Abstract
Influenza viruses are global pathogens that infect approximately 10–20% of the world’s population each year. Vaccines, including the live attenuated influenza vaccine (LAIV), are the best defense against influenza infections. The LAIV is a novel vaccine that actively replicates in the human nasal epithelium and elicits both mucosal and systemic protective immune responses. The differences in replication and innate immune responses following infection of human nasal epithelium with influenza seasonal wild type (WT) and LAIV viruses remain unknown. Using a model of primary differentiated human nasal epithelial cell (hNECs) cultures, we compared influenza WT and antigenically-matched cold adapted (CA) LAIV virus replication and the subsequent innate immune response including host cellular pattern recognition protein expression, host innate immune gene expression, secreted pro-inflammatory cytokine production, and intracellular viral RNA levels. Growth curves comparing virus replication between WT and LAIV strains revealed significantly less infectious virus production during LAIV compared with WT infection. Despite this disparity in infectious virus production the LAIV strains elicited a more robust innate immune response with increased expression of RIG-I, TLR-3, IFNβ, STAT-1, IRF-7, MxA, and IP-10. There were no differences in cytotoxicity between hNEC cultures infected with WT and LAIV strains as measured by basolateral levels of LDH. Elevated levels of intracellular viral RNA during LAIV as compared with WT virus infection of hNEC cultures at 33°C may explain the augmented innate immune response via the up-regulation of pattern recognition receptors and down-stream type I IFN expression. Taken together our results suggest that the decreased replication of LAIV strains in human nasal epithelial cells is associated with a robust innate immune response that differs from infection with seasonal influenza viruses, limits LAIV shedding and plays a role in the silent clinical phenotype seen in human LAIV inoculation.
Keywords: Influenza, live attenuated influenza vaccine (LAIV), Human nasal epithelial cells, innate immune response
INTRODUCTION
Influenza viruses are global pathogens that infect approximately 10–20% of the world’s population each year contributing to an excess in morbidity and mortality (1). Annual immunization, including the Live Attenuated Influenza Vaccine (LAIV) and the trivalent inactivated influenza vaccine (TIV), remain the world’s best defense against seasonal influenza.
LAIV strains are nasal vaccines that actively replicate in the human nasal epithelium and elicit a protective mucosal immune response without causing clinical disease. The initial LAIV was originally developed by cold passage of a strain of influenza (H2N2; A/Ann Arbor/6/60) in a primary chicken kidney cell line. The virus isolated from these conditions exhibited three novel phenotypes including the ability to replicate at cooler temperatures (cold adapted; CA), restricted replication at temperatures above 39°C (temperature sensitive; TS), and lack of clinical disease in animal models and human experiments (attenuation; att) (2, 3). Previous studies, comparing cold adapted LAIV strains and antigenically matched influenza viruses, in non-human and immortalized cell lines have identified mutations in the PB1, PB2, M, PA, and NP segments of the LAIV which have been associated with each of these phenotypes. While specific mutations have been associated with decreased replication or attenuation in animal models the actual mechanisms, including how the live attenuated influenza vaccine is able to elicit a protective immune response despite lower replication at the true site of infection – the human nasal epithelium remain unknown.
Human nasal epithelial cells are of particular importance in influenza virus and vaccine infection as they are not only the principle cells infected but are also key cells responsible for initiating an effective yet controlled immune response (4, 5). Influenza virus infection of nasal epithelial cells induces an innate antiviral response in addition to the recruitment of adaptive immune cells. An initial epithelial cell-specific innate immune response is elicited through the interaction of host cell pattern recognition receptors (PRRs; including TLR3, TLR7, and RIG-I) and viral RNA. The recognition of viral RNA by PRRs result in the production of Type I IFN that lead to induction of down stream signaling molecules such as STAT-I as well as antiviral interferon stimulated genes including myxovirus resistance gene A (MxA) (5). Together these innate immune mediators act to limit virus replication. Additionally, engagement of PRRs by influenza viral RNA also trigger the production and release of the neutrophil chemokine IL-8 and IP-10 which recruit Th1 activated T cells, NK cells, and dendritic cells into the airway (6). It is clear that the innate immune response directed by the nasal epithelium following influenza virus or vaccine infection plays a key role in limiting early virus replication and orchestrating an effective adaptive immune response.
In this study we characterize the differences in virus replication and the innate immune response to infection with a WT seasonal influenza virus and an antigenically-matched cold adapted (CA) LAIV strain in the principal cells infected by influenza viruses, human nasal epithelial cells, in order to begin to understand how LAIV viruses are able to replicate in the human nasal epithelium yet not cause clinical disease.
MATERIAL AND METHODS
Human Nasal Epithelial Cell Procurement and Culture
Primary differentiated human nasal epithelial cells (hNECs) were obtained and cultured as previously described (7–11). Briefly, nasal epithelial cells were obtained by gently stroking the inferior superficial surface of the inferior turbinate several times with a Rhino-Probe curette (Arlington Scientific, Arlington TX), which was inserted through a nasoscope. hNECs were obtained from healthy, non-smoking, volunteers (n=7) aged 18–55 years who identified themselves as healthy without a diagnosis of asthma or smoking related disorder. This protocol was approved by the Institutional Review Board for Biomedical Research of the University of North Carolina at Chapel Hill School of Medicine (IRB# 09-0716).
Primary hNECs were expanded to passage 2 in bronchial epithelial growth medium (Lonza, Walkersville, MD), plated on collagen-coated filter supports with a 0.4um pore size (Corning, Tewksbury, MA). Upon confluence, air-liquid-interface (ALI) conditions were created to promote differentiation. Muco-ciliary differentiation was achieved 21–28 days after ALI.
Viruses
Wild-type viruses were propagated on MDCK cells in Dulbecco’s Modified Eagle Medium (DMEM, Sigma, St. Louis, MO) containing 4ug/ml N-acetyltrypsin (Sigma), 100U/ml of penicillin, 100ug/ml of streptomycin, and 0.3% bovine serum albumin (BSA; Sigma). The viruses used in this study included a seasonal Wild Type (WT) virus, A/California/10/78 (H1N1), and an antigenically matched cold-adapted (CA) LAIV strain, A/California/10/78 CR37 CL1, which were kindly provided by K. Subbarao at the NIAID/NIH.
