2 labeled at positions 277, 320-324 and 328 in the absence of nucleotide and presence of Folch-SUVs (solid bars referring to the left Y-axis). Positive Φ values indicate membrane interaction. Based on calibrations with spin-labeled lipids, the membrane insertion depth of each residue was derived (open bars refer to the right Y-axis, see also Figure 1F).
F) Insertion depth calibration using power saturation experiments with Folch-SUVs containing 1% (w/w) of spin-labeled lipids mixed with nucleotide-free EHD2. For position 0, N-tempoylpalmitamide, for position 5, 7, 10 and 12, the respective 1-palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphocholines were used. Each position corresponds to an insertion depth taken from (Altenbach et al., 1994; Oh et al., 2010). The Φ–values could be fitted with a polynomial regression.
EHD2 can remodel liposomes into tubules and small unilamellar vesicles (SUVs). For our studies, we used pre-formed SUVs composed of bovine brain lipids (Folch) as a membrane-binding template for EHD2 for two reasons: First, EHD2 binds SUVs with high affinity, even in the absence of nucleotides (Daumke et al., 2007). Second, SUVs were stable over the time course of the experiments (~ 30 min) resulting in a reproducible membrane-bound conformation. In contrast, EHD2-decorated tubules generated from Folch large unilamellar vesicles (LUVs) were metastable and led to precipitation of EHD2 occluding further analysis.
First, we investigated the spectral changes of EHD2 spin labeled at position 322. Phe322 is directly located at the tip of the helical region and was previously proposed to interact with the membrane (Daumke et al., 2007). The continuous wave (CW) spectrum of F322R1 showed ordering upon addition of SUVs (Figure 1D). Addition of the non-hydrolyzable ATP analogue ATPγS did not induce further spectral changes (Figure 1D), in agreement with the nucleotide-independent interaction of EHD2 with SUVs. Next, we tested whether additional sites, which are further away from the tip region, also exhibit mobility changes. EHD2 derivatives containing single spin-labels at positions 320, 321, 323, 324 or 328 also revealed ordering upon addition of SUVs (Figure 1D). In contrast, no significant mobility changes upon addition of SUVs were observed in the spectra of an EHD2 variant labeled at residue 277, a site in the G domain not expected to interact with the membrane.
To identify residues directly inserting into the membrane, accessibilities of the spin labels to paramagnetic colliders O2 (partitions into the membrane) and NiEDDA (partitions into the aqueous phase) were measured. In the absence of membranes, the depth parameter Φ, defined as the logarithm of the ratio of accessibilities to O2 and NiEDDA, showed negative values for all EHD2 variants. This result was expected as negative Φ values (e.g. high accessibility to NiEDDA) are typically observed in solution containing 10 mM NiEDDA. In the presence of liposomes, the values of positions 320, 321, 322, 324 and 328 became positive due to enhanced O2 and reduced NiEDDA accessibilities (Figure 1E). Although all sites showed ordering with moderate reduction in mobility, none of the sites became severely immobilized by tight packing interaction. Thus, steric exclusion of NiEDDA, which can occur within protein cores (Isas et al., 2002), did not contribute to the positive Φ values. Together, these data indicated direct membrane insertion of the selected sites. Spin labels attached to position 323 and to position 277 in the G domain displayed negative values, and, thus, do not directly insert into the membrane.
Calibrating the Φ-values with spin labeled lipids revealed that the membrane insertion depth of the nitroxide radical was between 3 and 16 Å (Figures 1E and F), indicating that the entire tip region inserts into the outer leaflet of the membrane. These data support the notion that this region represents an important membrane-binding site; the membrane immersion depth suggests that this region may also be able to promote membrane curvature via a wedging mechanism (Campelo et al., 2008). Due to the essential nature of this region for membrane interaction and due to its important role in membrane curvature induction, we consider this region to be a primary membrane-binding site.
The N-terminus of EHD2 folds against the G domain
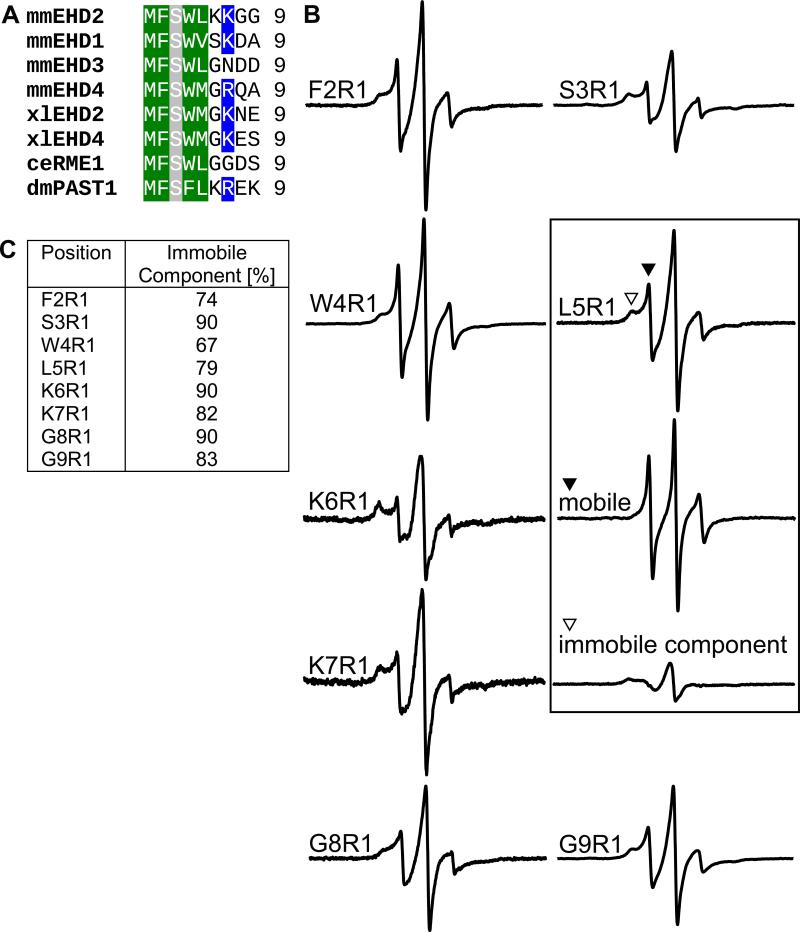
Our previous EHD2 model starts with residue 19. Residues 1-18 were not included since there was no connecting electron density before residue 19 (Figure 1B). Interestingly, the very N-terminal residues are highly conserved (Figure 2A) indicating a conserved functionality.
