Abstract
This article focuses on discoveries of the mechanisms governing the regulation of glycerolipid metabolism and stress response signaling in response to the phospholipid precursor, inositol. The regulation of glycerolipid lipid metabolism in yeast in response to inositol is highly complex, but increasingly well understood, and the roles of individual lipids in stress response are also increasingly well characterized. Discoveries that have emerged over several decades of genetic, molecular and biochemical analyses of metabolic, regulatory and signaling responses of yeast cells, both mutant and wild type, to the availability of the phospholipid precursor, inositol are discussed.
1. Introduction
In the yeast, Saccharomyces cerevisiae, as in other eukaryotes, regulation of lipid metabolism is extremely complex, involving coordination of the biosynthesis and turnover of an enormous number of lipid classes and species. All eukaryotic cells share the challenge of regulating and coordinating the complex and interconnected pathways of lipid metabolism across multiple, spatially distinct membrane compartments, adjusting for shifting precursor availability and membrane expansion in the course of cell division, growth and metabolism. The genes, enzymes and pathways of lipid metabolism in yeast share substantial homology with those in higher eukaryotes, including mammals, making yeast an attractive model system for biomedical research (Henry et al., 2012).
S. cerevisiae, as a free-living unicellular organism, must continuously monitor and coordinate endogenous metabolic activity in response to ever changing availability of precursors of lipid biosynthesis in the growth medium. Indeed, many insights into fundamental mechanisms of genetic regulation of phospholipid metabolism in yeast have come from studies of the cellular responses to the availability of exogenous precursors of phospholipid biosynthesis, especially inositol. The cellular consequences of inositol depletion have also been studied in mammalian cells and compared to yeast in the context of exposure to inositol depleting drugs lithium and valproic acid (Deranieh and Greenberg, 2009). Various aspects of regulation and signaling related to lipid and inositol metabolism in yeast have been extensively reviewed (Carman and Han, 2011; Carman and Henry, 1999; Chen et al., 2007; Dickson, 2008; Gaspar et al., 2007; Greenberg and Lopes, 1996; Henneberry and Sturley, 2005; Henry et al., 2012; Jesch, 2005; Majerus and York, 2009; Strahl and Thorner, 2007; Tsui and York, 2010). Thus it is not the intention of this article to provide a comprehensive coverage of the broader topics of lipid and inositol metabolism in regulation and signaling in eukaryotic cells in general. Rather, this article has a primary focus on the discoveries of mechanisms governing the regulation of glycerolipid metabolism and the signaling roles of specific lipids in yeast that have been made possible by genetic, molecular and biochemical analyses of the cellular response to the availability of the phospholipid precursor, inositol (Fig. 1).
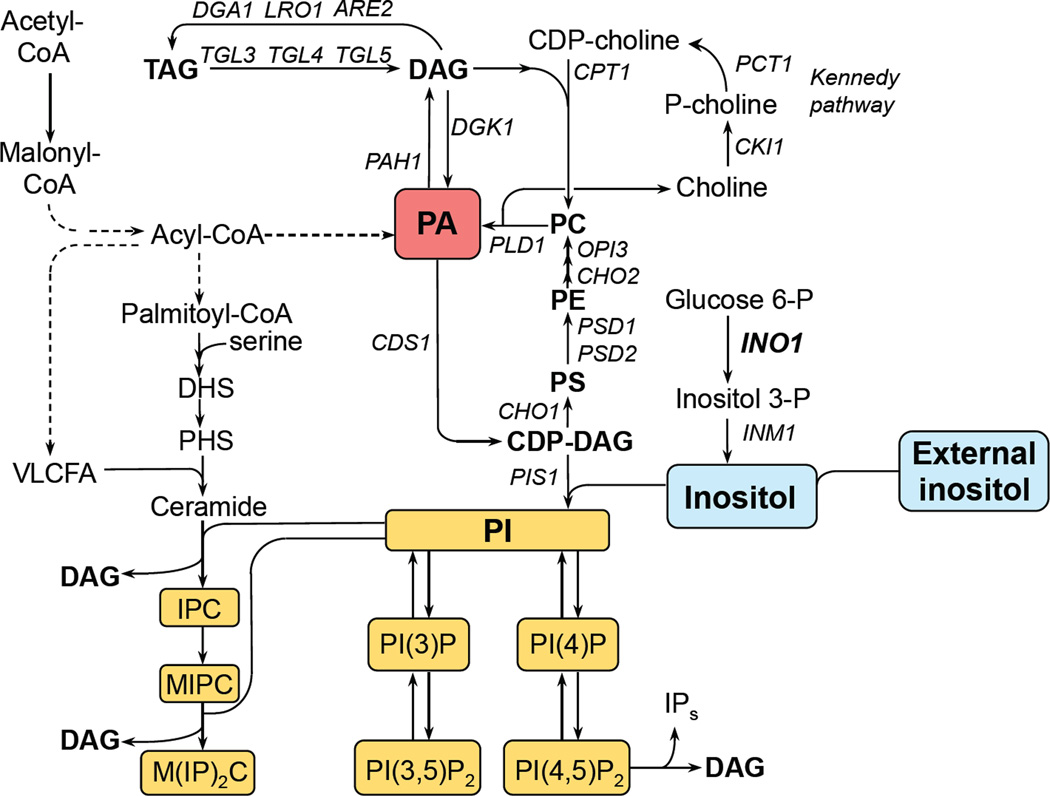
Figure 1. Major pathways for the synthesis of phospholipids, sphingolipids, phosphoinositides and triacylglycerols.
The names of the structural genes for enzymes discussed in the manuscript are shown adjacent to the arrows of the metabolic conversions that they catalyze. Solid arrows indicate direct enzymatic conversions. Dashed arrows indicate conversions that require more than one enzymatic step. Red box indicates the precursor and signaling lipid PA. Blue boxes indicate the phospholipid precursor inositol, which can be added or removed from the growth medium. Yellow boxes indicate the products from which inositol is precursor. DAG, diacylglycerol; CDP-DAG, cytidine diphosphate diacylglycerol; CDP-choline, cytidine diphosphate choline; PA, phosphatidic acid; PI, phosphatidylinositol; PI(4)P, phosphatidylinositol 4-phosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3)P, phosphatidylinositol 3-phosphate; PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate; inositol-phosphorylceramide, (IPC); mannosyl-inositol-phosphorylceramide, (MIPC); mannosyl-diinositol-phosphorylceramide, (M(IP)2C); PE, phosphatidylethanolamine; PS, phosphatidylserine; PC, phosphatidylcholine; TAG, triacylglycerols; FS, free sterols; FFA, free fatty acids; SE steryl esters; PL, phospholipids; VLCFA, very-long-chain fatty acids; DHS, dihydrosphingosine; PHS, phytosphingosine. IPs refers to the inositol soluble phosphates (Wilson et al., 2013; Ye et al., 2013).
Inositol serves as an essential precursor in yeast, as in other eukaryotic cells, for the synthesis of phosphatidylinositol (PI) (Fig. 1), which in turn serves as precursor to many important signaling molecules, including phosphoinositides, inositol polyphosphates (Carman and Han, 2011; Carman and Henry, 1999; Henry et al., 2012; Jesch, 2005; Majerus and York, 2009; Strahl and Thorner, 2007; Tsui and York, 2010) and inositol containing sphingolipids (Breslow and Weissman, 2010; Deranieh and Greenberg, 2009; Dickson, 2008), as well as glycosylphosphatidylinositol (GPI) anchor proteins (Pittet and Conzelmann, 2007). When inositol is added to the growth medium of actively proliferating yeast cells adapted to growth in its absence, the rate of PI synthesis and accumulation increases rapidly and dramatically (Gaspar et al., 2006; Gaspar et al., 2011; Loewen et al., 2004). Thus, inositol availability has the potential to influence many signaling pathways in yeast (Jesch et al., 2006; Jesch et al., 2005). Moreover, inositol availability also influences the synthesis of all lipids derived directly or indirectly from phosphatidic acid (PA) (Fig. 1), itself a powerful signaling lipid (Carman and Henry, 2007) (Henry et al., 2012).
In wild type cells under conditions of inositol limitation, hundreds of genes are activated, the most highly regulated of which is INO1, encoding inositol 3-phosphate synthase (Ino1p), the enzyme that catalyzes the rate limiting step in the de novo synthesis of inositol (Henry et al., 2012; Jesch et al., 2006; Jesch et al., 2005; Santiago and Mamoun, 2003). However, in addition to genes involved in inositol and phospholipid biosynthesis, the list of genes activated in response to inositol limitation also includes many that are known to be activated by stress response pathways, including the unfolded protein response (UPR) (Chang et al., 2004; Chang et al., 2002; Cox et al., 1993; Cox and Walter, 1996; Mori et al., 1993; Mori et al., 1992), the glucose response pathway (Shirra et al., 2001) and the protein kinase C (PKC) pathway (Jesch et al., 2010; Nunez et al., 2008).
Thus, the experimental exploitation of yeast mutants defective in diverse aspects of lipid metabolism and regulation, coupled with manipulation of the exogenous supply of phospholipid precursors, especially inositol, offers the potential to generate powerful insights into the diverse regulatory and signaling roles of eukaryotic lipids. This article will focus on the metabolism, genetics and molecular biology associated with these discoveries in yeast.
2. Biosynthesis of inositol in S. cerevisiae: biochemistry, genetics and regulation
2.1. Isolation and characterization of mutants defective in inositol biosynthesis and regulation
The rate-limiting step in synthesis of inositol in yeast (Donahue and Henry, 1981b), as in other eukaryotes, many archaea, and some hyperthermophilic bacteria (Majumder et al., 1997; Michell, 2007) involves the conversion of D-glucose 6-phosphate to D-myo-inositol-3-phosphate in the cytoplasm by a reaction catalyzed by the inositol 3-phosphate synthase (IP synthase, Ino1p). Inositol 3-phosphate is subsequently dephosphorylated by inositol 3-phosphate monophosphatase (Inm1p) (Murray and Greenberg, 1997). However, yeast inm1Δ mutants are not inositol auxotrophs and retain considerable inositol 3-phosphate phosphatase activity (Murray and Greenberg, 2000), suggesting that enzymes in addition to Inm1p are able to catalyze the dephosphorylation of inositol 3-phosphate. The activity of IP synthase is dramatically reduced in yeast cells grown in the presence of inositol, indicating that the enzyme is repressible (Culbertson et al., 1976). The isolation and characterization of S. cerevisiae mutants unable to grow in the absence of inositol (Ino− phenotype) permitted the identification of the structural gene (INO1), encoding IP synthase (Donahue and Henry, 1981b) and enabled its subsequent cloning (Klig and Henry, 1984) and sequencing (Dean-Johnson and Henry, 1989).
Two early screens for S. cerevisiae mutants unable to grow without inositol supplementation resulted in the isolation of about 150 independently generated inositol auxotrophs (Ino− mutants) (Culbertson and Henry, 1975; Donahue and Henry, 1981a). In comparison to wild type, cell extracts from the Ino− mutants were found to have little or no IP synthase activity (Culbertson et al., 1976). The mutants fell into genetic complementation groups, corresponding to 20 unlinked genes (Culbertson and Henry, 1975; Donahue and Henry, 1981a). However, only three of these genes were represented by more than one mutant allele, with ino1 mutants representing about 69% and ino2 and ino4 mutants each representing about 9% of the mutants isolated (Culbertson and Henry, 1975; Donahue and Henry, 1981a). INO1 was subsequently shown to be the structural gene encoding the subunit of IP synthase (Ino1p), while INO2 and INO4 were shown to encode positive regulatory proteins, essential, not only for expression and regulation of IP synthase, but also for regulation of a large number of co-regulated enzymes of phospholipid biosynthesis (Henry et al., 2012). Recent screenings of yeast genome wide collections of viable deletion mutants have revealed hundreds of additional individual viable yeast “gene knock out” mutants that exhibit Ino− phenotypes of varying strength. Many of these mutants are pleiotropic, exhibiting additional phenotypes on inositol free medium, such as temperature sensitivity and/or sensitivity to the presence of a second phospholipid precursor, choline (Villa-Garcia et al., 2011; Young et al., 2010). These recent findings indicate that the early genetic screens for inositol auxotrophy greatly underestimated the number of loci that are required for sustained growth in the absence of exogenous inositol under a variety of conditions, a topic to be discussed later in this review.