Virus Infections
High-multiplicity of infection (MOI) growth curves were generated by infecting hNEC cultures in 12-well plates at an estimated MOI of 10 TCID50/ml per cell. Inoculums were calculated based on known concentration of stock viruses to achieve an estimated MOI of 10 and then confirmed by back-titer in parallel with apical samples at designated time points. Inoculums and stock virus were also tittered by hemagglutination assay (data not shown). Viruses were diluted in DMEM (Sigma) containing penicillin, streptomycin, and 0.3% BSA on the apical surface for 1 hour at room temperature. The inoculum was then aspirated and the apical and basolateral chambers were washed twice with PBS. Basolateral media composed of equal proportions BEGM (Lonza) and DMEM (Gibco, Grand Island, NY) was added and the cultures were incubated at 33°C. Viral infections of hNEC cultures were performed at 33°C to faithfully represent the temperature of the human nasal epithelium (12, 13). At the indicated times, basolateral supernatants (for cytokine detection) were collected, apical chambers were washed with 200ul of DMEM containing penicillin, streptomycin, and 0.3% BSA (apical supernatant), and both were stored at −80°C until analysis.
Virus Titers
Infectious virus titers, at the indicated time points, were measured using a TCID50/ml assay as previously described using Madin Darby Canine Kidney Cells (MDCK) at 33°C (14).
Cytokine Quantification
Bi-directional hNEC secretion of cytokines was measured by sampling apical and basolateral chambers separately. Apical and basolateral supernatants were analyzed for IL-8 and IP-10 using commercially available ELISA kits (BD OptEIA, Biosciences, San Diego, CA). Lower limits of detection were as follows: IL-8 = 3.1 pg/ml and IP-10 = 7.8 pg/ml. Cytokines were assessed at 24 hours post infection in order to ensure that the innate immune response being measured was that to virus replication rather than inoculum.
Real-time Quantitative RT-PCR
Total RNA was isolated from hNEC cultures using TRizol (Invitrogen, Grand Island, NY) according to manufacturer’s instructions. cDNA synthesis and real-time quantitative PCR (qPCR) were performed as previously described (15–17). Differences in gene expression results were determined with the ΔΔCt method and normalized first to β-actin then to mock-infected cultures and presented as fold change in gene expression. Cell associated influenza viral mRNA was determined by qRT-PCR using primers specific for the influenza A virus M segment in paired samples. Differences in influenza A virus M segment detected were determined with the ΔΔCt method and normalized to a WT sample, since no RNA was detected in mock-infected samples.
Statistical Analysis
Experiments conducted in hNEC cultures from 7 healthy donors were paired for analysis. Replication kinetics for wild-type seasonal viruses and antigenically-matched cold-adapted vaccine strains were compared using two-way analysis of variance (ANOVA) with repeated measures. Virus titers, fold change of innate immune gene and fold change of chemokine expression at 24 hours post infection were compared using non-parametric, paired t tests (Wilcoxon signed rank test). All statistical analyses were done using Prism, version 5.0 (GraphPad Software Inc.). *p<.05; ***p<.001.
RESULTS
Live Attenuated Influenza Vaccine (LAIV) strains exhibit marked restriction in replication during infection of human Nasal Epithelial (hNECs) Cultures
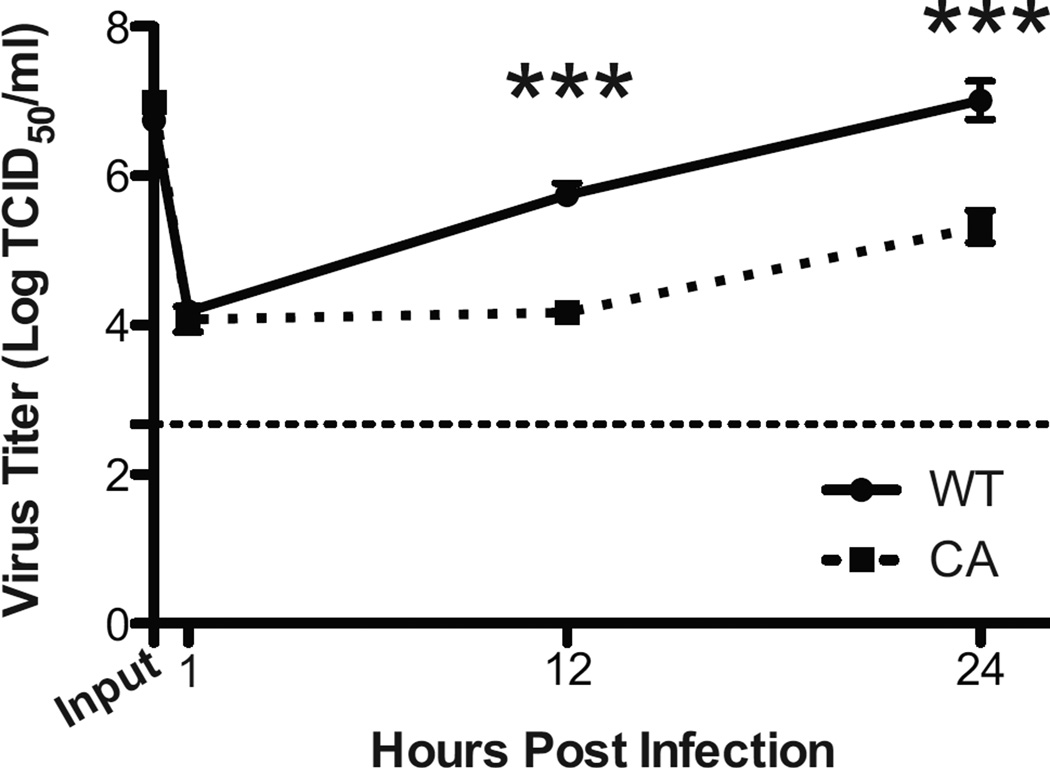
Live Attenuated Influenza Vaccine strains are cold-adapted viruses that elicit a protective mucosal immune response without causing clinical disease in contrast to wild-type (WT) seasonal viruses that result in varying phenotypes of upper and lower respiratory tract disease. In order to better understand how infection of human nasal epithelial cells with LAIV strains differ from infection with WT viruses at the true site of infection – the human nasal epithelium, we utilized hNEC cultures obtained from healthy non-smoking volunteers to compare both virus replication and the epithelial cell innate immune response to infection. At 33°C, LAIV viruses exhibited significantly reduced replication in hNEC cultures (Figure 1) with the WT viruses producing approximately 165 fold more infectious virus than the antigenically-matched LAIV strain at 24 hours post infection (HPI).