Figure 2. The N-terminus is in an ordered conformation in solution.
A) Sequence alignment of amino-terminal residues in the EHD family. Conserved hydrophobic residues are marked in green, positively charged residues in blue and serines in grey.
B) CW EPR measurements of nucleotide-free EHD2 spin-labeled at positions 2-9. As exemplified for L5R1 (boxed), all spectra can be separated into a small fraction of mobile component (filled triangle) and a predominant immobile component (open triangle).
C) Table listing the fraction of immobile spectral components of residue 2-9, based on CW EPR measurements from Figure 2B.
To study structural and functional properties of these residues, we used our newly established EPR spectroscopy approach with EHD2 variants singly spin labelled at residues 2-9. CW EPR spectra of these EHD2 variants in the absence of membranes and nucleotides showed predominantly immobile components indicating an ordered conformation (Figure 2B, open triangle). This was unexpected since the absence of the N-terminus in the X-ray derived model suggested a flexible conformation. Consistent with this notion, each site also revealed the presence of a small fraction of a highly mobile and disordered conformation (Figure 2B, closed triangle). Substitution of the hydrophobic amino acids Phe2, Trp4 and Leu5 with R1 resulted in larger percentages of mobile (Figure 2B) and smaller percentages of immobile components (Figure 2C) compared to the other N-terminal sites. Thus, the hydrophobic residues might contribute to the stabilization of the ordered conformation of the N-terminus, perhaps by interaction with a hydrophobic binding pocket.
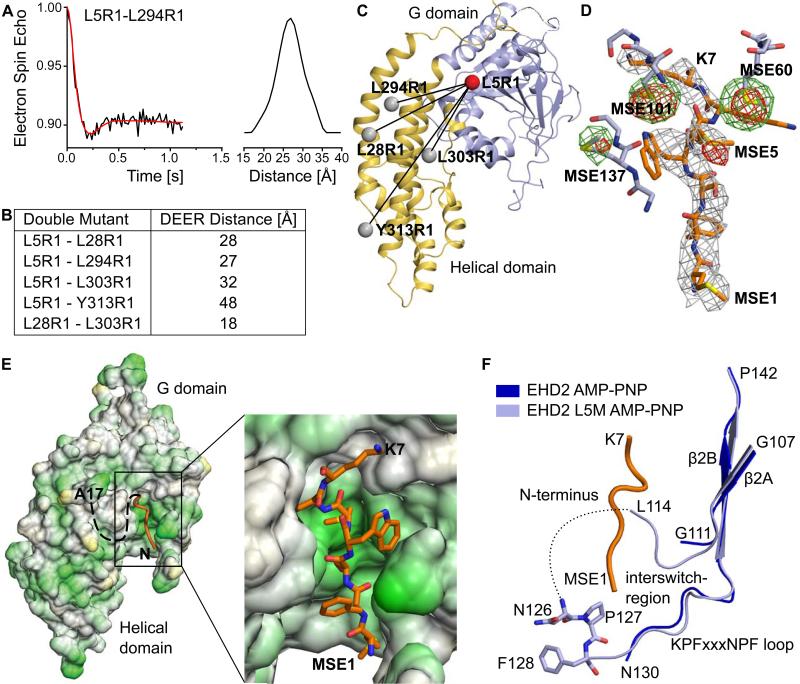
To locate the position of the N-terminus, double electron-electron resonance (DEER) distance measurements were performed using EHD2 variants spin labeled at position 5 in the N-terminus and an additional residue in the helical domain (Figures 3A and B, S1). Distances between residues 5-28, 5-294, 5-303 and 5-313 ranged from 27 Å to 48 Å (Figure 3B). As a control, the distance between residues 28 and 303 within the helical domain was determined to be 18 Å, which was consistent with the predicted distance derived from the crystal structure. The position of residue 5 was derived by a trilateration method (Figure 3C). This approach indicated that Leu5 is located in close proximity to the G domain.
Figure 3. The N-terminus binds in a hydrophobic pocket of the G domain.
A) Representative example of a distance measurement between Leu5R1 and Leu294R1, using pulsed EPR-measurements in the absence of nucleotides. See Figure S1 for other distance measurements.
B) Table listing distances between different pairs of spin labels, determined by DEER.
C) Based on the predicted position of the spin labels (grey balls) and four EPR distance pairs, the position of Leu5 (red ball) was determined to be close to the G domain.
D) The N-terminus (residues 1-7) was modeled in the 2Fo-Fc density, contoured at 1σ, derived from the AMP-PNP-bound EHD2 Leu5SeMet dataset. The anomalous signals (contoured at 4σ) derived from AMP-PNP-bound SeMet-labeled EHD2 Q410A (green) and EHD2 L5M Q410A (red) are superposed. Nearby residues from the G domain are shown as stick representations, without 2Fo-Fc density. An additional anomalous signal for SeMet5 is apparent.
E) Hydrophobic surface representation of the G and helical domain of EHD2. Green represents hydrophobic and yellow hydrophilic residues. The N-terminal sequence stretch (orange) binds into a hydrophobic pocket of the G domain. The boxed area is shown in more detail on the right.
F) Superposition showing the KPFxxxNPF loop from EHD2 Q410A (blue, pdb 2QPT) and EHD2 L5M Q410A (lightblue). The N-terminus is shown in orange. Seven previously unresolved residues in the KPFxxxNPF loop region were modeled in the refined electron density.