Purification and biochemical characterization of IP synthase from extracts of wild type yeast revealed that the enzyme is a tetramer consisting of four identical subunits (Donahue and Henry, 1981b), as later confirmed by X-ray crystallography (Geiger and Jin, 2006; Jin and Geiger, 2003). Immunological analysis of crude extracts of the original Ino− mutants conducted with antibody raised in response to purified IP synthase subunit (Ino1p) revealed that many ino1 mutants retained Ino1p cross-reacting material, consistent with their identification as structural gene mutants expressing an inactive mutant subunit (Donahue and Henry, 1981b). In contrast, Ino1p cross-reacting material was completely absent or greatly reduced in Ino− mutants representing the other loci, suggesting that their defects were most likely regulatory in nature (Donahue and Henry, 1981b). Based on these findings, the INO1 gene was identified as the structural gene encoding the subunit of IP synthase, the first IP synthase structural gene to be identified in any organism (Donahue and Henry, 1981b). Ino1p was also shown to be largely absent from extracts of wild type yeast cells grown in the presence of inositol, confirming that the enzyme is repressed at the level of expression of the protein subunit (Donahue and Henry, 1981b). Subsequent analysis of IP synthase enzymes and genes from a wide variety of organisms has revealed a high degree of sequence conservation, particularly among eukaryotes (Majumder et al., 2003). Significantly, IP synthase is a phosphoprotein in both yeast and humans (Deranieh et al., 2013) and the human IP synthase gene (hINO1) is able to rescue the inositol auxotrophy of the ino1Δ mutant when expressed in yeast (Ju et al., 2004). Analysis of phosphorylation defective and phosphomimetic mutations involving three phosphorylation sites, which are conserved in yeast and human IP synthases, suggests that that the serine residues at two of these sites play critical roles, given that phosphorylation at each of these sites inhibits enzyme activity in both yeast and humans (Deranieh et al., 2013).
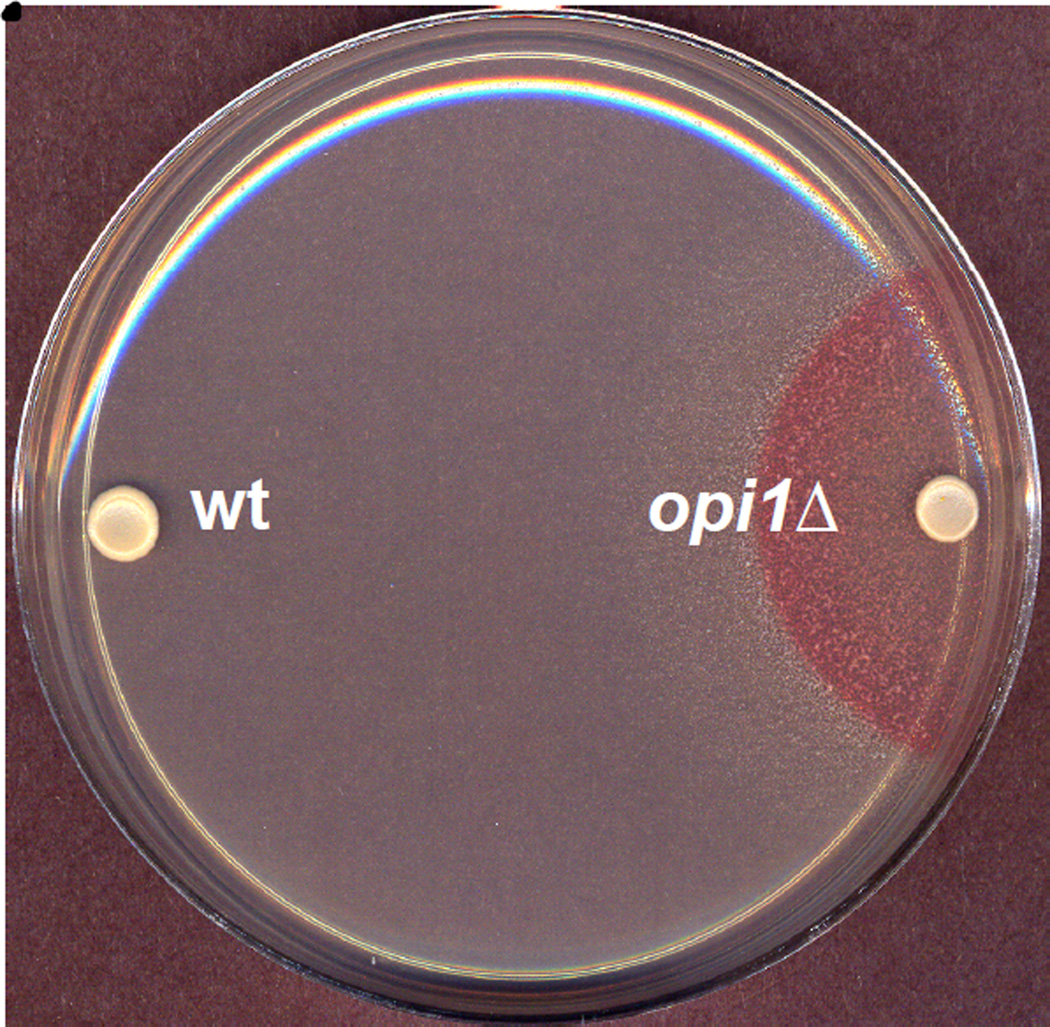
Yeast mutants defective in repression of Ino1p in response to inositol were first identified on the basis of a phenotype known as Opi−, overproduction and excretion of inositol (Greenberg et al., 1982b). The plate assay used in the original screen for mutants possessing this phenotype involved allowing mutagenized cells to grow into colonies on medium lacking inositol and then spraying the plates with a suspension of an inositol auxotrophic diploid strain (AID), which also carried a mutation in adenine biosynthesis (MATa/MATα, ade1/ade1, ino1-13/ino1-13, lys2/LYS2). Inositol excreting (Opi−) mutants were identified by the growth of the indicator strain in a red halo around inositol excreting colonies (Fig. 2) (Greenberg et al., 1982b; Swede et al., 1992). The opi1-1 mutant isolated in the initial screening for mutants with the Opi− phenotype (Greenberg et al., 1982b), exhibited about two-fold higher IP synthase activity and Ino1p subunit levels than in wild type cells grown under derepressing conditions (i.e. in the absence of inositol). The opi1-1 mutant also failed to repress IP synthase when grown in the presence of inositol. On this basis Opi1p was identified as a repressor of IP synthase expression (Greenberg et al., 1982a).
Figure 2. Overproduction of inositol (Opi−) phenotype of opi1Δ strain.
Wild type (wt) and opi1Δ cells were spotted on plates containing I− medium and incubated for 2 days at 30°C. A cell suspension of AID indicator strain, which grows only in the presence of inositol, was sprayed on the plates and incubated for a further 2 days at 30°C. Strains excreting inositol are visible as red halos around the strain being tested.
2.2. In yeast, enzymes of phospholipid biosynthesis are coordinately regulated in response to inositol and choline
An early indication that many enzymes of phospholipid biosynthesis in yeast are regulated coordinately with IP synthase in response to inositol emerged from the discovery that ino2 and ino4 mutants are pleiotropic. In addition to being unable to express IP synthase (Donahue and Henry, 1981a), ino2 and ino4 mutants exhibit reduced capacity to convert phosphatidylethanolamine (PE) to phosphatidylcholine (PC) (Fig. 1) (Loewy and Henry, 1984). The opi1-1 mutant also proved to be pleiotropic, exhibiting constitutive expression, not only of IP synthase, but also phosphatidylserine (PS) synthase and the phospholipid methyltransferases that convert PE to PC (Fig. 1) (Klig et al., 1985). In wild type cells, these same enzymes were shown to exhibit a unique pattern of coordinate regulation in response to inositol and choline. Full expression of these coordinately regulated enzymes of phospholipid biosynthesis is observed in wild type cells grown in the absence of inositol, whether choline is present or not. When inositol is present in the growth medium, the co-regulated enzymes are repressed and a further level of repression occurs when choline is present along with inositol. However, choline in the absence of inositol has little or no effect (Klig et al., 1985).
In related observations, Yamashita and Oshima (Yamashita and Oshima, 1980) reported a yeast mutant auxotrophic for choline, which exhibited low phospholipid methyltransferase activity in the presence of inositol. These investigators also observed that phospholipid methyltransferase activity was reduced in some wild type strains grown in the presence of inositol. Other early studies on the regulation of synthesis of PC via methylation of PE (Fig. 1) were conducted with yeast grown in synthetic complete medium containing yeast nitrogen base (YNB), which contains a low level of inositol (about 10µM). The concentration of inositol found in YNB medium was later found to cause partial repression of phospholipid biosynthetic genes (Hirsch and Henry, 1986). In early studies conducted in YNB media, the repression of PC biosynthesis was understandably attributed solely to the effect of additional phospholipid precursors, such as choline (Carson et al., 1982; Carson et al., 1984; Waechter and Lester, 1971, 1973). However, the presence of exogenous choline was subsequently shown to have little or no effect on the expression of INO1 and coregulated genes of phospholipid biosynthesis when inositol is absent from the medium (Hirsch and Henry, 1986; Jesch et al., 2005). Other enzymes catalyzing reactions involved in biosynthesis of PC via the pathway from CDP-diacylglycerol (CDP-DAG) (Fig. 1) are also repressed in a similar fashion in response to exogenous inositol in combination with choline or other phospholipid precursors, such as serine or ethanolamine (Homann et al., 1987; Homann et al., 1985; Klig et al., 1985; Klig et al., 1988a; Lamping et al., 1991; Poole et al., 1986).
2.3. Structural genes encoding enzymes of phospholipid biosynthesis are subject to complex transcriptional regulation
The isolation of the yeast INO1 gene, proved to be key to understanding the complex cellular transcriptional response to inositol (Henry et al., 2012). The presence of 75µM inositol in the growth medium was initially shown by slot blot analysis to repress INO1 transcription in wild type cells by more than 10 fold (Hirsch and Henry, 1986). However, 10µM inositol, a concentration similar to that found in standard yeast nitrogen base (YNB) medium, was shown to permit partial de-repression of INO1 during active growth (Hirsch and Henry, 1986). Under inositol limiting conditions, derepression of INO1 occurs when exogenous inositol is depleted, and as growth progresses, wild type yeast derepress INO1 and must rely on endogenous synthesis of inositol catalyzed by Ino1p to reach stationary phase. When exogenous inositol is completely absent or has been completely depleted during active growth, wild type yeast cells grow at a somewhat reduced rate in comparison to cells fully supplemented with inositol (Gaspar et al., 2011). Consistent with these observations, Hanscho et al., (Hanscho et al., 2012) recently reported that wild type yeast growing in YNB media, in the absence of any additional inositol supplementation, are unable to maintain rapid cell proliferation until the glucose in the medium is fully depleted. Recent studies using more sensitive RT-PCR methods, have shown that expression of the INO1 gene increases by 200–300 hundred fold within the first two to three hours following an abrupt shift of actively dividing cells from medium containing a fully repressing level of inositol (75–100µM) to medium lacking inositol. However, as cells acclimate to long-term growth in the absence of inositol, INO1 expression attenuates to a level of approximately 100 to 150 fold over fully repressed levels, depending on the precise growth conditions employed (Gaspar et al., 2011). In contrast to the several hours required for full derepression of INO1 following removal of inositol, almost complete repression is achieved within about 30 minutes following addition of inositol to cultures of wild type cells that have been fully acclimated to growth in its absence (Jesch et al., 2006). When a fully repressing concentration of inositol is present, the presence of choline (Fig. 1) results in a further several fold reduction in INO1 expression. However, as stated above, exogenous choline has little or no effect on INO1 expression when inositol is absent from the medium (Gaspar et al., 2011; Hirsch and Henry, 1986; Jesch et al., 2005).
Furthermore, even in the absence of inositol, INO1 and coregulated genes and enzymes are repressed as cells enter stationary phase (Homann et al., 1987; Lamping et al., 1994). However, in opi1Δ cells, INO1 expression continues at high, derepressed levels into stationary phase, both in the presence and absence of inositol (Griac et al., 1996; Jiranek et al., 1998). The INO1 gene was also shown to be transiently repressed when wild type cells, logarithmically growing in the absence of inositol in standard medium containing ammonium sulfate as a nitrogen source to medium containing a mixture of amino acids and bases as nitrogen sources. This result indicates that INO1 expression is sensitive to nitrogen limitation during active growth in the absence of inositol. In contrast, opi1Δ cells continued to express INO1 at high constitutive levels, without interruption, when shifted under identical conditions to medium lacking both ammonium sulfate and inositol, indicating that Opi1p is necessary for repression in response to nitrogen limitation, as well as in response to inositol (Griac and Henry, 1999).