Figure 1.
Attenuation of Infectious LAIV Virus Production during Infection of hNEC cultures at 33°C. hNEC cultures were infected with wild type (WT) and an antigenically-matched cold adapted (CA) LAIV strain at an MOI of 10 in triplicate at 33°C. Virus titers in apical supernatants were measured by calculating the TCID50/ml in MDCK cells at the indicated hours post infection (HPI). The inoculum (labeled input) was back tittered in parallel. The limit of detection is shown by the horizontal thin dotted line. Data are expressed as mean + SEM. *** p<.001 (n=7 different donors for hNECs).
Despite a decrease in infectious virus production, LAIV infection of hNEC cultures elicits an enhanced innate immune response
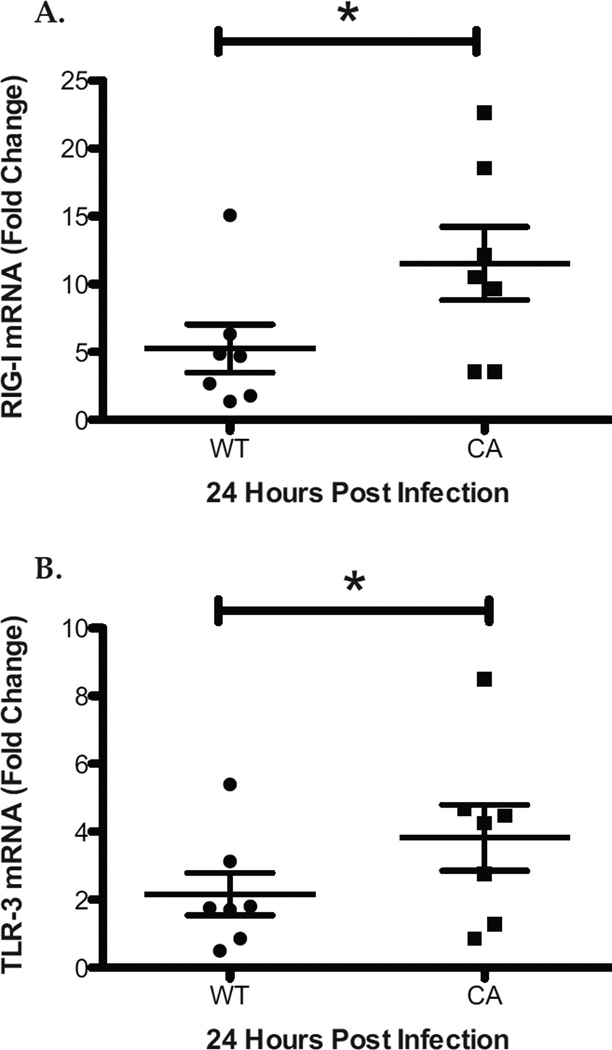
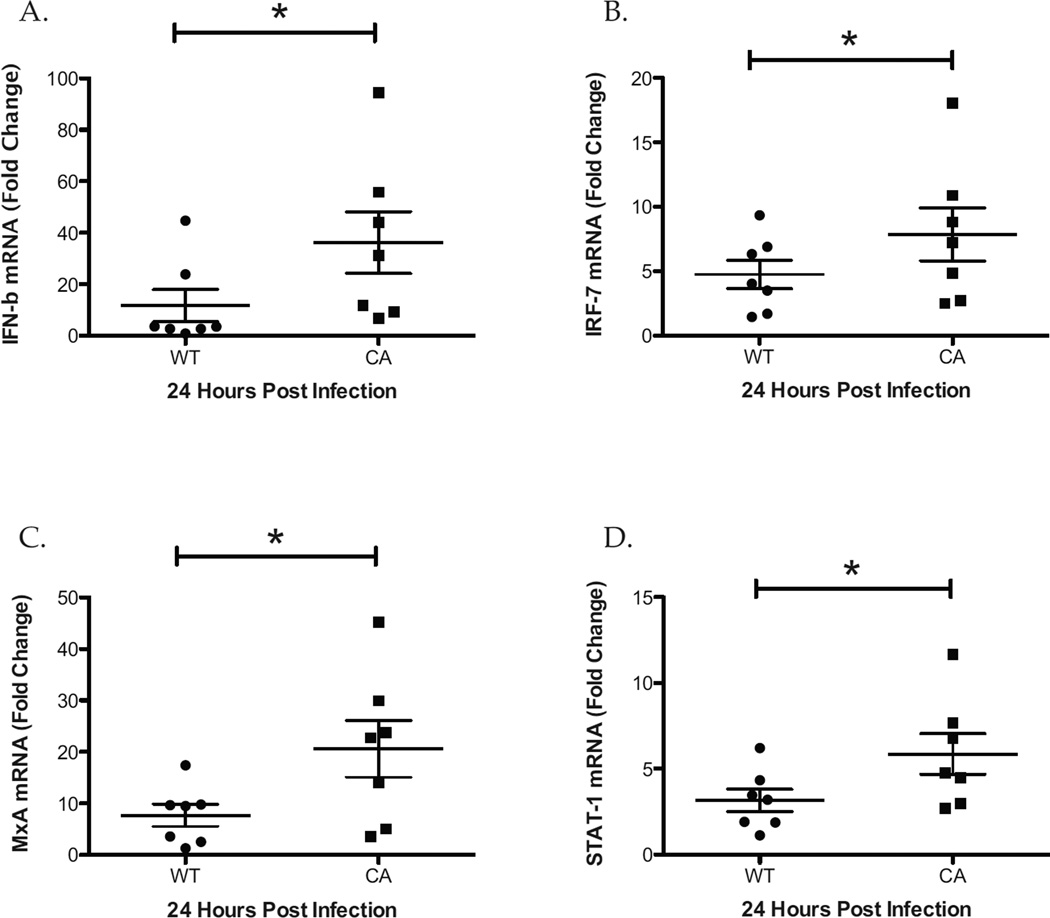
To compare the innate immune response during infection of human nasal epithelial cell cultures with LAIV and WT influenza viruses at 33°C we used quantitative RT-PCR to measure the expression of pattern recognition receptors (PRRs) including RIG-I and TLR-3. At 24 hours post infection, there was significantly greater expression of RIG-I and TLR-3 in LAIV, as compared with WT infected hNEC cultures (Figure 2 A and B). To confirm and expand these results we compared the down-stream expression of PRR signaling including Interferon-β (IFN-β) and interferon regulatory factor 7 (IRF-7). Consistent with increased PRR signaling, there was significantly greater levels of IFN-β and IRF-7 in LAIV-infected compared with WT-infected hNEC cultures (Figure 3 A and B). Similarly, IFN-stimulated genes including MxA and STAT-I were also expressed at significantly higher levels in LAIV-infected cultures (Figure 3 C and D). Collectively, these data reveal that there is a greater innate immune gene expression in LAIV infected hNEC cultures, as compared with WT infection, despite significantly restricted vaccine replication.