To accurately determine this position, Leu5 was mutated to methionine and a selenomethionine (SeMet)-derivatized EHD2 variant was crystallized in the presence of a non-hydrolyzable ATP analogue, adenylyl-imidodiphosphate (AMPPNP). SeMet5 gave rise to an additional signal in an anomalous difference Fourier map (Figure 3D, Table 1). Interestingly, the position of this signal was within a previously unexplained electron density patch in a conserved hydrophobic groove at the distal side of the G domain. Previously, the electron density could not be assigned to the EHD2 sequence, due to the limited resolution of the dataset, missing connectivity, only partial occupancy (as suggested by EPR) and the presence of two disordered loops in this region.
Table 1.
Data collection and refinement statistics
| mmEHD2 L5M Q410A SeMet | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 99.9, 134.7, 56.1 |
| α, β, γ (°) | 90.0, 106.1, 90.0 |
| Beamline | BESSY II MX-14.1 |
| Wavelength (Å) | 0.97969 |
| Total reflections* | 42,946 (4,095) |
| Unique reflections* | 26,426 (2,552) |
| Resolution (Å)* | 40–3.0 (3.1–3.0) |
| Robs (%)* | 4.7 (33.9) |
| Rmeas (%)* | 6.7 (47.8) |
| I/σI* | 10.5 (1.8) |
| Completeness (%)* | 95.9 (98.0) |
| Redundancy* | 1.6 (1.6) |
| Wilson B-factor (Å2) | 76 |
| Refinement | |
| Resolution (Å)* | 40–3.0 (3.078–3.0) |
| No. reflections* | 13,424 (989) |
| Rwork / Rfree (%)* | 23.0/27.9 (33.8/37.8) |
| Number of atoms | |
| Protein | 3,958 |
| Nucleotide | 31 |
| Metal Ions | 2 |
| Water | 4 |
| Averaged B-factor protein (Å2) | 84 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.027 |
Numbers in brackets represent values from the highest resolution shell.
Employing the refined phases, the improved resolution of this dataset and the additional SeMet signal, we assigned the N-terminal seven residues of EHD2 into this electron density (Figures 3D, E). As predicted by the DEER data, the N-terminus binds back to the G domain. Further in agreement with the EPR spectra, we found that the N-terminus packs into a mostly hydrophobic binding pocket (Figure 3E). In addition to the N-terminal residues, seven residues belonging to the spatially adjacent KPFxxxNPF loop were built in the refined electron density. Interestingly, the N-terminus is in close proximity to the NPF sequence of this loop and makes physical contact with amino acids that are flanking the loop region (Figure 3F). By bridging the KPFxxxNPF loop region to the G domain, the N-terminus may regulate the ability of the KPFxxxNPF region to interact with other binding partners (see below).
The N-terminus can insert into membranes, but is not essential for membrane-binding and oligomerization
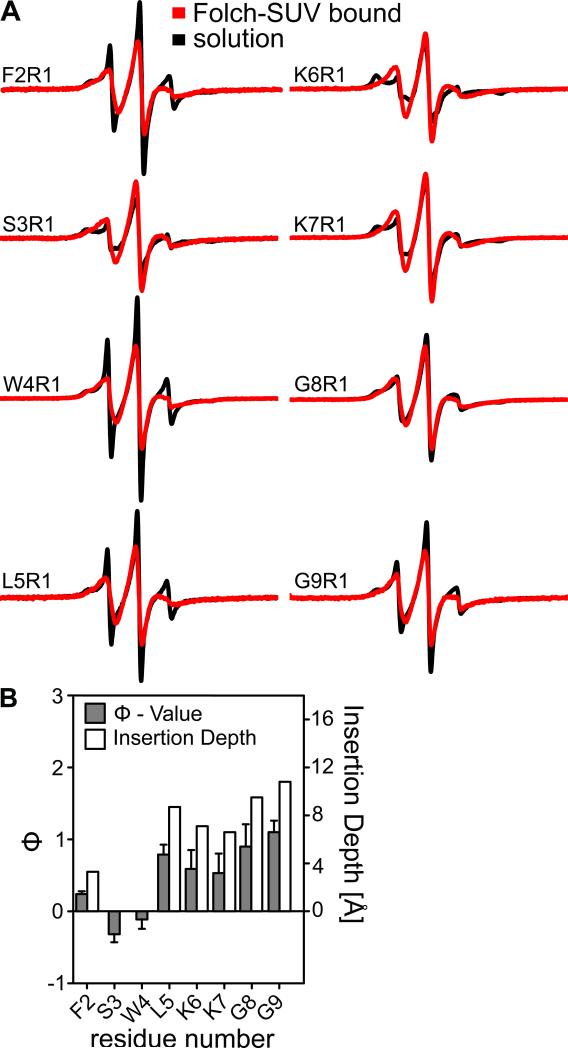
A recurring theme in membrane curvature inducing proteins is that N-terminal regions can undergo conformational reorganization and make contact with the membrane, for example in N-BAR proteins (Gallop et al., 2006), epsin (Boucrot et al., 2012) and Arf GTPases (Lee et al., 2005b). To test whether such a mechanism might also apply to EHD2, we used EPR spectroscopy of spin labeled EHD2 variants in the presence of membranes. Similar to residues in the primary membrane-binding site, N-terminally labeled EHD2 variants underwent spectral changes upon membrane-binding. N-terminal labeling sites (position 2-9) lost their highly mobile and highly immobile spectral components upon SUV binding, and the EPR spectra were instead dominated by spectral components with intermediate mobility (Figure 4A). This suggests an ordered, but not tightly packed conformation of this N-terminal sequence stretch in the presence of membranes. The lack of packing interactions also requires a release from the binding pocket in the G domain. Thus, the membrane-bound state of the N-terminus represents a third state that is ordered and distinctively different from the two states observed in solution. Spin labels attached at positions 2, 5, 6, 7, 8 and 9 showed positive Φ values upon liposome addition indicating membrane insertion (Figure 4B). However, spin labels attached to position 3 and 4 in the N-terminal membrane-binding site had negative values, indicating no membrane penetration. Based on our previous calibration (Figure 1F), we conclude that the N-terminus undergoes shallow insertion into the outer membrane monolayer.
Figure 4. The N-terminus can insert into membranes.
A) Continuous wave EPR spectra of nucleotide-free EHD2 spin-labeled at residue 2-9 in the absence (black) and presence (red) of Folch-SUVs.