Cloning of the CHO1 gene (Letts et al., 1983), encoding PS synthase (Atkinson et al., 1980a; Atkinson et al., 1980b; Kovac et al., 1980) and the CHO2 (PEM1) and OPI3 (PEM2) genes, encoding the two yeast phospholipid methyltransferases responsible for methylation of PE to form PC (Kodaki and Yamashita, 1987; Summers et al., 1988) (Fig. 1), permitted comparison of their transcriptional regulation to that of INO1. As in the case of INO1 (Hirsch and Henry, 1986), expression of CHO1, CHO2 and OPI3 is repressed when inositol is present in the growth medium and is further repressed when choline is also present (Bailis et al., 1992; Bailis et al., 1987; Gaynor et al., 1991; Hosaka and Kodaki, 1990; Kanipes and Henry, 1997; Kodaki et al., 1991b; Kodaki and Yamashita, 1987, 1989; Nikawa et al., 1987a; Summers et al., 1988). Many other genes related to lipid metabolism have subsequently been shown to show a similar pattern of regulation. For example, synthesis of S-adenosyl methionine (SAM), the methyl donor in conversion of PE to PC (Bremer and Greenberg, 1959; Gibson et al., 1961) is catalyzed by S-adenosyl methionine synthase. S. cerevisiae has two S-adenosyl methionine synthase genes, SAM1 and SAM2 (Thomas et al., 1988; Thomas and Surdin-Kerjan, 1987) and the SAM2 gene is regulated by inositol and choline, whereas SAM1 is not (Kodaki et al., 2003). The SAH1 gene, encoding S-adenosyl-L-homocysteine hydrolase is also repressed in a similar fashion in response to inositol and choline (Tehlivets et al., 2004). Moreover, down regulation of expression of SAH1 leads to decreased PC levels, increased accumulation of triacylglycerol levels and derepression of INO1 (Malanovic et al., 2008). Many additional genes encoding enzymes involved in phospholipid biosynthesis have been shown to exhibit a similar pattern of transcriptional regulation in response to inositol and choline. These co-regulated genes include: PSD1, encoding the mitochondrial PS decarboxylase, but not PSD2, encoding the ER localized PS decarboxylase (Griac, 1997; Lamping et al., 1991) and CDS1, encoding CDP-DAG synthase and PGS1, encoding phosphatidylglycerolphosphate synthase (Shen and Dowhan, 1998). A number of genes in the Kennedy pathway for PC and PE biosynthesis (Fig. 1), including CKI1, encoding choline kinase, PCT1, encoding cholinephosphotransferase, EPT1, encoding ethanolaminephosphotransferase and EKI1, encoding ethanolamine kinase, are also regulated in a similar fashion (Hosaka et al., 1990; Kim et al., 1999; McMaster and Bell, 1994b). The genes encoding the high and low affinity transporters of inositol, ITR1 and ITR2, also show this pattern of regulation (Lai and McGraw, 1994), as does the choline transporter HNM1/CTR1 (Li et al., 1991; Nikawa et al., 1986). Several genes involved in fatty acid synthesis, FAS1, FAS2, encoding the α and β, and ACC1/FAS3 encoding acetyl-CoA carboxylase, also exhibit repression in response to inositol and choline (Chirala, 1992; Chirala et al., 1987; Chirala et al., 1994; Hasslacher et al., 1993; Schuller et al., 1992a; Schuller et al., 1992b; Schwank et al., 1995).
However, not all genes involved in phospholipid metabolism in yeast are repressed in response to inositol. In contrast to INO1, the INM1 gene, encoding inositol 3-phosphate phosphatase and the DPP1 and PAH1 genes, encoding lipid phosphate phosphatases, are derepressed in the presence of inositol and in stationary phase (Murray and Greenberg, 1997, 2000; Oshiro et al., 2000; Pascual et al., 2013). Regulation of the PIS1 gene, encoding phosphatidylinositol synthase, (Nikawa et al., 1987a) is largely uncoupled from regulation by inositol and choline but requires Ino4p for full expression and is induced about two fold by inositol, a response requiring the pleiotropic regulatory protein Ume6p (Jani and Lopes, 2008). Importantly, altered expression of PIS1 has profound effects on the expression of INO1 and coregulated genes and on levels of PI, PA and PC (Gardocki et al., 2005; Jani and Lopes, 2009), as will be discussed below. Some genes encoding phospholipid biosynthetic enzymes, including PIS1, PAH1, CKI1 and EKI1, are also regulated by zinc in the absence of inositol. The regulation of these genes by zinc involves control of the level of PA by activation of PI synthase in the absence of inositol via the Zap1 transcription factor (Carman and Han, 2007; Henry et al., 2012).
A number of previous reviews (Carman and Han, 2011; Carman and Henry, 1999; Greenberg and Lopes, 1996; Henry et al., 2012; Paltauf, 1992) provide excellent and detailed coverage of the genes and enzymes subject to regulation by inositol and choline. Microarray analysis has also revealed many additional genes, genome wide, that are regulated in response to inositol and choline (Jesch et al., 2006; Jesch et al., 2005; Santiago and Mamoun, 2003). However, many of the genes identified as being activated in genome wide microarray studies of wild type cells growing in the absence of inositol are not involved in lipid metabolism and are not regulated in coordination with INO1 and coregulated genes of lipid metabolism. Rather, they are targets of stress response pathways that are activated when wild type cells are grown in the absence of inositol (Henry et al., 2012; Jesch et al., 2010; Jesch et al., 2006; Jesch et al., 2005; Lee et al., 2013; Nunez et al., 2008); a topic discussed below.
2.4. Regulation of INO1 and co-regulated genes involves the interaction of the Ino2p and Ino4p transcription factors, with each other, and with the UASINO promoter element and the Opi1p repressor
As described above, unlike wild type, ino2 and ino4 mutants fail to derepress INO1 when shifted to medium containing a concentration of inositol of 10µM inositol or less, leading to the identification of Ino2p and Ino4p as positive regulators of INO1 transcription (Hirsch and Henry, 1986). The INO2 and INO4 genes were cloned, sequenced and shown to encode proteins each containing a Helix-Loop-Helix (HLH) motif (Hoshizaki et al., 1990; Klig et al., 1988b; Nikoloff and Henry, 1994; Nikoloff et al., 1992; Schwank et al., 1995). The HLH motif is found in many eukaryotic regulatory proteins, including mammalian Myc and Max (Amati and Land, 1994), as well as a number of other yeast regulatory proteins (Robinson and Lopes, 2000). Ino2p and Ino4p were shown to bind as a heterodimer (Ambroziak and Henry, 1994; Schwank et al., 1995) to a repeated element, UASINO. UASINO is found in multiple copies in the promoter of the INO1 gene and in the promoters of other genes that are regulated in coordination with INO1 in response to inositol and choline (Bachhawat et al., 1995; Bailis et al., 1992; Lopes and Henry, 1991; Lopes et al., 1991). The genes shown to contain functional copies of this element include CHO1/PSS, encoding PS synthase (Bailis et al., 1992; Bailis et al., 1987; Kodaki et al., 1991b; Nikawa et al., 1987b) and the OPI3/PEM2 and CHO2/PEM1 genes, encoding the phospholipid methyltransferases required for conversion of PE to PC (Kodaki et al., 1991a; Koipally et al., 1996; Summers et al., 1988) (Fig. 1). The FAS1, FAS2 and ACC1/FAS3 genes involved in fatty acid biosynthesis, also contain functional copies of UASINO, alternately referred to as ICRE (inositol choline responsive element) (Schuller et al., 1992a; Schuller et al., 1992b; Schwank et al., 1995). The UASINO element, consensus sequence 5’CATGTGAAAT3’ (Bachhawat et al., 1995), has subsequently been found in the promoters of many other yeast genes (Carman and Han, 2011; Carman and Henry, 1999; Chen et al., 2007; Greenberg and Lopes, 1996; Henry et al., 2012; Kellis et al., 2003).
The INO1 gene, which carries multiple copies of UASINO, exhibits a higher repression/derepression ratio in response to inositol alone, or inositol plus choline, than any other UASINO containing gene (Jesch et al., 2006; Jesch et al., 2005; Santiago and Mamoun, 2003). The higher repression ratio of INO1 in comparison to other UASINO containing genes is in part attributable to the fact that its promoter also contains a copy of an upstream repression sequence, URS1, 5’ AGCCGCCCA 3’ (Lopes et al., 1993), found in a number of other genes in yeast not related to lipid metabolism. Reporter gene constructs derived from the native INO1 promoter lacking the URS element show increased expression under both repressing (presence of inositol and choline) and derepressing (absence of inositol and choline) conditions (Lopes et al., 1993). Mutations in the SIN3 gene also render the cell unable to fully repress the INO1, CHO1, CHO2 and OPI3 genes (Fig. 1) in the presence of inositol or inositol and choline, indicating that Sin3p plays a role in the full repression of these UASINO containing genes (Hudak et al., 1994). Sin3p reduces expression of INO1 by interaction, both with the URS1 element and with UASINO. Sin3p is a co-repressor and component of a negative regulatory complex, which also contains the Rpd3p histone deacetylase and Ume6p (Kadosh and Struhl, 1997, 1998; Kurdistani et al., 2002). While Ume6p negatively regulates INO1 through the URS1 element, it positively regulates CHO1, CHO2, and OPI3 (Jackson and Lopes, 1996) through an indirect effect on INO2 expression (Elkhaimi et al., 2000). Ume6p, as mentioned above, is also a positive regulator of PIS1 (Jani and Lopes, 2008). The Opi1p repressor interacts with Ino2p, as well as the pleiotropic corepressors, Sin3p and Ssn6p, at UASINO elements (Jaschke et al., 2011). These interactions result in recruitment of multiple histone deacetylases, including Rpd3p, Hda1p and Hos3p, which are collectively necessary for Opi1p mediated repression of UASINO containing genes by inositol and choline, (Grigat et al., 2012). Interestingly, global histone acetylation is regulated by acetyl CoA carboxylase, encoded by the ACC1 gene, (Galdieri and Vancura, 2012), which as described above contains UASINO element in its promoter. Galdieri and Vancura (Galdieri and Vancura, 2012) reported that attenuated expression of ACC1 results in increased bulk histone acetylation and altered transcriptional regulation, including increased INO1 expression. These results are consistent with the previous report by Shirra et al. (Shirra et al., 2001), which showed that reduced activity of Acc1p resulted in increased expression of INO1. Thus, histone acetylation/ deacetylation is, itself, regulated by the rate-limiting step in fatty acid biosynthesis.
However, full derepression of the INO1 gene also requires cooperation between the Ino2p/Ino4p heterodimer and a third HLH protein, Cbf1p (centromere binding factor 1), through a region distal to the INO1 promoter that encompasses an upstream open reading frame, SNA1. Binding of Cbf1 to upstream sites is required for binding of the ISW2 chromatin-remodeling complex, which is also required for full INO1 derepression (Shetty and Lopes, 2010). Cbf1p also interacts with Met4p, a transcriptional activator in the sulfur assimilation pathway, which is required for activation of genes, including SAM1 and SAM2, described above, which are required for maintaining levels of Sadenosyl methionine (SAM) (Hickman et al., 2011; Petti et al., 2012), the methyl donor in the phospholipid methylation reactions catalyzed by Cho2p and Opi3p in the synthesis of PC from PE (Fig. 1). Whereas Met4p is required for SAM2 activation, Opi1p is a direct repressor of SAM2 (Hickman et al., 2011).
The fact that the INO2 and INO4 genes also contain UASINO like elements in their own promoters and are subject to auto-regulation (Ashburner and Lopes, 1995a) introduces a further level of complexity in the regulation of INO1 and other UASINO containing genes. Thus, INO2 requires both Ino2p and Ino4p for expression and is regulated by inositol and choline. In contrast, INO4 is auto-regulated, requiring only Ino4p for its expression, and is constitutively expressed in the presence of inositol and choline (Ashburner and Lopes, 1995a). A further study, in which the INO2 gene was placed under the control of the GAL1 promoter, revealed that cells containing the GAL1 driven INO2 construct could still regulate expression of both INO1 and CHO1 in response to inositol. However, the level of expression of these two structural genes under the control of the GAL1 promoter was correlated, both in the absence and in the presence of inositol, to the relative level of expression of the INO2 gene. However, deletion of the OPI1 gene in a strain carrying INO2 under the control of GAL1 driven INO2 construct led to constitutive expression of INO1 in the presence of inositol (Ashburner and Lopes, 1995b). These results indicate that Opi1p, rather than Ino2p, is the sensor for the regulatory signal that is generated in the presence of inositol and is responsible for repression of UASINO containing genes. This insight was later validated, as will be discussed below, when Opi1p was found to interact in the endoplasmic reticulum (ER) with the phospholipid precursor, phosphatidic acid (PA), which is highly elevated in cells grown in the absence of inositol (Loewen et al., 2004). However, a low residual level of expression and regulation of INO1 in response to inositol and choline was observed in strains carrying various combinations of deletions of the OPI1, INO2, INO4 and SIN3 genes, namely: opi1Δ ino4Δ, opi1Δ ino2Δ ino4Δ, sin3Δ ino2Δ ino4Δ and opi1Δ sin3Δ ino2Δ ino4Δ (Graves and Henry, 2000), suggesting that the full complexity of this regulation has yet to be completely described.