Figure 2.
Increased Expression of Pattern Recognition Receptors during LAIV Infection of hNEC cultures. Total RNA from WT and LAIV infected hNECs at 24HPI was analyzed for expression of RIG-I (A) and TLR-3 (B) by real time qRT-PCR and compared using the ΔΔ Ct method. Data are represented as the fold change in expression from Mock infected cultures. *p<.05 (n=7 different donors for hNECs).
Figure 3.
hNEC cultures Infected with LAIV Express an Augmented Antiviral Innate Immune Response. Total RNA from hNECs collected at 24 hours post infection with WT and CA viruses was analyzed for the expression of antiviral innate immune gene expression including Interferon-β (A), IRF-7 (B), MxA (C), and STAT-1 (D) by real time qRT-PCR and compared using the ΔΔ Ct method. Data are represented as the fold change in expression from Mock infected cultures. *p<.05 (n=7 different donors for hNEC).
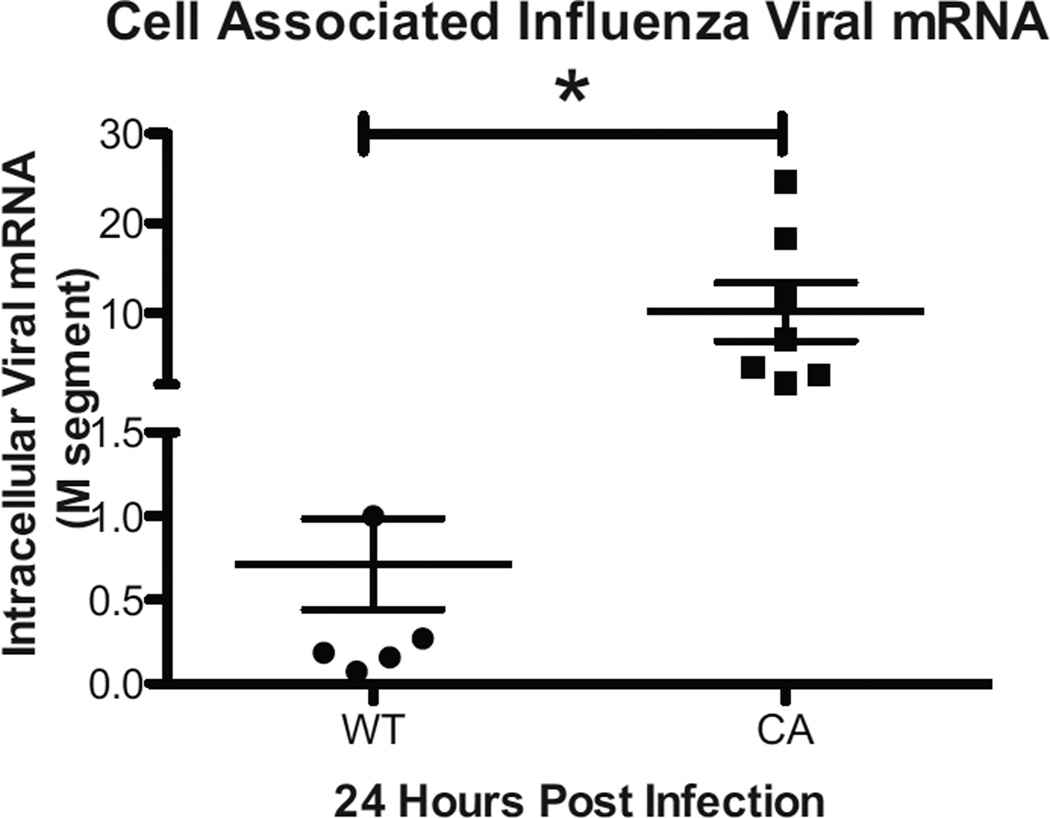
LAIV infection of hNEC cultures results is significantly greater levels of cell-associated viral RNA
To explore the etiology of the increased innate immune gene expression during LAIV infection of hNEC cultures we compared the level of viral RNA inside infected hNECs. Using primers specific for the M segment, viral RNA was amplified from total RNA and compared using the ΔΔCT method as previously described (15). At 24 hours post infection, there was significantly greater levels of viral RNA in LAIV- as compared with WT infected hNEC cultures despite a 165 fold higher level of infectious virus production in the latter (Figure 4). Together these data demonstrate that although inoculation with LAIV results in decreased replication in hNEC cultures it elicits a greater epithelial cell-specific innate immune response, likely due to increased levels of viral RNA within infected nasal epithelial cells.
Figure 4.
LAIV infected Nasal Epithelial Cell Cultures Contain Greater Levels of Viral RNA. Total RNA from WT and LAIV infected hNEC cultures collected at 24HPI were analyzed for the presence of viral RNA using qRT-PCR with primers specific for the Influenza M-segment. Levels of viral RNA were compared using the ΔΔ Ct method and represent the fold-change in expression relative to a WT-infected sample. p<.05 (n=7 different donors for hNEC).