B) Similarly as in Figure 1E, Φ and the insertion depth for EHD2 spin labeled at positions 2-9 was determined. At position 6 and 7, the spin label might insert deeper into the membrane compared to the corresponding lysines, which may snorkel in the membrane.
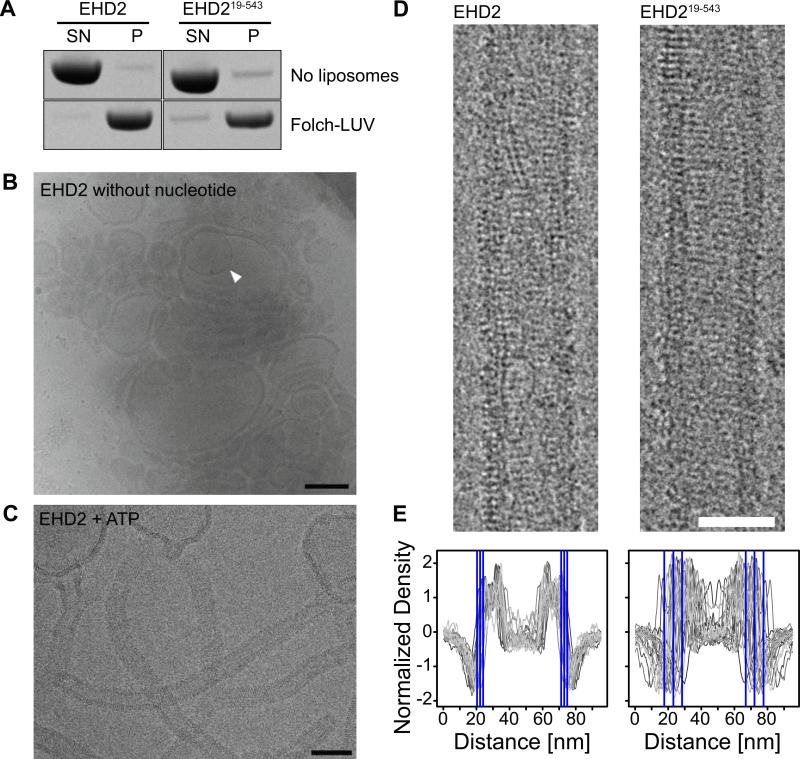
Although the N-terminus could bind to membranes, it was not essential for the interaction with liposomes. In liposome co-sedimentation assays, EHD2 bound with similar efficiency to LUVs as an EHD2 variant without the N-terminal 18 residues (EHD219-543) (Figure 5A). We therefore consider the N-terminal region as a secondary membrane-binding site that may modulate membrane interaction.
Figure 5. The N-terminus does not affect membrane-binding and oligomerization.
A) Co-sedimentation assays of EHD2 and EHD219-543 in the absence (upper panel) and presence (lower panel) of 800 nm filtered Folch-LUVs, without addition of nucleotides.
B, C) CryoEM of EHD2 in the presence of 800 nm Folch LUVs and absence of nucleotide (B) and in the presence of ATP (C). Inner vesicles of occasional multi-lamellar vesicles (B, white arrow) were shielded from the EHD2-containing buffer and showed typical bilayer structures, indicating that the surrounding liposomes were densely coated by EHD2. Similar to cryo electron micrographs of dynamin (Danino et al., 2004), we did not observe an accumulation of small vesicles, suggesting that EHD2 is not fragmenting liposomes under these conditions. Scale bar in B) 200 nm, in C) 100 nm.
D) CryoEM images of membrane tubules decorated with EHD2 and EHD219-543 were prepared by incubating EHD2 with Folch-LUVs in the presence of ATP. Regular patterns, most likely corresponding to ordered assemblies of the protein on the lipid tubule surface, were observed for both constructs. Scale bar is 50 nm.
E) 1D density profiles of membrane tubules decorated with EHD2 and EHD219-543. In case of EHD2, the average outer diameter of protein-coated tubules was similar (d=51±4 nm, n=2,156), as shown by the small standard deviation (SD). Deletion of the N-terminus had little effect on the average diameter of the tubules, but significantly increased the spread (d=49 ±11 nm, n=1,081). The blue lines in the 1D profile indicate the average outer limit of the tubes ± 1 SD.
To probe the tubulation potential of EHD2, cryoEM was employed. In the absence of ATP, EHD2 efficiently bound to Folch-LUVs. However, this resulted only in weak liposome tubulation (Figure 5B). Only in the presence of ATP, EHD2 significantly tubulated liposomes and formed a highly regular oligomeric EHD2 scaffold (Figures 5C and D). Quantification of the average tubule diameter indicated a narrow size distribution of EHD2-coated tubules (51 nm ± 3.8 nm) (Figure 5E).
In the presence of ATP, EHD219-543 also efficiently deformed liposomes into tubules of similar diameter, which were decorated by an ordered protein coat (Figure 5D). This indicates that the N-terminus of EHD2 is not required for membrane tubulation and oligomerization. However, the N-terminally truncated variant showed a broader size distribution of the tubule diameter (Figure 5E). This may point to a role of the N-terminus in the formation of defined oligomeric assemblies on membranes. Regardless of these subtle structural differences, a specific highly oligomeric protein coat could be observed in both cases supporting the notion that scaffolding is an important aspect of EHD2's ability to induce membrane curvature.