As described above, the INO1 gene shows a much higher repression/derepression ratio in response to availability of inositol than any other gene in the yeast genome, including all other UASINO containing genes such as OPI3 and CHO1 (Jesch et al., 2006; Jesch et al., 2005; Santiago and Mamoun, 2003). Inositol auxotrophy (Ino− phenotype) (Villa-Garcia et al., 2011; Young et al., 2010), and/or reduced INO1 transcription, has been observed in many other mutants, including mutants defective in components of RNA polymerase II (Berroteran et al., 1994; Hampsey, 1997; Nonet and Young, 1989; Scafe et al., 1990), the RNA polymerase II SRB/mediator complex and the Paf1 and CCR4-NOT complexes (Betz et al., 2002; Koleske et al., 1992). Mutants defective in components of many other complexes involved in transcriptional regulation, including SWI/SNF (Peterson and Herskowitz, 1992), Ino80 (Ebbert et al., 1999; Fernandez-Murray et al., 2009; Shen et al., 2003a), ADA and SAGA complexes (Gansheroff et al., 1995; Horiuchi et al., 1997; Roberts and Winston, 1996) and subunits of the Set3p deacetylase complex (Cohen et al., 2008) exhibit Ino− phenotypes. In addition, mutants defective in several steps in the production of the inositol polyphosphates (Fig. 1), also exhibit Ino− phenotypes (Villa-Garcia et al., 2011; Ye et al., 2013). This phenotype is likely due to misregulation of INO1 transcription (Shen et al., 2003b; Steger et al., 2003; Ye et al., 2013). Inositol polyphosphates regulate transcription by controlling the activity of chromatin remodeling complexes. Other nuclear functions of inositol polyphosphates also include regulation of mRNA export as well as telomere maintenance (York, 2006). To date, eight distinct inositol polyphosphates have been detected in yeast (Wilson et al., 2013), which are produced by the phospholipase C mediated turnover of PI(4,5)P2, and the subsequent phosphorylation of IP3 (Fig. 1). Overall, these phenotypes suggest that transcription of the INO1 gene is more sensitive to perturbations in the cellular transcription apparatus than other UASINO containing genes. In addition, screening of genome wide collections of viable deletion mutants for the Ino− phenotype has revealed many additional mutants with the Ino− phenotype, which are not directly involved in lipid metabolism or in RNA II polymerase mediated transcription. Many of these additional mutants have defects in various stress response pathways or in membrane trafficking (Villa-Garcia et al., 2011; Young et al., 2010), topics discussed in the final sections of this review.
3. Mutations in structural genes encoding a number of enzymes involved in phospholipid biosynthesis affect regulation of INO1
Yeast does not synthesize choline de novo, but rather in the absence of exogenous choline, relies on synthesis of PC via the CDP-DAG pathway, starting with the synthesis of PA and passing through the intermediates CDP-DAG, PS and PE (Fig. 1) (McMaster and Bell, 1994a). Surprisingly, strains carrying mutations in structural genes encoding enzymes involved in biosynthesis of PC via the CDP-DAG pathway also exhibit phenotypes indicative of misregulation of INO1. These regulatory phenotypes are not seen in mutants defective in the Kennedy pathway for PC biosynthesis via diacylglycerol (DAG) (Fig. 1) for PC biosynthesis (Henry et al., 2012; Henry and Patton-Vogt, 1998). Specifically, mutants defective in the CDP-DAG pathway exhibit regulatory phenotypes including elevated expression of INO1, even in the presence of inositol, as well as the Opi− phenotype in the absence of inositol. In cho1, cho2 and opi3 mutants, these regulatory phenotypes are “conditional”. In these mutants, constitutive overexpression of INO1 and coregulated genes is eliminated when the mutant in question is provided with a soluble precursor of PC that can enter the Kennedy pathway, bypassing the mutated step in the CDP-DAG pathway, thus restoring PC synthesis. The conditional regulatory phenotypes in these mutants led to the hypothesis that a signal necessary for repression/derepression of INO1 and co-regulated genes is generated in the course of active phospholipid biosynthesis (Henry and Patton-Vogt, 1998). As described below, these insights, ultimately and collectively, led to the discovery that build up of PA, precursor to all phospholipids, as well as DAG and TAG (Fig. 1), is the metabolic signal responsible for derepression of the UASINO containing genes (Henry et al., 2012; Henry and Patton-Vogt, 1998; Loewen et al., 2004).
3.1. Some mutations in structural genes encoding enzymes of PC biosynthesis exhibit misregulation of INO1
The first structural gene mutant defective in PC biosynthesis to be identified as having altered regulation of Ino1p was the opi3 mutant, which was isolated on the basis of its Opi− phenotype in the same genetic screen as the original opi1-1 mutant (Greenberg et al., 1982b). The opi3 mutant was subsequently shown to be defective in synthesis of PC via PE methylation (Fig. 1) (Greenberg et al., 1983). The original opi3 mutant synthesized only 2–3% of the level of PC found in wild type cells and yet did not require choline supplementation for growth. The opi3Δ mutant (McGraw and Henry, 1989), similar to the original opi3 mutant isolated by Greenberg et al. (Greenberg et al., 1983), also does not require choline for growth. When grown in the absence of inositol and choline, opi3 mutants accumulate high levels of the intermediates phosphatidylmonomethylethanolamine (PMME) and phosphatidyldimethylethanolamine (PDME) (Fig. 1). In comparison, wild type strains produce only trace amounts of these compounds (Greenberg et al., 1983). The OPI3 gene (also known as PEM2) was shown to encode the phospholipid methyltransferase that is primarily responsible for catalyzing the final two methylations in the conversion of PE to PC (i.e. the conversion of PMME to PDME and PDME to PC, Fig. 1) (Kodaki and Yamashita, 1987, 1989; McGraw and Henry, 1989; Summers et al., 1988). However, the presence of choline in the growth medium restores of PC biosynthesis in opi3 mutants by the Kennedy pathway (Fig. 1), and simultaneously eliminates the Opi− phenotype and restores repression of INO1 in response to inositol (Greenberg et al., 1983; McGraw and Henry, 1989). The cho2Δ (pem1Δ) mutant, defective in the methyltransferase, which is primarily responsible for the first of the three methylations required for the synthesis of PC from PE (Fig. 1), exhibits similar conditional regulatory phenotypes (Summers et al., 1988). Similar to opi3Δ, the cho2Δ mutant has an Opi− phenotype and exhibits constitutive expression of INO1. However, in the case of cho2Δ mutant, these regulatory phenotypes are eliminated not just by the presence of exogenous choline, but also by MME and DME, precursors that can enter the Kennedy pathway, bypassing the block in PE methylation, thus restoring PC synthesis (Fig. 1) (Summers et al., 1988).
The cho1 mutants, defective in PS synthase (Fig. 1), cannot synthesize PS under any growth condition. Despite being unable to synthesize PS under any growth condition, cho1 mutants, including cho1Δ, are able to grow if provided with ethanolamine or choline (ethanolamine/choline auxotrophy) (Atkinson et al., 1980a; Atkinson et al., 1980b; Kovac et al., 1980). However, cho1 mutants express Ino1p even in the presence of inositol when shifted to medium lacking both ethanolamine and choline. cho1 mutants also overproduce and excrete inositol (Opi− phenotype) when shifted to medium lacking ethanolamine and choline as well as inositol (Letts and Henry, 1985). cho1, cho2 and opi3 mutants all accumulate metabolic precursors below the specific metabolic “bottleneck” resulting from their individual mutations in the CDP-DAG pathway leading through PS and PE to PC (Fig. 1). Moreover, down-regulation of expression of SAH1, encoding S-adenosyl-L-homocysteine hydrolase, which is necessary for the degradation of S-adenosyl-L-homocysteine, a potent inhibitor of adenosyl-methionine dependant methyltransferases, including Cho2p and Opi3p, leads to decreased PC synthesis and also results in derepression of INO1 (Malanovic et al., 2008). In the cho1, cho2 and opi3 mutants, the provision of an exogenous precursor that enters the Kennedy pathway above the mutational block specific to each mutant enables PC biosynthesis via the Kennedy pathway. In each case, the restoration of PC biosynthesis simultaneously eliminates the Opi− phenotype and restores INO1 repression in response to inositol. Thus, ethanolamine and choline restore INO1 regulation and PC synthesis in cho1 mutants, while MME, DME or choline suffice in cho2 mutants, but only choline can restore INO1 regulation in response to inositol in opi3 mutants (Henry and Patton-Vogt, 1998).
3.2. Effects on INO1 regulation of combinations of mutations in the Kennedy and CDP-DAG pathways for PC biosynthesis
In wild type cells, however, exogenous choline influences INO1 regulation only if inositol is also present (Hirsch and Henry, 1986) and mutations in the Kennedy pathway, including and pct1 have little effect by themselves on INO1 regulation (Griac et al., 1996). In a cho2Δ pct1Δ ept1Δ strain, however, INO1 expression is impervious to repression by every combination of inositol, choline and ethanolamine supplementation, indicating that an active Kennedy pathway is required for restoration of regulation by inositol in the cho2Δ mutant (Griac et al., 1996). The EPT1 and PCT1 (originally named CPT1) are also UASINO containing genes and are regulated in response to inositol and choline following the same general pattern as INO1. Morash et al. (Morash et al., 1994) studied the influence of the Kennedy pathway on various aspects of phospholipid metabolism and regulation in response to exogenous inositol and choline in pct1Δ, ept1Δ and pct1Δept1Δ strains. They showed that pct1Δ strains do not repress EPT1 in response to inositol and choline, whereas the ept1Δ mutation has no effect on regulation of PCT1. McMaster and Bell (McMaster and Bell, 1994b) used a variety of constructs and strains to dissect the influence of Ept1p and Cpt1p on both choline uptake and regulation of PC biosynthesis in response to inositol. They observed that the rate incorporated at which labeled serine was into PC at which labeled serine was reduced in wild type, pct1Δ and ept1Δ strains when inositol was present. Serine enters the phospholipid biosynthesis via the reaction catalyzed by PS synthase (Cho1p, Fig 1). However, the pct1Δ strain showed reduced incorporation of 32P into PC in both wild type and ept1Δ strains in the presence of inositol, despite showing no change in the rate of incorporation of labeled serine into PC under these same conditions. McMaster and Bell (McMaster and Bell, 1994b) concluded that this result could only be explained by turnover of PC produced via the CDP-DAG pathway, thereby releasing free choline, which is reincorporated into PC via the Kennedy pathway (Fig. 1) in both wild type and ept1Δ strains. However, reincorporation of choline produced by PC turnover is blocked in the pct1Δ mutant (Fig. 1), thus explaining the reduced 32P incorporation into PC in this mutant. Indeed, these results reported by McMaster and Bell (McMaster and Bell, 1994b) foreshadowed the subsequent observation by Patton-Vogt et al. (Patton-Vogt et al., 1997) that mutants in the Kennedy pathway, which are unable to reincorporate free choline produced via PC turnover, excrete it into the medium.
Using combinations of mutants defective in the CDP-DAG and Kennedy pathways for PC biosynthesis, Griac et al. (Griac et al., 1996) showed that the ability of such mutants to repress INO1, and by inference all other UASINO containing genes, is not specific to the route by which synthesis of PC occurs (i.e. via the Kennedy versus CDP-DAG pathways, Fig. 1). Nor is any single intermediate in the Kennedy pathway or in the pathway from CDP-DAG to PC responsible for the regulatory signal that controls repression/derepression of these genes in response to inositol. Rather, the evidence suggested that the overall ability of the cell to synthesize PC somehow created a signal necessary for the cell to be able to sense inositol (Griac et al., 1996). This hypothesis, however, proved to be only partially correct. In mutants blocked in PC synthesis via CDP-DAG, the signal for repression of INO1 is not PC synthesis itself, but rather the build up of PA, the immediate precursor of CDP-DAG (Fig. 1). As discussed below, under certain growth conditions, mutations in any of the steps from PA to PC via CDP-DAG can produce metabolic bottlenecks that result in the accumulation of PA. PA accumulation is the signal responsible for derepression of INO1 and other UASINO containing genes in the absence of inositol, as well as the constitutive over-expression these genes and the Opi− phenotype in the mutants defective in the CDP-DAG pathway (Henry et al., 2012; Henry and Patton-Vogt, 1998; Loewen et al., 2004; Sreenivas et al., 1998).