LAIV infected hNEC cultures produce equivalent or greater pro-inflammatory chemokines despite significantly reduced replication
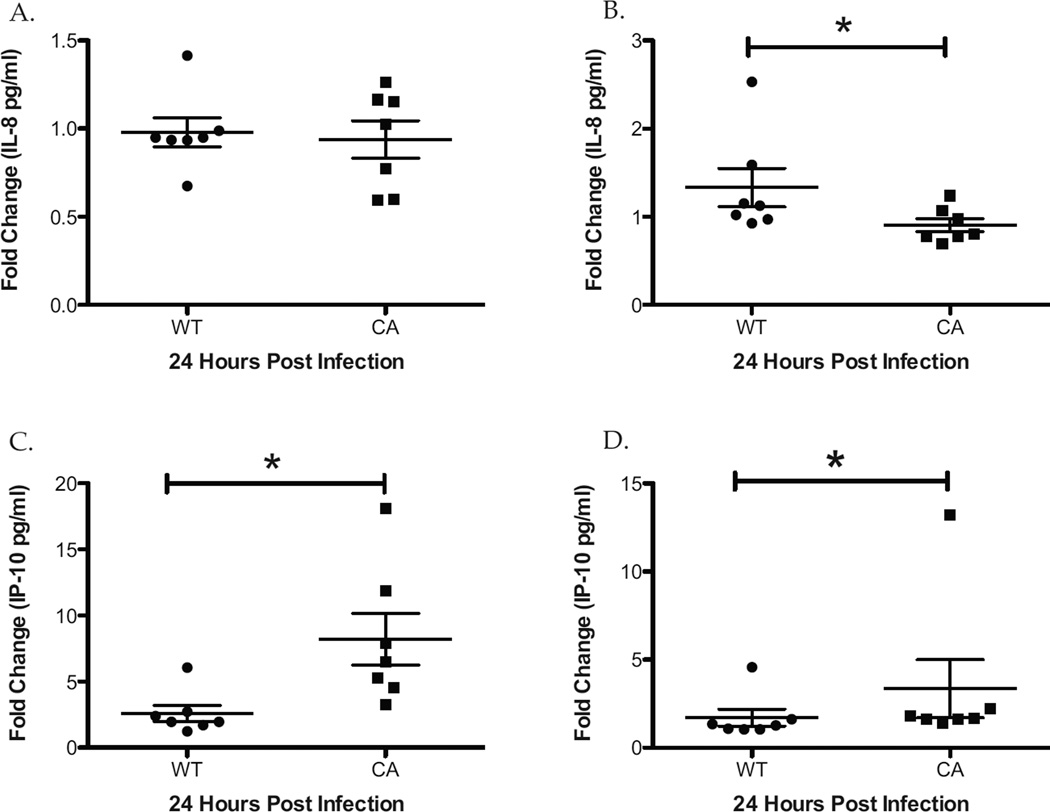
Given the enhanced intracellular innate immune signaling in LAIV infected hNEC cultures, chemokine expression was compared to determine if differential external cell signaling occurs during vaccination and seasonal influenza infection of nasal epithelial cells. While there was no significant difference in the apical secretion of IL-8 (Figure 5A), WT-infected hNEC cultures secreted 46% more IL-8 basolaterally compared with LAIV infected cultures (Figure 5 B). However this increase in WT induced basolateral IL-8 secretion was not proportional to the 165 fold greater infectious virus production in WT compared with LAIV-infected cultures (Figure 1).
Figure 5.
WT and LAIV-Infected hNEC cultures Result in a Differential Chemokine Response. Apical and basolateral supernatants were collected 24hours after hNEC infection with WT and LAIV viruses and analyzed for the presence of IL-8 (A and B) and IP-10 (C and D) by ELISA and are displayed as fold change in expression relative to Mock-infected cultures. *p<.05 (n=7 different donors for hNEC).
In contrast to IL-8, there was significantly increased expression of IP-10 detected apically (Figure 5 C) and basolaterally (Figure 5 D) in LAIV infected hNEC cultures compared with WT-infected cultures at 33°C. Taken together these results suggest that infection of human nasal epithelial cells with LAIV triggers the expression of a different chemokine profile than seen with WT infection.
DISCUSSION
The Live Attenuated Influenza Vaccine represents an important and innovative advancement in the prophylaxis against seasonal influenza. In contrast to the inactivated parenteral influenza vaccine, the LAIV replicates at the site of influenza infection and is thus able to elicit both a mucosal and systemic immune response (18). As a result, LAIV strains have been shown to be more effective than the inactivated influenza vaccine in pediatric populations and among hosts of all ages during years of poor homology between the circulating strain and the vaccine strain (19–26). While in vitro and in vivo studies have demonstrated significantly reduced LAIV infectious virus production, as compared with WT viruses, the mechanisms underlying the LAIV’s ability to elicit a protective immune response despite a deficient replication is unknown (27–31). In this study we compare the replication and innate immune response during infection of primary differentiated human nasal epithelial cells with WT and antigenically-matched cold adapted LAIV viruses at 33°C in order to better understand how infection with the live attenuated influenza vaccine differs from seasonal viruses at the level of the human nasal epithelium.
Data from this study indicate that, despite significantly less infectious virus production (Figure 1), during LAIV compared with WT hNEC infection, LAIV strains elicit an enhanced innate immune response as evidenced by the significantly greater transcription of RIG-I, TLR-3, IFN-beta, STAT-1, IRF-7 and MxA genes. While the reduced replication of LAIV viruses are consistent with human clinical trials and recent in vitro infections of human adenoid epithelial cells using the same viruses, we believe this is the first report of a differential epithelial cell-specific innate immune response induced by WT influenza viruses and antigenically-matched LAIV strains (30, 32). Previous reports have identified important mutations in the LAIV backbone that are associated with the 3 novel phenotypes of LAIV including cold adaptation, temperature sensitivity, and attenuation in animal models and human studies, however the enhanced intracellular antiviral response following LAIV infection likely also plays an important role in the control of virus replication and thus in the differing clinical phenotypes elicited by LAIV and WT viruses (27–29, 31, 33). Comparison of WT and LAIV inoculums revealed a greater hemagglutination unit titer (HAU) from the LAIV (1:80) compared with the WT stock (1:20) despite equivalent TCID50/ml titers. This increased HAU to TCID50/ml ratio in the LAIV inoculums, suggests the presence of greater non-infectious virus particles in the LAIV inoculums compared with the WT inoculums, and may contribute to the increased innate immune expression in AIV infected hNEC cultures.