The N-terminal residues control the localization and stability of EHD2 oligomers in cells
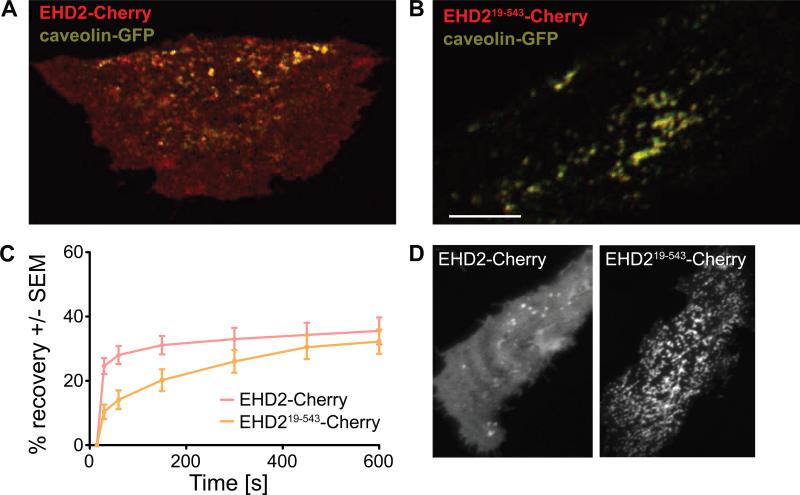
Having established that the N-terminus of EHD2 undergoes a major conformational change upon membrane interaction that is not essential for membrane binding and tubulation in vitro, we next asked for the functional significance of the N-terminus in living cells. We previously showed that EHD2 fused to an N-terminal EGFP-tag extensively co-localized with caveolae when overexpressed in 3T3-L1 fibroblasts (Morén et al., 2012). Due to the spatial proximity and its size, we suspected that an N-terminal tag might mask the functionality of the N-terminal residues. To overcome this problem, we over-expressed a C-terminally Cherry-tagged EHD2 construct (EHD2-Cherry) in 3T3-L1 cells. In agreement with previous data (Stoeber et al., 2012), this construct showed a more diffuse and cytoplasmic distribution than N-terminally tagged EHD2 and only partly associated with GFP-tagged caveolin (Figure 6A). These results indicate that N- and/or C-terminal tags influence membrane recruitment and/or oligomerization of EHD2. In contrast to EHD2-Cherry, an N-terminally truncated EHD2 variant (EHD219-543-Cherry) was found in big clusters and tubes positive for caveolin, with almost no cytoplasmic pool (Figure 6B). This suggested that EHD219-543-Cherry associates more stably with caveolin-positive structures than EHD2-Cherry. This was subsequently confirmed in fluorescence recovery after photobleaching (FRAP) experiments (Figure 6C).
Figure 6. The N-terminus of EHD2 directs caveolar targeting.
A, B) Fluorescent confocal micrographs of 3T3-L1 cells expressing EHD2-Cherry (A) or EHD219-543-Cherry (B) together with caveolin-GFP. Scale bar is 10 μm.
C) Quantification of FRAP microscopy of 3T3-L1 fibroblasts from (A) and (B), respectively. For this, distinct caveolae enriched in EHD2-Cherry and caveolin-GFP were bleached, and the time-dependent recovery of EHD2 fluorescence in this area was traced over 10 minutes. The graphs show recovery of fluorescence, as quantified from three independently bleached regions in 3-5 cells. Error bars represent standard error of the mean (SEM).
D) Fluorescent confocal micrographs of HeLa cells expressing EHD2-Cherry or EHD219-543-Cherry. Scale bar is 10 μm.
In HeLa cells, EGFP-EHD2 has been shown to tubulate membranes, probably due to the low abundance of caveolin in these cells and the tendency of EGFP-EHD2 to oligomerize at membrane surfaces (Morén et al., 2012). Here, we used HeLa cells to study the role of the N-terminus in membrane recruitment. Similarly as in 3T3-L1 fibroblasts, EHD2-Cherry localized to the cytoplasm and to discrete punctae representing caveolae (Figure 6D, left panel). In contrast, EHD219-543-Cherry was hardly found in the cytosol, but formed an interconnected tubular membrane network at the cell surface, indicative of more stable membrane association (Figure 6D, right panel). These data suggest that the N-terminus constitutes a switch region that controls membrane recruitment of EHD2.
Discussion
The large size, multi-domain nature and oligomerization of dynamin superfamily proteins make their structural and functional characterization challenging. In particular, structures of membrane-bound oligomerized forms of these proteins are difficult to characterize. To overcome these hurdles and elucidate structural features of membrane-bound EHD2, we used a combination of methods including EPR, X-ray crystallography, cryoEM and cellular imaging. This study represents the first application of EPR to a member of the dynamin superfamily, whose proteins are significantly larger than most other proteins typically studied by EPR. Our data suggest that analogous approaches can be applied for structural studies of other membrane-bound protein systems of similar complexity.
Our structural analysis of soluble EHD2 revealed a previously unknown three state nature of the N-terminal switch region. When EHD2 is in solution, the predominant conformation of this region is a highly ordered state that is stabilized by specific interactions with the G domain. In addition, the N-terminus can also exist in a highly dynamic, unfolded state in solution. Finally, a third conformational state with intermediate conformational flexibility was observed in the presence of membranes. This conformational flexibility of the N-terminus, as identified by a combination of EPR and X-ray crystallography, is likely a prerequisite for its function as a cellular regulatory switch. Intriguingly, point mutations in the N-terminal region of C. elegans Rme-1 prevent the exit of proteins from the endocytic recycling compartment showing the functional importance of this region (Grant et al., 2001).
To characterize the membrane-bound state of EHD2, we also performed cryoEM studies of EHD2-decorated membrane tubules. This approach resolved a highly defined and periodic coat of EHD2 proteins wrapped around the tubular membrane. These data support the previously proposed notion (Daumke et al., 2007) that EHD2 oligomers act as scaffolds that induce membrane curvature, similarly as reported for BAR proteins (Frost et al., 2008; Mim et al., 2012; Mizuno et al., 2010). It was previously found that residues at the tip of the helical domain are essential for membrane interaction in vitro and in vivo (Daumke et al., 2007). Interestingly, our EPR data demonstrate that these residues do not simply anchor the protein to the membrane but that a number of residues directly penetrate into the membrane. Based upon theoretical considerations (Campelo et al., 2008), this insertion may help to further promote membrane curvature through a wedging mechanism. The concerted use of scaffolding and wedging is not without precedent as amphipathic helices and scaffolding domains are thought to act together in the N-BAR proteins endophilin and amphiphysin (Gallop et al., 2006). Due to this importance of the tip of the helical domain in membrane binding and curvature induction, we consider this region to be a primary membrane-binding site.