While the cho2 and opi3 mutations slow metabolism from PE to PC, exhibit decreased PC synthesis and misregulation of INO1, resulting in the Opi− phenotype (Griac et al., 1996; Kodaki and Yamashita, 1987, 1989; McGraw and Henry, 1989; Summers et al., 1988), these mutants, unlike cho1 mutants, are not choline auxotrophs. The lack of choline auxotrophy in cho2 and opi3 mutants is presumably due, both to the overlapping substrate specificities of the phospholipid methyltransferase enzymes encoded by the OPI3 and CHO2 genes and the ability of PDME, but not PMME, to substitute for PC to some degree in membrane biogenesis and function (Griac et al., 1996; McGraw and Henry, 1989; Summers et al., 1988). While the cho2Δ opi3Δ double deletion strain is auxotrophic for choline, it continues to grow for some time after a shift to medium lacking choline, finally ceasing growth when PC composition drops below 2% (Boumann et al., 2006). During the process of PC depletion in this strain, both PE and PI accumulate and considerable acyl chain remodeling of PE occurs, leading to shortening and increased saturation of the acyl chains, suggesting a regulatory mechanism that compensates for the nonbilayer propensity of PE. However, Choi et al. (Choi et al., 2004) showed that the auxotrophic requirement of a cho2Δ (pem1Δ) opi3Δ (pem2Δ) strain could also be satisfied in glucose grown cells by propanolamine (Prn), which is incorporated into phospholipid to form phosphatidylpropanolamine (PPrn). This leads to formation of cellular membranes essentially devoid of methylated phospholipids, with no compensatory shift in saturation of the acyl chains of PE or PPrn. Since PPrn is a hexagonal phase forming phospholipid like PE, these data suggest that the functions of PC, and methylated phospholipids in general, are nonessential in glucose grown cells of S. cerevisiae (Choi et al., 2004). However, expression and regulation of INO1 and coregulated genes has not been examined in this strain under these conditions.
As additional structural genes encoding the enzymes that catalyze the reactions in phospholipid biosynthesis were identified and characterized, it became possible to examine the effect on regulation of INO1 and co-regulated genes of mutations in each step along the CDP-DAG pathway leading to PC. However, the identification and cloning of the two genes in yeast encoding PS decarboxylases proved especially challenging, as neither of the single deletion mutants ultimately proved to have an auxotrophic requirement for ethanolamine or choline and strains carrying the single deletions retain considerable PS decarboxylase activity (Clancey et al., 1993; Trotter et al., 1993; Trotter et al., 1995; Trotter and Voelker, 1995). The PSD1 gene, encoding the mitochondrial form of PS decarboxylase, was ultimately isolated in two different laboratories virtually simultaneously each using somewhat different strategies (Clancey et al., 1993; Trotter et al., 1993). The yeast psd1Δ strain was then used to isolate ethanolamine auxotrophs, thereby identifying psd1Δ psd2 double mutants that retain virtually no PS decarboxylase activity and thus, similar to cho1 mutants, require ethanolamine or choline for growth (Trotter et al., 1995; Trotter and Voelker, 1995). The ethanolamine auxotrophy of the psd1Δ psd2 strain enabled the cloning of the PSD2 gene and the creation of a double deletion strain, psd1Δ psd2Δ, which retains no detectable PS decarboxylase activity and is an ethanolamine/choline auxotroph (Trotter et al., 1995). In wild type strains, transcription of PSD1 is regulated by inositol and ethanolamine, in a fashion similar to the CHO1 gene, while transcription of the PSD2 gene is not affected by any of these phospholipid precursors (Griac, 1997). The psd1Δ strain also fails to repress INO1 in the absence of ethanolamine and exhibits INO1 overexpression in the absence of both inositol and ethanolamine. However, the disruption of the PSD2 gene has no effect on either INO1 or PSD1 expression and PSD2 itself is not regulated in response to inositol or ethanolamine (Griac, 1997).
3.3. Mutations in the structural genes encoding PI synthase and CDP-DAG synthase also affect INO1 expression
A subsequent screen for mutants with Opi− phenotypes resulted in the identification of a mutant, initially named cdg1, which exhibited a very strong inositol excretion (Opi−) phenotype and very high constitutive INO1 expression, phenotypes that could not be corrected by choline supplementation. The cdg1 mutant exhibited changes in phospholipid synthesis consistent with a defect in CDP-DAG synthase, including very reduced synthesis of CDP-DAG and high PA levels (Klig et al., 1988a). This mutant was later shown by Shen et al. (Shen et al., 1996) to carry a point mutation in the structural gene for CDP-DAG synthase (CDS1), an essential gene. By modulating expression of the wild type CDS1 gene, Shen and Dowhan (Shen and Dowhan, 1997) showed that INO1 and CHO1 mRNA levels increased as expression of CDS1 decreased. These findings are consistent with the phenotype of the original cdg1 point mutant (Klig et al., 1988a), as well as the model of regulation of these genes in response to PA accumulation, as described above.
However, expression of the PIS1 gene, encoding phosphatidylinositol (PI) synthase was found to be unaffected in strains with modified CDS1 expression (Shen and Dowhan, 1997). The PIS1 gene is regulated by carbon source (Anderson and Lopes, 1996) and under anaerobic conditions PIS1 is also regulated by Rox1p, which is a repressor of hypoxic genes (Gardocki and Lopes, 2003). While PIS1 expression was originally believed to be completely uncoupled from inositol/choline regulation (Anderson and Lopes, 1996; Gardocki and Lopes, 2003), Jani and Lopes (Jani and Lopes, 2008) later showed that expression of PIS1 is induced about two fold in response to inositol, by a mechanism that is independent of the Ino2p and Ino4p transcription factors. Regulation of PIS1 does, however, involve Ume6p, a general regulator of INO1 and other phospholipid biosynthetic genes (Jackson and Lopes, 1996), as discussed above. Indeed, both Ume6p and Ino4p are positive regulators of PIS1 (Jani and Lopes, 2008). The PIS1 gene is also regulated by zinc through the interaction of the Zap1p transcription factor with the UASZRE element in the PIS1 promoter under conditions of zinc depletion (Carman and Han, 2007; Han et al., 2005). The DPP1 gene encoding diacylglycerol pyrophosphatase is also among the genes induced by Zap1p under zinc depletion (Carman and Han, 2007; Han et al., 2001; Henry et al., 2012).
Importantly, the level of PIS1 expression affects expression of UASINO containing genes. When PIS1 expression was placed under the control of the GAL1 promoter allowing PI synthesis to be reduced, the INO1 and CHO1 genes were derepressed and an Opi− phenotype was observed (Jani and Lopes, 2009). Thus, a reduction in PI synthesis in response to reduced PIS1 expression mimics the reduction of PI synthesis that occurs in the absence of exogenous inositol supplementation in wild type cells. Thus, both reduced expression of PIS1 and growth of wild type cells in the absence of inositol lead to decreased PI synthesis and INO1 derepression (Jani and Lopes, 2009) (Gaspar et al., 2006; Henry and Patton-Vogt, 1998; Loewen et al., 2004), but decreased PI synthesis itself is not the signal for activation of INO1. This is made clear by the fact that mutants, such as cho2 and opi3, which are defective in synthesis of PC via the CDPDAG pathway, have elevated PI levels, while expressing INO1 constitutively and exhibiting Opi− phenotypes (Henry and Patton-Vogt, 1998).
3.4. PA provides the metabolic signal for derepression/repression of UASINO containing genes in response to inositol
In each of the studies described above, in which derepression of INO1 and/or an Opi− phenotype are observed, PA levels were either directly shown to have increased or could be predicted to have increased on the basis of metabolic flux (Henry and Patton-Vogt, 1998). Indeed, PA metabolism is influenced by the rates of synthesis and turnover of essentially all lipids that are derived directly or indirectly from it (Carman and Henry, 2007; Henry et al., 2012). However, none of the studies cited above specifically perturbed PA metabolism without having the consequence of also affecting the flow and levels of intermediates from PA to PC along the CDP-DAG pathway (Fig. 1), making it difficult to determine the specific metabolic signal involved. A breakthrough in “pinpointing” PA as the actual metabolic signal responsible for INO1 derepression came as the result of discovery of both Opi− and choline excretion (Opc−) phenotypes (Patton-Vogt et al., 1997) in strains carrying a temperature sensitive mutation in the essential SEC14 gene, encoding the yeast phosphatidylinositol transfer protein (PITP) (Bankaitis et al., 1990; Bankaitis et al., 2005).
The sec14ts mutant itself was isolated in the original screen for temperature sensitive (Sec−) mutants, defective in secretion of invertase, a periplasmic enzyme (Novick et al., 1980). The SEC14 gene was shown to encode an essential, cytoplasmic protein, which functions to promote protein export from the Golgi (Bankaitis et al., 1989). Sec14p was subsequently identified as a phosphatidylinositol transfer protein (PITP) (Bankaitis et al., 1990), involved in regulation of diverse cellular functions (Bankaitis et al., 2005). The complex and diverse roles of Sec14p and related proteins in yeast include coordination of PI and PC metabolism, regulation of PC metabolism, phosphoinositide signaling and promotion of Golgi secretory and vesicular transport from the Golgi and have been extensively described in a number of comprehensive reviews (Bankaitis et al., 2005; Ghosh and Bankaitis, 2011; Griac, 2007; Howe and McMaster, 2006; LeBlanc and McMaster, 2010). Second site mutations, referred to as “bypass suppressors”, that suppress sec14 lethal growth defects were isolated and shown to include the cki1, cct1 and pct1 mutations in the Kennedy pathway for PC biosynthesis (Cleves et al., 1991). Skinner et al. (Skinner et al., 1995) subsequently reported that Sec14p influences the activity of the Kennedy pathway through inhibition of choline-phosphate cytidylyltransferase (Pct1p) (Fig. 1), the rate-limiting enzyme in the pathway.
The sec14ts strains carrying Kennedy pathway bypass suppressors exhibit high levels of PA and INO1 expression, coinciding with elevated PC synthesis and turnover (Patton-Vogt et al., 1997; Sreenivas et al., 1998). These observations provided the first clear evidence that increased accumulation of PA, rather than decreased PC synthesis, is responsible for the signal for activation of INO1 expression (Sreenivas et al., 1998). The sec14tscki1Δ, sec14tscct1Δ and sec14tspct1Δ strains all possess inositol excretion (Opi−) phenotypes and very strong choline excretion (Opc−) phenotypes when growing at the sec14ts restrictive temperature of 37°C (Patton-Vogt et al., 1997). While wild type cells do not excrete detectable choline in the Opc− assay, the cki1Δ, cct1Δ and pct1Δ mutants all exhibit mild Opc− phenotypes, because they turn over PC via a phospholipase D (Fig. 1) mediated route, liberating both free choline and PA (Fig. 1). Yet, unlike wild type, mutants with defects in the Kennedy pathway are unable to reincorporate choline produced during PC turnover catalyzed by phospholipase D (Spo14/Pld1p) back into PC and, thus, excrete it into the medium (Patton-Vogt et al., 1997). However, sec14tsckiΔ, sec14tscctΔ and sec14tspct1Δ strains all exhibit enormously increased choline excretion rings at the sec14ts restrictive temperature in comparison to strains carrying only the respective Kennedy pathway mutations (Patton-Vogt et al., 1997). Sreenivas et al. (Sreenivas et al., 1998) demonstrated that both the Opi− and elevated Opc− phenotypes of sec14ts strains carrying bypass suppressors in the Kennedy pathway are dependent on activation of the phospholipase D encoded by the SPO14 (PLD1) gene. High PC turnover catalyzed by phospholipase D produces both excess choline, resulting in the Opc− phenotype, and high PA levels and derepression of INO1, leading to the Opi− phenotype (Fig. 2). Deletion of the SPO14 gene, encoding phospholipase D, simultaneously eliminated the Opi− and the Opc− phenotypes in sec14 “bypass” strains at a temperature semipermissive for sec14ts growth. High PA levels produced by Spo14p (Pld1p) turnover of PC, in turn, proved to be responsible for high INO1 expression and the Opi− phenotype in these strains (Sreenivas et al., 1998). These phenotypes provided the pivotal clue that elevated PA, rather than decreased PC synthesis, generates the signal controlling derepression of INO1 and other UASINO containing genes (Henry et al., 2012; Henry and Patton-Vogt, 1998; Sreenivas et al., 1998). Moreover, the activity of Spo14p proved to be essential for bypass suppression of sec14ts growth phenotypes by all classes of bypass suppressors of sec14, not just the suppressor mutations associated with the Kennedy pathway (Xie et al., 1998).