The increased level of intracellular viral RNA (Figure 4) in LAIV-infected hNEC cultures, however indicates that defective replication, rather than non-infectious virus particles offers a more likely mechanism for the augmented innate immune response to LAIV infection. Detection of Viral RNA by RIG-I and TLR-3 leads to the induction of antiviral gene expression including Type I interferon as well as interferon stimulated genes (ISGs) which are critical factors in the innate immune response to influenza infection (34, 35). Our data demonstrate that, in addition to the up-regulation of these pattern recognition receptors (Figure 2), transcription of IFN-beta, STAT-1, IRF-7, and MxA genes (Figure 3) is also significantly increased in LAIV as compared with WT-infected hNECs. The increased expression of the PRRs, RIG-I and TLR-3 are either the result of increased viral RNA directly or more likely due to increased Type I-IFN as ISGs. Whether the increased intracellular viral RNA is a result of defective replication due to mutations in the LAIV backbone or a consequence of this heightened LAIV-specific epithelial cell antiviral immune response is not clear and the subject of ongoing investigation.
The differential chemokine production that results from hNEC infection with WT and LAIV viruses indicates that vaccination likely results in a substantially different immune cell recruitment than wild type infection. WT-infected hNEC cultures produce significantly greater levels of the neutrophil chemokine IL-8 whereas LAIV-infected hNEC cultures release significantly greater levels of IP-10 (Figure 5), a chemokine that preferentially recruits activated Th1 lymphocytes and NK cells (36). In RSV infected mice, neutralization of IP-10 led to an increase in disease pathogenesis and impaired virus clearance highlighting the importance of IP-10 in promoting viral clearance and generating a protective immune response (6). The differential immune cell recruitment during infection of hNEC cultures by LAIV and WT viruses may contribute to the decrease in LAIV replication in the human host and explain the absent clinical phenotype during LAIV vaccination. A greater understanding of the differences in immune cell recruitment between vaccination and wild-type infection is needed to better understand the differential determinants of eliciting a protective immune response versus generating a pathologic immune reaction.
CONCLUSIONS
The results of this study advance our understanding of how seasonal WT influenza viruses and antigenically matched LAIV strains differ in their ability to replicate and elicit an immune response in the nasal epithelium. The Live Attenuated Influenza Vaccines are nasal vaccine strains that are able to replicate in the nasal epithelium and elicit a protective mucosal immune response without causing clinical disease. We postulate that the decreased replication of these nasal vaccines is due to both mutations inherent in the LAIV backbone as well as an enhanced epithelial cell specific immune response. Improved understanding of how LAIV strains differ from WT viruses in nasal epithelial cells offers important insights in how virus-host cell interactions can result in either a protective or pathologic response and may also provide a way to rationally improve current LAIV strains in order to maximize the protective immune response while limiting the expression of clinical disease.
HIGHLIGHTS.
We compare the immune response to and replication of influenza viruses and live vaccines in hNECs
Despite lower replication, the LAIV viruses elicit a more robust innate immune response
LAIV infection of hNECs results in a differing chemokine expression profile immune response
The increased innate immune response is likely due to an increase in the level of intracellular viral RNA
ACKNOWLEDGEMENTS
The authors wish to thank Martha Almond and Carole Robinette for their assistance with volunteer recruitment and screening.
FINANCIAL SUPPORT:
This work was supported by grants from the National Institutes of Health (AG045088 WF) and (ES013611, HL095163 IJ), the Infectious Disease Society of America Young Investigator Award in Geriatrics (WF), Association of Specialty Providers T. Franklin Williams Award Supported by a T. Franklin Williams Scholarship Award [Funding provided by: Atlantic Philanthropies, Inc, the John A. Hartford Foundation, the Alliance for Academic Internal Medicine-Association of Specialty Professors, and the Infectious Disease of Society of America] (WF), and the Flight Attendant Medical Research Institute (IJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Although the research described in this article has been funded in part by the U.S. Environmental Protection Agency through cooperative agreement CR83346301 with the Center for Environmental Medicine, Asthma and Lung Biology at the University of North Carolina-Chapel Hill, it has not been subjected to the agency's required peer and policy review and therefore does not necessarily reflect the views of the agency, and no official endorsement should be inferred. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: WF contributed to study design, analysis plan, data analysis, interpretation, and writing and editing of the report. IJ contributed to study design, analysis plan, interpretation, data analysis and editing of the report, and was responsible for data analysis.