The shallow insertion of the N-terminus seen by EPR depth measurements suggests that this region could also act as a wedge or a membrane curvature sensor. However, in contrast to the membrane insertion of the primary membrane-binding site, the N-terminal region is not essential for tubulation and membrane remodeling of liposomes. This makes it unlikely that the N-terminus plays an important wedging function. Instead, our cellular data indicate that the N-terminus plays a regulatory role in caveolar targeting. In the absence of the N-terminus, EHD2 forms an interconnected membranous network at the cell surface, which might result from altered properties of its recruitment, oligomerization or membrane remodeling activity. The physiological significance of this network, if any, is unclear.
What could be the mechanism for such functions? In the absence of membranes, the predominant, ordered form of the N-terminus is at a significant distance from the membrane-interacting tip of the helical domain. Nonetheless, our data indicate that the N-terminus can move away from its binding pocket in the G domain toward the membrane. This movement will disrupt the contact between the N-terminus and the KPFxxxNPF loop and its flanking region that is formed in solution. It has previously been suggested that oligomerization is promoted by an interaction of the EH domains from one dimer with the KPFxxxNPF loops of neighboring dimers (Daumke et al., 2007; Morén et al., 2012). Accordingly, deletions of the EH domain or mutations in the KPFxxxNPF motif reduce oligomerization of EHD2. We propose that a membrane-mediated movement of the N-terminus induces a conformation of the KPFxxxNPF loop that makes it competent to bind the neighboring EH domain. In this case, the N-terminus would act as a switch that senses membrane proximity and/or membrane curvature and its movement would help to promote oligomerization. Additionally, the N-terminus might directly interact with partner molecules at caveolae. Further experiments will be required to test this hypothesis.
The N-terminus of EHD2 has similarities but also some differences when compared to small G proteins of the Arf family (Lee et al., 2005b; Pucadyil et al., 2009; Lundmark et al., 2008; Krauss et al., 2008). It binds into a hydrophobic groove of the G domain in both cases. However, in small G proteins, GTP binding triggers the release of the N-terminal amphipathic helix into the membrane. This helix promotes membrane curvature, likely via a wedging mechanism. In contrast to small G proteins, our current analysis of EHD2 did not provide evidence for an interplay between nucleotide-binding and release of the N-terminus, and did not indicate a function as membrane wedge. Moreover, unlike in the N-terminal regions of membrane-bound epsin (Lai et al., 2012) and endophilin (Jao et al., 2010), the Φ-values of labeled N-terminal sites of EHD2 did not reveal a clear periodicity that would correspond to a stable α-helical structure. This finding may not be too surprising as the N-terminus of EHD2 contains two glycine residues at positions 8 and 9 that would likely destabilize an α-helix. Also PACSINs interact with the membrane via a wedge loop (Wang et al., 2009), a region also lacking distinctive secondary structure.
Similar to EHD2, most members of the dynamin superfamily interact with membranes via a primary membrane-binding motif located at the tip of their helical domains. This was shown, for example, for the pleckstrin homology domain of dynamin, and for other membrane-binding motifs in dynamin-like myxovirus resistance (Mx) protein A (von der Malsburg et al., 2011), bacterial dynamin-like protein (Low et al., 2009) and the membrane fusion GTPase atlastin (Liu et al., 2012). However, additional N-terminal membrane-binding sites might also be present in other dynamin superfamily members. For example, a long isoform of dynamin-like optic atrophy 1 (OPA1) has a predicted transmembrane helix at the N-terminus, which might act as a secondary membrane-binding site. Also Mx proteins, guanylate binding proteins and BDLP, but not dynamin, have long N-terminal extensions of unknown function, which may contribute to membrane interaction (Praefcke et al., 2004). Thus, we envisage that the mechanism of N-terminal membrane-inserting sequences is a common theme in dynamin superfamily proteins.
Experimental Procedures
Protein expression and purification
Full-length mouse EHD2 and all mutant constructs, including the SeMet substituted protein, were expressed as N-terminal His6-tag fusions in E. coli Rosetta 2 (DE3) (Novagen) and purified as described (Daumke et al., 2007). EGFP- and Cherry-tagged variants of EHD2 were cloned into pEGFP-C3 and pmCherry-N1 (Clontech), respectively. The mRFP- and EGFP-tagged caveolin1 plasmids were purchased from Addgene (# 14434 and # 27704).
Crystallization and structure determination
Crystallization trials by the sitting-drop vapor-diffusion method were performed at 4 °C using a Hydra-II pipetting robot (Thermo Scientific). 300 nL of SeMet-substituted mmEHD2 L5M Q410A at a concentration of 20 g/L in the presence of 2 mM AMPPNP (Jena Bioscience) and 4 mM MgCl2 were mixed with an equal volume of reservoir solution containing 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.5, 4% PEG3350, 5% 2-Methyl-2,4-pentanediol (MPD) and 20 mM MgCl2. Tetragonal crystals grew within 4 days. Crystals were cryo-protected in two steps by transferring them into buffer containing 50 mM MES, pH 6.5, 75 mM NaCl, 4 mM MgCl2, 5 mM AMPPNP and 14% or 27% MPD and flash-frozen in liquid nitrogen. Diffraction data from a single crystal were recorded at BL14.1 at BESSY II, and processed using the XDS program suite (Kabsch, 2010). Crystals were isomorphous to those previously obtained for the mmEHD2 Q410A mutant (PDB Code 2QPT) (Daumke et al., 2007). Experimental phases were calculated from single wavelength anomalous diffraction (SAD) using Sharp (Bricogne et al., 2003). Model building was done in COOT (Emsley et al., 2010). TLS and restrained refinement was carried out with Refmac5 (Murshudov et al., 2011). All residues were in the allowed regions of the Ramachandran-plot, as assessed by COOT. Figures were prepared with PyMol (Schrödinger, LLC) and sequences were aligned using ClustalW (Larkin et al., 2007).