3.5. Discovery of the mechanism of regulation of INO1 and other UASINO genes in response to changing PA levels
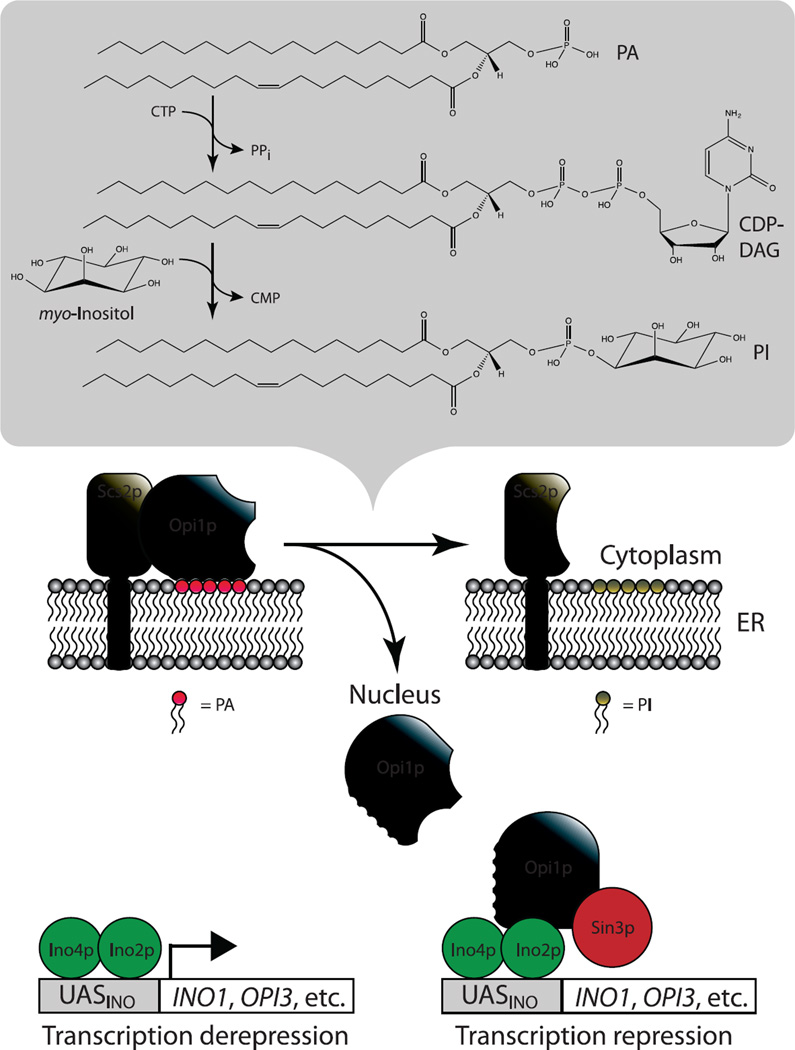
The question then remained; how does the level of PA communicate a signal to the transcription apparatus to control expression of INO1 and coregulated genes? Loewen et al. (Loewen et al., 2004) provided an answer to this question, demonstrating that Opi1p is a PA binding protein, which interacts with PA in the perinuclear ER. In addition to PA binding domains, Opi1p also contains a motif, FFAT (two phenylalanines in an acidic tract), shown by Loewen et al. (Loewen et al., 2003) to interact with an integral ER associated protein, Scs2p. Scs2p is a homologue of mammalian VAP (VAMP associated protein) which interacts with a number of additional lipid associated proteins in the ER. In the presence of exogenous inositol, PA remains at a low level as a consequence of being rapidly consumed as a precursor in the synthesis of high levels of PI (Fig. 1). Conversely, when wild type cells are growing in the absence of inositol, PI levels are low and PA levels remain high (Gaspar et al., 2006; Gaspar et al., 2011). Since Opi1p tethering to the ER requires interaction with both PA and Scs2p, Opi1p remains in the ER when cells are growing in the absence of inositol (Loewen et al., 2004) (Fig. 3). However, when inositol is abruptly added to the medium of cells acclimated to growth in its absence, PI synthesis increases dramatically, causing PA levels drop rapidly. The drop in PA results in loss of Opi1p binding to the perinuclear ER and, consequently, it rapidly translocates to the nucleus, where it represses INO1 and other Opi1p target genes (Loewen et al., 2004) (Fig. 3). Conversely, growth in the absence of inositol results in increased build up of PA, resulting in retention of Opi1p in the perinuclear ER. This in turn results in derepression of INO1 and coregulated UASINO containing genes (Jesch et al., 2006; Jesch et al., 2005; Loewen et al., 2004). Since the de novo synthesis of inositol supported by Ino1p does not support a rate of PI synthesis comparable to that in cells provided with exogenous inositol, in wild type cells growing in the absence of inositol, PI levels remain low and PA levels remain elevated (Gaspar et al., 2006; Gaspar et al., 2011).
Figure 3. Model for the regulation of UASINO-containing genes by changing PA levels.
In cells grown in the absence of inositol, PI levels are low due to the limited availability of endogenously produced inositol, which leads to the accumulation of PA. Under these growth conditions, the Opi1p transcriptional repressor localizes to the cytoplasmic face of the ER though dual interactions with PA and Scs2p, and the transcription of UASINO-containing genes, regulated by the Ino2p-Ino4p complex, are maximally derepressed. Upon the addition of inositol, PI synthesis dramatically increases, which causes the rapid consumption of the ER pool of PA. This drop in PA levels releases Opi1p, which translocates to the nucleus, where it binds to Ino2p and represses the transcription of UASINO-containing genes, which is mediated by the general transcriptional repressor Sin3p.
4. The critical roles of Pah1p and TAG metabolism in regulation of glycerolipid homeostasis in yeast
Opi1p is not the only protein that recognizes PA in the ER. Pah1p, a homolog of mammalian lipin 1 (Pascual and Carman, 2013), is a cytoplasmic Mg2+-dependent PA phosphatase that interacts with PA in the perinuclear ER (Han et al., 2007; Han et al., 2006). In mice overexpression of lipin 1 causes obesity, while its loss causes lipodystrophy (Peterfy et al., 2001; Phan and Reue, 2005). The fact that Pah1p shares sequence homology with lipin 1 at its N-terminal end within a HAD-like (haloacid dehydrogenase like) domain led to the hypothesis that lipin 1 is also a PA phosphatase enzyme (Han et al., 2006). Subsequent studies using purified human lipin 1 protein confirmed that lipin 1 is indeed a PA phosphatase enzyme (Han and Carman, 2010). While interaction of Opi1p with PA plays a pivotal role in regulating the expression of phospholipid biosynthetic genes (Henry et al., 2012; Loewen et al., 2004), Pah1p controls PA homeostasis (Han et al., 2006). In addition to Pah1p, yeast contains three additional enzymes with PA phosphatase activity; Lpp1p, Dpp1p, App1p (Chae et al., 2012; Faulkner et al., 1999; Toke et al., 1998a; Toke et al., 1998b; Wu et al., 1996). However, it is PA dephosphorylation by Pah1p that generates the pool of DAG used in the synthesis of TAG (Han et al., 2006), as well as the pool of DAG used in the synthesis of PE and PC via the Kennedy pathway (Carman and Han, 2006, 2009) (Fig. 1). Consequently, the pah1Δ mutant exhibits low TAG levels and elevated levels of PA and PI (Han et al., 2007; Han et al., 2006). The pah1Δ mutant also exhibits abnormal expansion of the nuclear membrane (Santos-Rosa et al., 2005).
Consistent with its elevated PA levels in comparison to wild type (Han et al., 2006), the pah1Δ mutant also exhibits elevated INO1 expression, both in the presence and absence of inositol (Han et al., 2007). However, unlike opi1Δ, the pah1Δ mutant does not exhibit an Opi− phenotype (Han et al., 2006) and INO1 is still subject to repression in response to inositol in the pah1Δ strain, although not to the same degree as in the wild type (Han et al., 2007). Pah1p is found in both cytoplasmic and membrane fractions in wild type yeast (Han et al., 2006). The association of Pah1p with the membrane, where its substrate PA resides, is essential to its function in vivo, and membrane association of Pah1p depends on the phosphorylation state of the enzyme (Choi et al., 2012; Choi et al., 2011; Karanasios et al., 2010; O'Hara et al., 2006). Dephosphorylation of Pah1p by the Nem1p-Spo7p phosphatase complex (Santos-Rosa et al., 2005) promotes both membrane anchoring of Pah1p via an amino-terminal amphipathic helix and its activity as a PA phosphatase (Karanasios et al., 2010). A short carboxy-terminal acidic peptide on Pah1p mediates its interaction with the Nem1p-Spo7p phosphatase complex that is localized to the nuclear membrane and ER. The Nem1p-Spo7p phosphatase complex is consequently important for Pah1p membrane translocation and production of DAG and TAG, as well as lipid droplet biogenesis (Karanasios et al., 2013). The unconventional diacylglycerol kinase, Dgk1p, catalyzes the formation of PA from DAG, utilizing CTP, instead of ATP, as the phosphate donor in the reaction (Han et al., 2008a; Han et al., 2008b). Dgk1p counteracts the function of Pah1p (Han et al., 2008a; Han et al., 2008b) but deletion of the DGK1 gene does not itself result in major changes in lipid metabolism or in INO1 expression in proliferating cells (Han et al., 2008a).
5. TAG synthesis and breakdown are interdependent with ongoing membrane lipid synthesis in actively growing cells
Because of its rapid and profound effects on the levels of both PI and PA in actively growing cells, inositol availability influences the levels and rates of synthesis and turnover of all lipids derived directly or indirectly from PA and PI, including inositol phospholipids and sphingolipids, DAG and TAG (Gaspar et al., 2006; Gaspar et al., 2011; Gaspar et al., 2008; Jesch et al., 2010). Accumulation of TAG, as cells enter into stasis, as well as TAG turnover as cells exit stasis and reenter active growth, has been extensively documented in yeast (Gray et al., 2004; Kurat et al., 2006; Kurat et al., 2009). However, changes in the availability of inositol also lead to dramatic changes in the rates of turnover and synthesis of TAG even during active growth (Gaspar et al., 2006; Gaspar et al., 2011).
5.1. The availability of inositol affects TAG accumulation and turnover in logarithmically growing wild type cells
When exogenous inositol is supplied to actively growing yeast cells that have been acclimated to growth in inositol free medium, PI synthesis increases rapidly and dramatically, while TAG synthesis declines (Gaspar et al., 2006; Gaspar et al., 2011). This is due to rapid channeling of PA through CDP-DAG into PI synthesis leading to a rapid decrease in PA levels (Loewen et al., 2004), making PA less available as a precursor for TAG synthesis (Gaspar et al., 2006; Gaspar et al., 2011). Conversely, lack of inositol supplementation results in a substantial reduction in synthesis of PI and an increase in TAG synthesis (Gaspar et al., 2006; Kelley et al., 1988). When choline is present in addition to inositol, PC synthesis also increases via the Kennedy pathway, consuming DAG, the immediate precursor to TAG (Fig. 1). Thus the presence of choline also acts in concert with inositol to counteract the build up of TAG during active growth. Thus, even during logarithmic growth, a complex competition between TAG and phospholipid synthesis is ongoing (Gaspar et al., 2011).
The cellular response to addition of inositol to wild type cells previously acclimated to growth in its absence includes a five-to six-fold increase in cellular PI content within a period of 30 minutes (Gaspar et al., 2006). This increase in PI content following addition of inositol is dependent upon fatty acids derived from multiple sources, including de novo fatty acid synthesis, PC turnover and TAG hydrolysis (Gaspar et al., 2006; Gaspar et al., 2011). The rapid increase in turnover of PC that occurs following inositol addition requires participation of Nte1p (Gaspar et al., 2006), an ER-localized phospholipase B (Zaccheo et al., 2004). Nte1p was also shown to be responsible for PC turnover in yeast when choline is present in the medium and/or when cells are grown at 37°C (Dowd et al., 2001).
As described above, TAG is also consumed in logarithmically growing wild type cells upon inositol reintroduction, coincident with the increase of PI content (Gaspar et al., 2011). Thus, upon shift to inositol free medium, the storage lipids in lipid droplets are also required to maintain normal rates of phospholipid synthesis in proliferating yeast cells (Gaspar et al., 2011). TAG degradation occurs via the activity of lipid hydrolases encoded by the TGL3, TGL4 and TGL5 genes (Athenstaedt and Daum, 2005; Kohlwein, 2010; Kurat et al., 2009). At the cellular level, TAG degradation is upregulated by Cdc28p/Cdk1p-dependent phosphorylation of the Tgl4p lipase (Kurat et al., 2009). Lipolysis contributes to bud formation, presumably by providing precursors for synthesis of lipids involved in membrane biogenesis or signaling (Kurat et al., 2009). The DAG generated from lipolysis is converted to PA by Dgk1p (Han et al., 2008a) and, consequently, dgk1Δ cells fail to resume growth from stationary phase in the presence of the fatty acid synthesis inhibitor cerulenin (Fakas et al., 2011). However, when choline is present, DAG can be used directly for PC synthesis (Fig. 1), bypassing the requirement of Dgk1p for growth resumption, thereby linking the Kennedy pathway for phospholipid synthesis to the mobilization of TAG (Fakas et al., 2011).