Financial/nonfinancial disclosures:
Dr. Fischer has no conflicts of interest. Dr. Jaspers has no conflicts of interest. Ms. Brighton has no conflicts of interest
WORKS CITED
- 1.Fischer WF, Hayden F. Novel Antiviral Agents and Approaches in Pandemic Influenza. Humana Press; 2008. [Google Scholar]
- 2.Maassab HF. Adaptation and growth characteristics of influenza virus at 25 degrees c. Nature. 1967;213(5076):612–614. doi: 10.1038/213612a0. Epub 1967/02/11. PubMed PMID: 6040602. [DOI] [PubMed] [Google Scholar]
- 3.Murphy BR, Coelingh K. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunol. 2002;15(2):295–323. doi: 10.1089/08828240260066242. Epub 2002/06/26. doi: 10.1089/08828240260066242. PubMed PMID 12081014. [DOI] [PubMed] [Google Scholar]
- 4.Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol. 2012;12(4):295–305. doi: 10.1038/nri3166. Epub 2012/03/10. doi: 10.1038/nri3166. PubMed PMID: 22402670; PubMed Central PMCID: PMC3364025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oslund KL, Baumgarth N. Influenza-induced innate immunity: regulators of viral replication, respiratory tract pathology & adaptive immunity. Future Virol. 2011;6(8):951–962. doi: 10.2217/fvl.11.63. Epub 2011/09/13. PubMed PMID: 21909336; PubMed Central PMCID: PMC3168512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindell DM, Lane TE, Lukacs NW. CXCL10/CXCR3-mediated responses promote immunity to respiratory syncytial virus infection by augmenting dendritic cell and CD8(+) T cell efficacy. Eur J Immunol. 2008;38(8):2168–2179. doi: 10.1002/eji.200838155. Epub 2008/07/16. doi: 10.1002/eji.200838155. PubMed PMID: 18624292; PubMed Central PMCID: PMC2743117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaspers I, Samet JM, Reed W. Arsenite exposure of cultured airway epithelial cells activates kappaB-dependent interleukin-8 gene expression in the absence of nuclear factor-kappaB nuclear translocation. J Biol Chem. 1999;274(43):31025–31033. doi: 10.1074/jbc.274.43.31025. Epub 1999/10/16. PubMed PMID: 10521501. [DOI] [PubMed] [Google Scholar]
- 8.Jaspers I, Horvath KM, Zhang W, Brighton LE, Carson JL, Noah TL. Reduced expression of IRF7 in nasal epithelial cells from smokers after infection with influenza. Am J Respir Cell Mol Biol. 2010;43(3):368–375. doi: 10.1165/rcmb.2009-0254OC. Epub 2009/11/03. doi: 10.1165/rcmb.2009-0254OC. PubMed PMID: 19880818; PubMed Central PMCID: PMC2933552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath KM, Brighton LE, Zhang W, Carson JL, Jaspers I. Epithelial cells from smokers modify dendritic cell responses in the context of influenza infection. Am J Respir Cell Mol Biol. 2011;45(2):237–245. doi: 10.1165/rcmb.2010-0190OC. Epub 2010/10/12. doi: 10.1165/rcmb.2010-0190OC. PubMed PMID: 20935192; PubMed Central PMCID: PMC3175553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciencewicki JM, Brighton LE, Jaspers I. Localization of type I interferon receptor limits interferon-induced TLR3 in epithelial cells. J Interferon Cytokine Res. 2009;29(5):289–297. doi: 10.1089/jir.2008.0075. Epub 2009/02/24. doi: 10.1089/jir.2008.0075. PubMed PMID: 19231996; PubMed Central PMCID: PMC2956593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller L, Brighton LE, Jaspers I. Ozone exposed epithelial cells modify cocultured natural killer cells. Am J Physiol Lung Cell Mol Physiol. 2013;304(5):L332–L341. doi: 10.1152/ajplung.00256.2012. Epub 2012/12/18. doi: 10.1152/ajplung.00256.2012. PubMed PMID: 23241529; PubMed Central PMCID: PMC3602740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keck T, Leiacker R, Heinrich A, Kuhnemann S, Rettinger G. Humidity and temperature profile in the nasal cavity. Rhinology. 2000;38(4):167–171. Epub 2001/02/24. PubMed PMID: 11190750. [PubMed] [Google Scholar]
- 13.Keck T, Leiacker R, Riechelmann H, Rettinger G. Temperature profile in the nasal cavity. Laryngoscope. 2000;110(4):651–654. doi: 10.1097/00005537-200004000-00021. Epub 2000/04/14. doi: 10.1097/00005537-200004000-00021. PubMed PMID: 10764013. [DOI] [PubMed] [Google Scholar]
- 14.McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol. 2005;79(6):3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. Epub 2005/02/26. doi: 10.1128/JVI.79.6.3595-3605.2005. PubMed PMID: 15731254; PubMed Central PMCID: PMC1075690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer RN, Brighton LE, Mueller L, Xiang Z, Rager JE, Fry RC, et al. Influenza enhances caspase-1 in bronchial epithelial cells from asthmatic volunteers and is associated with pathogenesis. J Allergy Clin Immunol. 2012;130(4):958–967. e14. doi: 10.1016/j.jaci.2012.07.013. Epub 2012/10/02. doi: 10.1016/j.jaci.2012.07.013. PubMed PMID: 23021143; PubMed Central PMCID: PMC3470476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaspers I, Zhang W, Fraser A, Samet JM, Reed W. Hydrogen peroxide has opposing effects on IKK activity and IkappaBalpha breakdown in airway epithelial cells. Am J Respir Cell Mol Biol. 2001;24(6):769–777. doi: 10.1165/ajrcmb.24.6.4344. Epub 2001/06/21. doi: 10.1165/ajrcmb.24.6.4344. 10.1165/ajrcmb.24.6.4344. PubMed PMID: 11415944. [DOI] [PubMed] [Google Scholar]
- 17.Jaspers I, Ciencewicki JM, Zhang W, Brighton LE, Carson JL, Beck MA, et al. Diesel exhaust enhances influenza virus infections in respiratory epithelial cells. Toxicol Sci. 2005;85(2):990–1002. doi: 10.1093/toxsci/kfi141. Epub 2005/03/18. doi: 10.1093/toxsci/kfi141. PubMed PMID: 15772371. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol. 2007;81(1):215–228. doi: 10.1128/JVI.01957-06. Epub 2006/10/20. doi: 10.1128/JVI.01957-06. PubMed PMID: 17050593; PubMed Central PMCID: PMC1797237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashkenazi S, Vertruyen A, Aristegui J, Esposito S, McKeith DD, Klemola T, et al. Superior relative efficacy of live attenuated influenza vaccine compared with inactivated influenza vaccine in young children with recurrent respiratory tract infections. Pediatr Infect Dis J. 2006;25(10):870–879. doi: 10.1097/01.inf.0000237829.66310.85. Epub 2006/09/29. doi: 10.1097/01.inf.0000237829.66310.85. PubMed PMID: 17006279. [DOI] [PubMed] [Google Scholar]