Liposome preparation
Folch fraction I bovine brain lipids (Sigma-Aldrich) were dissolved in chloroform, dried under gentle argon stream and desiccated over night. EPR experiments were performed with small unilamellar vesicles (SUVs). SUVs at a final concentration of 20 g/L were obtained by hydrating 5 mg Folch lipids in 250 μL 20 mM HEPES, pH 7.5, 300 mM NaCl and tip-sonication until the solution turned clear. For the liposome co-sedimentation assay, LUVs at a final concentration of 4 g/L were obtained by hydrating 1 mg Folch lipids in 250 μL 10 mM HEPES, pH 7.5, 2.4 mM KCl, 1 mM EDTA and filtering eleven times through 800 nm pore-size polycarbonate membranes in a mini-extruder (Avanti Polar Lipids). For CryoEM, Folch-LUVs were prepared by hydration to a final concentration of 4 g/L in 10 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 2.5 mM DTT. The hydrated lipids were vigorously vortexed, then subjected to five freeze/thaw cycles and filtered 21 times through 800 nm pore-size polycarbonate membranes.
Spin labeling
Dithiothreitol (DTT) was removed from single or double cysteine mutants using PD-10 columns (GE Healthcare) equilibrated with 20 mM HEPES, pH 7.5, 300 mM NaCl. Five-time molar excess of spin label MTSL (Toronto Research Chemicals, Canada) was added and reacted with the cysteines at 4 °C overnight, resulting in residue R1. Excess spin label was removed using PD-10 columns and spin labeled proteins were flash-frozen and stored at −80 °C.
CW EPR experiments
CW EPR spectra of EHD2 in solution at a concentration of approximately 2 g/L were recorded on a Bruker X-band CW EPR spectrometer fitted with an ER4119HS resonator at 12.7 mW incident microwave power at room temperature. Liposome-bound samples were produced by incubating 50 μg of spin labeled protein with 1.5 mg Folch-SUV (1:30 w/w protein to lipid ratio) at room temperature (RT) for 20 min. SUV-bound EHD2 was separated from unbound EHD2 by centrifugation (152,800 g, 20 min, 22 °C). The pellet was resuspended in 20 mM HEPES, pH 7.5, 300 mM NaCl and spectra were recorded as described above. Accessibilities to O2 (from air, ΠOx) and 10 mM NiEDDA (ΠNiEDDA) were obtained from power saturation experiments using a Bruker ER4123D dielectric resonator. The depth parameter Φ was calculated from Φ = ln[ΠOx/ΠNiEDDA] (Altenbach et al., 1994). The membrane insertion depth was obtained by calibrating Φ using N-tempoylpalmitamide (gift from Kyoung Joon Oh) for position 0 and 1-palmitoyl-2-stearoyl-(n-DOXYL)-sn-glycero-3-phosphocholine (Avanti Polar Lipids) with spin labels attached at carbon 5, 7, 10 and 12 (Altenbach et al., 1994).
Pulse EPR and distance measurements
To obtain the distance between spin labels, four pulse DEER (Pannier et al., 2000) experiments were performed at 78 K on a Bruker Elexsys E580 X-band pulse EPR spectrometer fitted with a 3-mm split ring (MS-3) resonator, a continuous-flow cryostat (CF935, Oxford Instruments), and a temperature controller (ITC503S, Oxford Instruments). Samples of EHD2 Leu5R1 Leu28R1, EHD2 Leu5R1 L294R1, EHD2 Leu5R1 Leu303R1, EHD2 Leu5R1 Tyr313R1 and EHD2 Leu28R1 Leu303R1 were mixed with an equal amount of cysteine-less EHD2 to reduce background from intermolecular distances, cryo protected in 15 – 20% sucrose, flash frozen and measured. The data were fitted using Tikhonov regularization (Chiang et al., 2005) as implemented in DEERAnalysis 2011 (Jeschke et al., 2007).
Determining the position of residue 5 by EPR
Residue R1 can adopt several conformations. We predicted sterically and stereochemically allowed positions of R1 using PRONOX (Hatmal et al., 2012). The position of nitroxide radical was calculated by averaging the probability-weighted predicted positions. The unknown position of Leu5R1 was determined as the intersection of 4 spheres with the predicted spin label position as midpoints and the DEER distances as radii with a python script (see Supplementary Information).
Liposome co-sedimentation assay
10 μM EHD2 was incubated with 1 g/L Folch LUVs in 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM MgCl2 for 10 min at RT and centrifuged (10 min, 25 °C, 70,000 rpm, TLA100). Pellet and supernatant were analyzed by SDS-PAGE.
Cryo electron microscopy and diameter measurements
160 μM of mmEHD2 or mmEHD219-543 were equilibrated in 10 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 2.5 mM DTT either supplemented with 2.25 mM ATP (Carl Roth) or without added nucleotide for 5 min at RT, then mixed 1:1 (v/v) with Folch LUVs. The resulting mixture (1.125 mM ATP; 80 μM mmEHD2; 2 g/l Folch liposomes) was incubated for 15 min at RT before vitrification by flash freezing in liquid ethane on carbon-coated grids (Quantifoil) using a Vitrobot device (FEI). Images were automatically collected under minimal dose conditions using the Leginon system (Suloway et al., 2005) on a 120 kV Tecnai Spirit (FEI) transmission electron microscope equipped with an Eagle 2k CCD-camera (FEI) or on a Tecnai G2 Polara (FEI) microscope operating at 300 kV equipped with a TemCam-F416 4k CMOS-camera (TVIPS). Nominal magnifications were 42,000x and 39,000x, respectively. To determine diameters, individual straight and non-overlapping tubules were manually selected using e2helixboxer from EMAN2 (Tang et al., 2007). To account for the low persistence length, each tubule was segmented into overlapping square windows with a length of 95 Å. Each segment was then individually aligned parallel to the y-axis based on its layer lines and centered on the x-axis based on its 1D density profile. Both steps were performed iteratively using the vector from tubule selection as the starting point until convergence was reached. Final rotations and shifts were applied and manually verified before a combined 1D density profile along the x-axis was calculated for each selected tubule from its segments. All alignment steps were performed in SPARX using custom scripts (Hohn et al., 2007).