5.2. Cells unable to synthesize TAG exhibit an Ino− phenotype, despite being able to derepress INO1
The synthesis of TAG is catalyzed mainly by the diacylglycerol transferases encoded by the DGA1 and LRO1 genes (Oelkers et al., 2002; Oelkers et al., 2000; Sorger and Daum, 2002), whereas the enzymes encoded by ARE1 and ARE2 primarily mediate the esterification of ergosterol and its precursors leading to steryl ester production (Jensen-Pergakes et al., 2001; Sandager et al., 2000; Sandager et al., 2002). These four enzymes, Dga1p, Lro1p, Are1p and Are2p, account for TAG and steryl ester biosynthesis in yeast. Synthesis of these storage lipids begins during exponential growth and reaches its peak as cells enter stationary phase (Mullner and Daum, 2004). The dga1Δlro1Δare1Δare2Δ quadruple mutant is an inositol auxotroph (Ino− phenotype) at 37°C, a phenotype that is enhanced when choline is present (Gaspar et al., 2011). However, unlike many other strains exhibiting an Ino− phenotype, the dga1Δlro1Δare1Δare2Δ strain does not display a defect in INO1 derepression when shifted to inositol free medium (Gaspar et al., 2011). However, upon reintroduction of inositol into such inositol-deprived cultures, the dga1Δlro1Δare1Δare2Δ mutant strain exhibits slow recovery of PI content compared to wild type cells. This is primarily due to the absence of TAG as a source of fatty acids, which in wild type cells serve as precursors for PI synthesis upon recovery following inositol deprivation (Gaspar et al., 2011). Moreover, synthesis of phospholipids, especially PI, is dramatically reduced in the dga1Δlro1Δare1Δare2Δ strain even when it is growing continuously in the presence of inositol. The mutant also utilizes a greater proportion of newly synthesized PI than wild type for the synthesis of inositol containing sphingolipids, especially in the absence of inositol (Gaspar et al., 2011). Thus, storage lipid synthesis actively influences membrane phospholipid metabolism in logarithmically growing cells. Another indication of crosstalk between membrane biogenesis versus lipid storage is the observation that following a block in membrane trafficking, yeast cells channel lipid metabolism from phospholipid synthesis into synthesis of TAG and other neutral lipids to form lipid droplets (Gaspar et al., 2008).
6. Inositol starvation in an ino1 mutant leads to rapid cell death
As discussed above, S. cerevisiae, mutants carrying loss-of-function mutations in a large number of genes exhibit inositol auxotrophy (Culbertson and Henry, 1975; Donahue and Henry, 1981b; Villa-Garcia et al., 2011; Young et al., 2010). Inositol auxotrophs of Neurospora (Lester and Gross, 1959) and several inositol-requiring species of yeast (Ridgway and Douglas, 1958) were shown to die logarithmically when deprived of inositol, a trait referred to by Ridgeway and Douglas (Ridgway and Douglas, 1958) as “unbalanced growth”. A similar phenomenon of rapid exponential loss of viability was observed in the ino1-13 mutant of S. cerevisiae, defective in IP synthase, when starved for inositol (Henry et al., 1977). A similar phenomenon was also observed in fatty acid auxotrophs (fas1) mutants, defective in fatty acid synthase, but not in ole1 mutants defective in fatty acid desaturase, when starved for their respective fatty acid requirements (Henry, 1973; Henry and Horowitz, 1975). In the case of the ino1 and fas1 mutants, cell death could be largely prevented by simultaneously interrupting protein synthesis, either by starvation for an amino acid required by the strain in question or by treating with cycloheximide. The fact that the phenomenon of rapid cell death was observed in mutants defective in both inositol and fatty acid biosynthesis starved for their respective requirements, suggested that rapid and profound loss of viability might be a general phenomenon in mutants defective in membrane lipid biosynthesis (Henry et al., 1977). However, an ino1 cho1 strain, auxotrophic both for inositol and for ethanolamine/choline, lost viability at a rate comparable to the ino1 single mutant strain when starved for inositol, but showed little viability loss when starved for ethanolamine/choline, despite its abnormal membrane phospholipid composition (Atkinson et al., 1980b). Thus, the rapid cell death observed in the ino1 and fas1 mutants is not universally shared by all mutants possessing defects in membrane lipid synthesis.
6a. Inositol starvation in ino1 mutants leads to rapid cessation of PI synthesis
In the original study of the ino1-13 mutant undergoing inositol starvation, cells continued to double for the first two hours and then commenced to die logarithmically such that only 10% or less survived after 5 hours. Under these circumstances, ino1-13 cells arrested in all stages of budding, indicating that they did not undergo cell cycle arrest (Henry et al., 1977). By 4 hours of inositol starvation, PI synthesis had virtually ceased, while synthesis of PS, PE and PC continued at levels similar to the inositol supplemented control (Henry et al., 1977). Becker and Lester (Becker and Lester, 1977) examined the change in lipid content during inositol starvation of both the ino1-13 strain and a double mutant strain, ino1-13 ino4-8, and reported that PI levels dropped to virtually undetectable levels within 2 hours following a shift to inositol free medium. The drop in PI content was accompanied by a build up of PA and CDP-DAG. This early observation by Becker and Lester (Becker and Lester, 1977) of PA and CDP-DAG accumulation during inositol deprivation in inositol auxotrophs foreshadowed later experiments, discussed above, which showed that wild type cells grown in the absence of inositol also accumulate elevated levels of both PA and CDP-DAG (Gaspar et al., 2006; Gaspar et al., 2011; Loewen et al., 2004). Hanson and Lester (Hanson and Lester, 1980) subsequently reported that the rate of PI synthesis was drastically affected as early as 5–20 minutes following removal of inositol and was reduced by more than 90% after 1–2 hours of inositol starvation in the ino1-13 ino4-8 strain. They also observed a decrease of 50% in the rate of synthesis of the inositol containing sphingolipids within 5–20 minutes and 75% by 1–2 hours following removal of inositol in this strain. The synthesis of di- and tri-phosphoinositides, while affected less drastically than sphingolipid synthesis, also decreased measurably within 1–2 hrs following removal of inositol, as did cell wall glycan synthesis. Thus, inositol starvation was shown to rapidly affect the rate of synthesis of all inositol-containing lipids, as well as cell wall glycan, in a mutant completely lacking Ino1p.
6b. Starvation of ino1, ino2 and ino4 mutants for inositol leads to greater viability loss than starvation of other classes of auxotrophs
The question then arises; is “inositol-less death” in inositol auxotrophs a unique phenomenon related to the vital functions of inositol in metabolism and cellular function in yeast or is it triggered by a generic mechanism(s) similar to those operating during starvation of other classes of auxotrophic mutants? It has been postulated that yeast and other free living organisms have necessarily evolved mechanisms for survival during starvation for “natural nutrients”, such as carbon, phosphorous, nitrogen and sulfur, which vary in natural environments and which cells cannot manufacture (Boer et al., 2008). In contrast, mutants unable to synthesize an essential compound, such as leucine or uracil, which must be synthesized endogenously from available “natural” nutrients, have not had the benefit of natural selection to evolve protective mechanisms. Such mechanisms include concerted cell cycle arrest and conservation of energy stores when cells are confronted with the absence of a nutrient. Consequently, under starvation for a compound, such as leucine or uracil, for which auxotrophic cells lack an evolved protective mechanism, such auxotrophs may undergo exponential loss of viability with a half-life of several days (Boer et al., 2008).
Recent studies of starvation of methionine requiring mutants of S. cerevisiae defective in methionine biosynthesis (met6Δ and met13Δ), suggest that these mutants react to methionine starvation, as if starved for a natural nutrient such as phosphate, and undergo cell cycle arrest and consequently survive (Petti et al., 2011). In contrast, methionine auxotrophs having defects in the transcription factors Met31p or Met32p, which are involved in regulation of a broad array of metabolic functions, were found by Petti et al. (Petti et al., 2011) to respond to methionine starvation by undergoing logarithmic cell death in a fashion more similar to the response of leucine and uracil auxotrophs starved for their respective requirements (Boer et al., 2008). During inositol less death, however, cells of ino1, ino2 or ino4 mutant strains lost 3–5 log orders of viability within 24 hrs (Culbertson and Henry, 1975; Henry et al., 1975). This rate of viability loss is significantly more rapid and severe than that described for other classes of auxotrophs starved for their specific requirements, including the methionine, leucine and uracil auxotrophs (Boer et al., 2008; Petti et al., 2011) starved for their specific requirements. Inositol starved ino1-13 cells lost 4–5 log orders of viability within 24 hrs following inositol removal. Furthermore, “inositol-less death” can be used to enrich and select for spontaneous mutants exhibiting auxotrophy for leucine, methionine, tryptophan, lysine, histidine, uracil and adenine (Henry et al., 1975).
6c. Inositol starved ino1 mutants share some characteristics in common with temperature sensitive Sec− mutants raised to their restrictive temperature
While effects of inositol starvation availability are more severe than starvation for other types of auxotrophic requirements, similarities do exist in certain of characteristics of inositol starved ino1 mutants and temperature sensitive secretion (Sec−) mutants raised to their restrictive temperature. In both cases, metabolism continues while cell surface and membrane expansion ceases. When ino1-13 cells were shifted to inositol free medium, they underwent approximately one doubling in cell number and then arrested in all stages of the cell cycle. However, after cell number stopped increasing and both plasma membrane and cell wall ceased expanding, the cells continued to metabolize (Henry et al., 1977). This ongoing metabolism in cells no longer dividing or expanding in volume resulted in an osmotic imbalance (Atkinson et al., 1977) and cell density increased, causing the inositol starved cells to sediment more rapidly in a ludox gradient than the inositol-supplemented control cells (Henry et al., 1977). The sec1-1 mutant, the first yeast Sec− mutant to be isolated and characterized, was shown to be blocked in cell surface growth as well as secretion (Novick and Schekman, 1979). When a mixture sec1-1ts and wild type cells, both of which had been shifted to the sec1-1ts restrictive temperature for 3 hours, was applied to a ludox gradient similar to the one used in the inositol starvation experiment described above, the sec1-1 cells separated completely from wild type cells (Novick et al., 1980). This procedure was then used to enrich (select) for other classes of temperature sensitive Sec− mutants, leading to the isolation of the mutants representing the 23 original complementation groups of Sec− mutants isolated by Novick et al. (Novick et al., 1980).
Thus, by inference, all of the original Sec− mutants isolated in the screen by Novick et al. (Novick et al., 1980) share in common with inositol starved ino1 cells, the property of cessation of membrane synthesis and cell surface expansion, uncoupled from cessation of macromolecular synthesis. Moreover, Sec− strains shifted to restrictive growth temperatures and inositol deprived wild type cells both exhibit a build up of neutral lipids, especially TAG (Gaspar et al., 2011; Gaspar et al., 2008). The sec13-1 mutant, isolated in the original screen by Novick et al. (Novick et al., 1980), was later shown to be defective in COPII vesicle formation and secretory protein exit from the ER (Pryer et al., 1993; Salama et al., 1993). This mutant also exhibits an Ino− phenotype at a semi-permissive growth temperature (Gilstring et al., 1999) and pronounced TAG accumulation when shifted to its restrictive temperature (Gaspar et al., 2008). Moreover, the introduction of mutations in the TAG synthase genes into the sec13-1 mutant background caused a reduction in the permissive growth temperature at which the mutant could grow and resulted in the appearance of the Ino− phenotype at still lower growth temperatures (Gaspar et al., 2008). These findings suggest that secretory stress and inositol starvation both result in the channeling of lipid metabolism from membrane phospholipid synthesis into TAG and do so in a synergistic manner. Of the original Sec− mutants isolated by Novick et al. (Novick et al., 1980), only the sec13-1 (Gilstring et al., 1999) and sec14-1 mutants (Kearns et al., 1997) have been reported to exhibit inositol auxotrophy at a semi-permissive growth temperature. However, a screen of a genome-wide viable deletion collection for mutants exhibiting inositol auxotrophy identified an additional 54 non-essential genes encoding products involved in membrane trafficking that confer an Ino− phenotype (Villa-Garcia et al., 2011). These 54 mutants represent about 13% of the total known nonessential genes that exhibit the Ino− phenotype when mutated.
7. Stress response signaling is triggered by changes in lipid metabolism during inositol starvation
As described in previous sections, cells grown in the absence of inositol contain PI levels that are 4–5 times lower than cells grown in the presence of inositol, which leads to changes in the metabolism lipids derived from PI, including phosphoinositides (Jesch et al., 2010), sphingolipids (Alvarez-Vasquez et al., 2005; Jesch et al., 2010), as well as GPI anchors (Doering and Schekman, 1996). These lipid metabolic changes are accompanied by the activation of several stress response signaling pathways, most notably the UPR pathway (Cox et al., 1993; Cox and Walter, 1996; Kohno et al., 1993; Mori et al., 1993; Mori et al., 1992; Nikawa and Yamashita, 1992), as discussed above, and the cell integrity pathway controlled by the highly conserved protein kinase C-MAP kinase (PKC-MAPK) signal transduction pathway (Jesch et al., 2010; Nunez et al., 2008). Moreover, deletion of components of these and other stress response pathways leads to an Ino− phenotype (Villa-Garcia et al., 2011), indicating that activation of these stress responses is required for growth in the absence of inositol and that growth of wild type yeast in the absence of inositol is a stress inducing growth condition. Indeed, an early indication that stress response signaling is triggered by inositol starvation was the finding that mutants in the UPR pathway are inositol auxotrophs (Cox et al., 1993; Nikawa et al., 1996; Nikawa and Yamashita, 1992; Sidrauski et al., 1996). In fact, IRE1, the gene encoding the transmembrane kinase that senses ER stress (Cox and Walter, 1996; Mori et al., 2000) was originally identified in a screen for inositol auxotrophs, and its name — Inositol REquiring — indicates this fact (Nikawa and Yamashita, 1992). Initially it was believed that the UPR pathway regulates the expression of INO1 and other UASINO-regulated genes (Cox et al., 1997), suggesting that the UPR directly controls the transcription of phospholipid biosynthetic gene expression. However, it has been demonstrated that neither INO1 nor UASINO-containing genes, in general, are directly controlled by the UPR (Chang et al., 2002; Jesch et al., 2006).