- 20.Belshe RB, Ambrose CS, Yi T. Safety and efficacy of live attenuated influenza vaccine in children 2–7 years of age. Vaccine. 2008;26(Suppl 4):D10–D16. doi: 10.1016/j.vaccine.2008.06.083. Epub 2008/07/10. doi: 10.1016/j.vaccine.2008.06.083. PubMed PMID: 18611422. [DOI] [PubMed] [Google Scholar]
- 21.Belshe RB, Edwards KM, Vesikari T, Black SV, Walker RE, Hultquist M, et al. Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med. 2007;356(7):685–696. doi: 10.1056/NEJMoa065368. Epub 2007/02/16. doi: 10.1056/NEJMoa065368. PubMed PMID: 17301299. [DOI] [PubMed] [Google Scholar]
- 22.Bridges CB, Thompson WW, Meltzer MI, Reeve GR, Talamonti WJ, Cox NJ, et al. Effectiveness and cost-benefit of influenza vaccination of healthy working adults: A randomized controlled trial. JAMA. 2000;284(13):1655–1663. doi: 10.1001/jama.284.13.1655. Epub 2000/10/04. PubMed PMID: 11015795. [DOI] [PubMed] [Google Scholar]
- 23.Fleming DM, Crovari P, Wahn U, Klemola T, Schlesinger Y, Langussis A, et al. Comparison of the efficacy and safety of live attenuated cold-adapted influenza vaccine, trivalent, with trivalent inactivated influenza virus vaccine in children and adolescents with asthma. Pediatr Infect Dis J. 2006;25(10):860–869. doi: 10.1097/01.inf.0000237797.14283.cf. Epub 2006/09/29. doi: 10.1097/01.inf.0000237797.14283.cf. PubMed PMID: 17006278. [DOI] [PubMed] [Google Scholar]
- 24.Halloran ME, Piedra PA, Longini IM, Jr, Gaglani MJ, Schmotzer B, Fewlass C, et al. Efficacy of trivalent, cold-adapted, influenza virus vaccine against influenza A (Fujian), a drift variant, during 2003–2004. Vaccine. 2007;25(20):4038–4045. doi: 10.1016/j.vaccine.2007.02.060. Epub 2007/03/31. doi: 10.1016/j.vaccine.2007.02.060. PubMed PMID: 17395338; PubMed Central PMCID: PMC2883284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferson T, Rivetti A, Harnden A, Di Pietrantonj C, Demicheli V. Vaccines for preventing influenza in healthy children. Cochrane Database Syst Rev. 2008;(2):CD004879. doi: 10.1002/14651858.CD004879.pub3. Epub 2008/04/22. doi: 10.1002/14651858.CD004879.pub3. PubMed PMID: 18425905. [DOI] [PubMed] [Google Scholar]
- 26.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. Epub 2011/10/29. doi: 10.1016/S1473-3099(11)70295-X. PubMed PMID: 22032844. [DOI] [PubMed] [Google Scholar]
- 27.Chan W, Zhou H, Kemble G, Jin H. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology. 2008;380(2):304–311. doi: 10.1016/j.virol.2008.07.027. Epub 2008/09/05. doi: 10.1016/j.virol.2008.07.027. PubMed PMID: 18768193. [DOI] [PubMed] [Google Scholar]
- 28.Jin H, Bin L, Zhou H, Kemble G. Genetic studies of Flumist influenza vaccines derived from cold-adapted A/Ann Arbor/6/60. International Congress Series. 2004;1263:153–156. [Google Scholar]
- 29.Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang CF, et al. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist) derived from cold-adapted A/Ann Arbor/6/60. Virology. 2003;306(1):18–24. doi: 10.1016/s0042-6822(02)00035-1. Epub 2003/03/07. PubMed PMID: 12620793. [DOI] [PubMed] [Google Scholar]
- 30.Murphy BR, Clements ML, Madore HP, Steinberg J, O'Donnell S, Betts R, et al. Dose response of cold-adapted, reassortant influenza A/California/10/78 virus (H1N1) in adult volunteers. J Infect Dis. 1984;149(5):816. doi: 10.1093/infdis/149.5.816. Epub 1984/05/01. PubMed PMID: 6726007. [DOI] [PubMed] [Google Scholar]
- 31.Snyder MH, Clements ML, De Borde D, Maassab HF, Murphy BR. Attenuation of wild-type human influenza A virus by acquisition of the PA polymerase and matrix protein genes of influenza A/Ann Arbor/6/60 cold-adapted donor virus. J Clin Microbiol. 1985;22(5):719–725. doi: 10.1128/jcm.22.5.719-725.1985. Epub 1985/11/01. PubMed PMID: 4056002; PubMed Central PMCID: PMC268513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, et al. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. J Virol. 2012;86(21):11725–11734. doi: 10.1128/JVI.01477-12. Epub 2012/08/24. doi: 10.1128/JVI.01477-12. PubMed PMID: 22915797; PubMed Central PMCID: PMC3486302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder MH, Betts RF, DeBorde D, Tierney EL, Clements ML, Herrington D, et al. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol. 1988;62(2):488–495. doi: 10.1128/jvi.62.2.488-495.1988. Epub 1988/02/01. PubMed PMID: 3336068; PubMed Central PMCID: PMC250559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Sastre A. Antiviral response in pandemic influenza viruses. Emerg Infect Dis. 2006;12(1):44–47. doi: 10.3201/eid1201.051186. Epub 2006/02/24. doi: 10.3201/eid1201.051186. PubMed PMID: 16494716; PubMed Central PMCID: PMC3291409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang JP, Kurt-Jones EA, Finberg RW. Innate immunity to respiratory viruses. Cell Microbiol. 2007;9(7):1641–1646. doi: 10.1111/j.1462-5822.2007.00961.x. Epub 2007/05/18. doi: 10.1111/j.1462-5822.2007.00961.x. PubMed PMID: 17506818. [DOI] [PubMed] [Google Scholar]
- 36.Korpi-Steiner NL, Bates ME, Lee WM, Hall DJ, Bertics PJ. Human rhinovirus induces robust IP-10 release by monocytic cells, which is independent of viral replication but linked to type I interferon receptor ligation and STAT1 activation. J Leukoc Biol. 2006;80(6):1364–1374. doi: 10.1189/jlb.0606412. Epub 2006/10/06. doi: 10.1189/jlb.0606412. PubMed PMID: 17020930. [DOI] [PubMed] [Google Scholar]