Cell Biology and microscopy
3T3-L1 and HeLa cells were grown in DMEM medium (GIBCO) supplemented with 10% fetal calf serum and transfected using Lipofectamine 2000 (life technologies) for transient protein expression. For immunofluorescence analysis, 3T3-L1 cells were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at RT, then washed and blocked in 5% goat serum with 0.05% saponin in PBS before staining with the appropriate antibodies in 1% goat serum, 0.05% saponin in PBS using standard protocols. For live cell confocal microscopy, cells were grown and transfected according to standard protocols on uncoated MatTek dishes, then placed in a temperature-controlled chamber at 37 °C with 95% air / 5% CO2 and 100% humidity (Okolab). Live cell imaging data were acquired using a fully motorized inverted microscope (Nikon A1 R Laser Scanning Confocal Microscope) using a 60x lens (Plan Apochromat VC Oil, Nikon) under control of the NIS-Elements Microscope Imaging Software. For FRAP experiments, the region of interest was photobleached for 10 s using a 488 nm laser. Single images were taken every 10 s before and after photobleaching, and recovery intensity was measured for a total of 10 min. Time points were analysed using NIS-Elements Microscope Imaging Software and Graphpad Prism.
Supplementary Material
Highlights.
Establishment of an EPR approach to study membrane interaction of EHD2
The helical domain of EHD2 is inserting into the membrane
The N-terminus of EHD2 can switch from the G domain into the membrane
The N-terminus regulates caveolar targeting of EHD2
Acknowledgments
This project was supported by grants of the Deutsche Forschungsgemeinschaft (SFB740 and SFB958) and a Career Development Fellowship of “The International Human Frontier Science Program Organization” to OD, by a grant of the Leibnitz Graduate School to CS, by NIH grant GM063915 to RaL and the Swedish Medical Research Council and Swedish Foundation for Strategic Research to RiL. We are grateful for technical assistance by S. Werner, for help with electron microscopy data collection by J. Bürger and with X-ray data collection by the BESSY staff.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The Protein Data Bank accession number for the crystal structure of EHD2 L5M Q410A bound to AMP-PNP is 4CID.
Supplemental Information
Supplemental Information includes one figure and one Python script and can be found with this article online at xxx.
References
- Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon HT, Kozlov MM. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J Struct Biol. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- de Beer T, Carter RE, Lobel-Rice KE, Sorkin A, Overduin M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science. 1998;281:1357–1360. doi: 10.1126/science.281.5381.1357. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, Camilli PD, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Hatmal MM, Li Y, Hegde BG, Hegde PB, Jao CC, Langen R, Haworth IS. Computer modeling of nitroxide spin labels on proteins. Biopolymers. 2012;97:35–44. doi: 10.1002/bip.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct. Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Isas JM, Langen R, Haigler HT, Hubbell WL. Structure and dynamics of a helical hairpin and loop region in annexin 12: a site-directed spin labeling study. Biochemistry (Mosc) 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- Jao CC, Hegde BG, Gallop JL, Hegde PB, McMahon HT, Haworth IS, Langen R. Roles of Amphipathic Helices and the Bin/Amphiphysin/Rvs (BAR) Domain of Endophilin in Membrane Curvature Generation. J Biol Chem. 2010;285:20164–20170. doi: 10.1074/jbc.M110.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, Jia JY, Roux A, Beck R, Wieland FT, De CP, Haucke V. Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J Biol Chem. 2008;283:27717–27723. doi: 10.1074/jbc.M804528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CL, Jao CC, Lyman E, Gallop JL, Peter BJ, McMahon HT, Langen R, Voth GA. Membrane binding and self-association of the epsin N-terminal homology domain. J Mol Biol. 2012;423:800–817. doi: 10.1016/j.jmb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lasiecka ZM, Yap CC, Caplan S, Winckler B. Neuronal early endosomes require EHD1 for L1/NgCAM trafficking. J. Neurosci. 2010;30:16485–16497. doi: 10.1523/JNEUROSCI.3127-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Zhao X, Scarselletta S, Schweinsberg PJ, Eisenberg E, Grant BD, Greene LE. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J Biol Chem. 2005a;280:17213–17220. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005b;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc Natl Acad Sci U S A. 2012;109:E2146–E2154. doi: 10.1073/pnas.1208385109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Löwe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Howard G, Mendoza-Topaz C, Deerinck T, Mackey M, Sandin S, Ellisman MH, Nichols BJ. Molecular composition and ultrastructure of the caveolar coat complex. PLoS. Biol. 2013;11:e1001640. doi: 10.1371/journal.pbio.1001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Doherty GJ, Vallis Y, Peter BJ, McMahon HT. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J. 2008;414:189–194. doi: 10.1042/BJ20081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Jao CC, Langen R, Steven AC. Multiple modes of endophilin-mediated conversion of lipid vesicles into coated tubes: implications for synaptic endocytosis. J. Biol. Chem. 2010;285:23351–23358. doi: 10.1074/jbc.M110.143776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morén B, Shah C, Howes MT, Schieber NL, McMahon HT, Parton RG, Daumke O, Lundmark R. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol. Biol. Cell. 2012;23:1316–1329. doi: 10.1091/mbc.E11-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S. EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 2011;21:122–131. doi: 10.1016/j.tcb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KJ, Singh P, Lee K, Foss K, Lee S, Park M, Lee S, Aluvila S, Park M, Singh P, Kim RS, Symersky J, Walters DE. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J Biol Chem. 2010;285:28924–28937. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- Praefcke GJK, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 2009;325:1217–1220. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwefel D, Fröhlich C, Eichhorst J, Wiesner B, Behlke J, Aravind L, Daumke O. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2). Proc Natl Acad Sci U S A. 2010;107:20299–20304. doi: 10.1073/pnas.1010322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Akmentin W, Toledo-Aral JJ, Rosenbaum J, Valdez G, Cabot JB, Hilbush BS, Halegoua S. Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J Cell Biol. 2002;157:679–691. doi: 10.1083/jcb.200201063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber M, Stoeck IK, Hanni C, Bleck CK, Balistreri G, Helenius A. Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. EMBO J. 2012;31:2350–2364. doi: 10.1038/emboj.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- von der Malsburg A, Abutbul-Ionita I, Haller O, Kochs G, Danino D. Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop. J Biol Chem. 2011;286:37858–37865. doi: 10.1074/jbc.M111.249037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MVAS, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci U S A. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.