7a. Inositol deprivation triggers the UPR in wild type cells
The UPR is thought to maintain ER homeostasis by balancing protein folding in the luminal ER with secretory capacity (Gardner et al., 2013; Korennykh and Walter, 2012; Walter and Ron, 2011). In yeast, activation of the UPR is solely controlled by Ire1p, which senses ER stress produced by inositol starvation, secretory stress, or accumulation of unfolded ER luminal proteins (Walter and Ron, 2011). Current models for UPR activation in yeast suggest that Ire1p, a highly conserved transmembrane ER resident protein containing both luminal and cytosolic domains, directly binds to unfolded proteins in the lumen of the ER, which induces its oligomerization and activation (Credle et al., 2005). Activated Ire1p subsequently catalyzes the unconventional splicing of HAC1 mRNA, leading to translation of the Hac1p transcription factor and up-regulation of UPR target genes, including ER resident protein-folding chaperones and oxidoreductases (Travers et al., 2000), which together function to maintain ER homeostasis (Walter and Ron, 2011). Moreover, ire1Δ and hac1Δ mutants both exhibit inositol auxotrophy (Cox et al., 1993; Nikawa et al., 1996; Nikawa and Yamashita, 1992).
Despite a wealth of studies investigating the mechanism of UPR activation, it remains unclear how inositol deprivation triggers ER stress. Paradoxically, although inositol starvation is a potent activator of the UPR (Chang et al., 2002; Cox et al., 1997; Jesch et al., 2006; Pincus et al., 2010), unfolded proteins do not appear to build up in the ER of cells starved for inositol. In a recent study using a pelleting assay to measure unfolded protein aggregates in the ER lumen, Promlek et al. (Promlek et al., 2011) did not detect accumulated unfolded proteins in inositol-depleted cells, suggesting that inositol depletion leads to membrane lipid aberrations that are sensed by Ire1p in the absence of unfolded proteins. This proposal is supported by the observation of Promlek et al. (Promlek et al., 2011) that ire1 mutant cells, lacking a critical unfolded protein interaction domain in the luminal domain of Ire1p, exhibited identical activation kinetics during inositol starvation compared to wild type cells deprived of inositol. Notably, UPR activation by treatment of these same ire1 mutants with DTT or tunicamycin, both of which induce the accumulation of unfolded proteins in the ER, was significantly reduced (Promlek et al., 2011). These results suggest that the cytoplasmic or transmembrane domain of Ire1p senses membrane aberrancy, a conclusion which is in agreement with recent experiments in mammalian cells showing that accumulation of saturated fatty acids triggers UPR (Volmer et al., 2013). Therefore, inositol starvation may trigger the UPR by producing ER stress that is caused by membrane- or lipid-related changes in the ER and is distinct from a build up of unfolded proteins.
One possible source of ER stress produced by inositol starvation may be through its effects on sphingolipid metabolism. As described above, inositol induces changes in the synthesis and levels of numerous lipids, including the sphingolipids (Alvarez-Vasquez et al., 2005; Jesch et al., 2010). Moreover, several recent studies indicate that sphingolipid metabolism may regulate ER membrane homeostasis. Mutant cells lacking ORM1 and ORM2, which are members of the conserved ORMDL family of ER membrane proteins that negatively regulate sphingolipid metabolism (Breslow et al., 2010; Han et al., 2010), exhibit a constitutive UPR (Han et al., 2010). Mutant orm1Δ orm2Δ cells contain elevated levels of sphingolipids (Breslow et al., 2010; Han et al., 2010) and are hypersensitive to stress, including inositol starvation (Han et al., 2010). Similarly, isc1Δ mutant cells, which contain elevated sphingolipids due to a block in sphingolipid turnover (Sawai et al., 2000), also exhibit constitutive UPR (Gururaj et al., 2013). Conversely, inhibiting sphingolipid synthesis with myriocin suppresses the activation of the UPR by inositol starvation, but not by DTT treatment (Promlek et al., 2011). These results suggest the intriguing possibility that elevated sphingolipids in the ER lead to membrane perturbations that activate the UPR independent of unfolded protein accumulation. However, much additional work will be required to substantiate this hypothesis.
Another possible source of ER stress produced by inositol starvation may be through its effects on membrane trafficking. Similar to inositol deprivation in wild type cells, secretory stress caused by slowing or blocking membrane trafficking at multiple steps in the secretory pathway in various Sec− mutants also leads to activation of the UPR (Chang et al., 2004; Chang et al., 2002; Jonikas et al., 2009). However, membrane trafficking does not appear to be affected in wild type cells deprived of inositol, since wild type cells, unlike cells of Ino− mutants, continue to grow indefinitely in the absence of inositol. Moreover, Doering et al. (Doering and Schekman, 1996) demonstrated that the rates of trafficking of prototypical secretory proteins in yeast including invertase, alkaline phosphatase, and carboxypeptidase Y, were largely unaffected by inositol starvation of an ino1-13 ino2-8 strain.
However, the studies of Doering et al. (Doering and Schekman, 1996) revealed that trafficking of Gas1p, a glycosyl phosphatidylinositol (GPI)-anchored protein, was inhibited between the ER to the plasma membrane (PM) in inositol starved ino1-13 ino2-8 cells. Moreover, the block in Gas1p ER-to-PM trafficking under these conditions was shown to be due to cessation of synthesis the GPI anchor (Doering and Schekman, 1996), for which PI serves as a precursor. The pathway of GPI anchor synthesis is highly conserved between yeast and mammals (Pittet and Conzelmann, 2007) with an estimated 60 GPI-anchored proteins in yeast (Fujita and Kinoshita, 2012). The GPI anchor is synthesized from PI in the ER, where newly synthesized proteins are added to the fully constructed GPI anchor en bloc. It is only after proteins are added to the GPI anchor that these proteins are able to exit the ER through packaging into COPII vesicles. Gas1p trafficking was shown to be inhibited by at least 90% within 95 min and was essentially completely blocked by 135 min following removal of inositol from the medium of ino1-13 ino2-8 cells. Nevertheless, under these conditions, trafficking of Gas1p was completely reversible upon addition of inositol even after 180 min (Doering and Schekman, 1996). Like Sec− mutants raised to their restrictive temperature, mutants that are defective in the synthesis of GPI anchors also exhibit induction of the UPR (Castillon et al., 2011; Jonikas et al., 2009). Thus, activation the UPR in wild type cells deprived of exogenous inositol may result from general slowing in GPI anchor synthesis caused by the reduced rate of PI synthesis that occurs in wild type cells relying on de novo synthesis of inositol (Gaspar et al., 2006; Loewen et al., 2004).
7b. Genome wide studies have revealed many more genes regulated by inositol
Genome wide expression studies of cells grown in the presence and absence of inositol and/or choline have expanded the number of genes identified as being regulated by inositol, with or without choline (Jesch et al., 2006; Jesch et al., 2005; Nunez et al., 2008; Santiago and Mamoun, 2003). One initial study (Jesch et al., 2005), which compared the changes in gene expression in cells grown continuously in the presence of inositol versus those grown in its absence, revealed a surprisingly small number of genes, which are differentially regulated under these two steady state growth conditions. Indeed, only 29 genes, including both UASINO-containing genes and UPR targets, were found to be significantly upregulated in cells grown continuously in the absence of inositol, versus in its presence and only 3 genes were observed to be significantly upregulated in the presence of inositol (Jesch et al., 2005). While, the inclusion of choline in the media increased the number of genes observed to be differentially expressed in presence versus the absence of inositol, choline, on its own, had little effect on global gene expression in the absence of inositol (Jesch et al., 2005).
Gene expression profiling following the addition of inositol to cells acclimated to growth in the absence of inositol identified additional signaling pathways that respond rapidly to the changes in lipid metabolism induced by inositol addition (Gaspar et al., 2006; Loewen et al., 2004). As described in previous sections, addition of inositol leads to a rapid and profound change in PI metabolism as well as other changes in phospholipid and TAG metabolism. In all over 700 genes showed a change in expression over a 30 min time course following inositol addition (Jesch et al., 2006). Six distinct expression responses were detected by this analysis, suggesting that multiple ER-to-nucleus signaling pathways are regulated by changes in lipid metabolism that are induced by inositol addition.
7c. PKC-MAPK signaling is activated transiently during inositol starvation of wild type cells
The UPR was the only stress response pathway observed to be activated in cells acclimated to “steady state” growth in the absence of inositol (Jesch et al., 2005). However, subsequent studies have revealed that, while the PKC-MAPK pathway is not activated in cells grown continuously in the absence of inositol (Jesch et al., 2006; Jesch et al., 2005), it is activated in actively dividing cells shifted to inositol free medium for a period of 3 hrs (Jesch et al., 2010; Nunez et al., 2008). These data suggest that some stress response signaling is only transiently activated following depletion of exogenous inositol from the growth medium of wild type cells. Wild type cells, unlike ino1, ino2 and ino4 mutants are able to depress INO1, thus enabling the de novo synthesis of inositol. However, under these conditions, as described above, wild type cells still exhibit a somewhat reduced growth rate, slowing of PI synthesis and many other changes in lipid metabolism and composition. Indeed, recent gene expression profiling conducted using logarithmically growing wild type cells, shifted from medium containing inositol to medium lacking inositol, has revealed that numerous additional stress response pathways are transiently activated in wild type cells as they adjust to growth in the absence of inositol (S. Jesch, S. Henry unpublished). Moreover, the genome-wide screen for inositol auxotrophy study conducted by Villa et al. (Villa-Garcia et al., 2011), revealed Ino− phenotypes associated with mutants defective in many of these same stress response pathways. Besides the UPR and PKC-MAPK pathways discussed above, pathways found to be activated following an abrupt shift of actively growing wild type cells to inositol free medium include the high osmolarity glycerol (HOG)/p38 and calcineurin pathways (S. Jesch, S. Henry unpublished). Interestingly, Ino− phenotypes in mutants defective in UPR and PKC-MAPK pathways are alleviated by overexpression of the NTE1 gene (Fernandez-Murray et al., 2009; Nunez et al., 2008), encoding a phospholipase B, which controls PC turnover in response to high temperature and the presence of choline (Fernandez-Murray and McMaster, 2005; Zaccheo et al., 2004). Moreover, mutants lacking the MAPK of the PKC pathway, Slt2p, show severe defects in lipid metabolism, including aberrant turnover of phosphatidylcholine as well as accumulation of PC, DAG, TAG, and free sterols, suggesting that PKC-MAPK plays a crucial role in control of lipid homeostasis (Nunez et al., 2008).
While the precise mechanism for activation of the UPR pathway during inositol starvation remains elusive, a clue for the activation of PKC-MAPK signaling during inositol starvation of wild type cells came from the finding that inhibition of the synthesis of inositol sphingolipid synthesis by pharmacological agents, such as myriocin or aureobasidin A, leads to robust activation of PKC-MAPK signaling. Several studies have shown that inositol sphingolipid synthesis is sensitive to inositol supplementation. As discussed above, inositol sphingolipid synthesis was shown to be inhibited during inositol starvation both in the ino1-13 ino4-8 mutant (Hanson and Lester, 1980) and in wild type strains (Jesch et al., 2010). Conversely, synthesis of inositol sphingolipids increases upon provision of inositol to wild type cells adapted to continuous growth in its absence (Alvarez-Vasquez et al., 2005). Hansen and Lester (Hanson and Lester, 1980) also showed that inositol starvation of an ino1-13 ino4-8 mutant strain led to a decrease in the rate of synthesis of phosphoinositides. However, the relative levels of all four phosphoinositides were observed to increase and a biosensor, which detects PI4P, was seen to accumulate on the plasma membrane during inositol starvation of wild type cells starved for inositol (Jesch et al., 2010). The accumulation of the PI4P biosensor on the plasma membrane correlated with PKC-MAPK activation, suggesting that sphingolipid levels may control access of PI4P effector proteins required for PKC-MAPK activation on the plasma membrane (Jesch et al., 2010). This model is consistent with the finding that Stt4p-dependent formation of PI4P is necessary for PKC-MAPK activation (Audhya and Emr, 2002). The activation the PKC-MAPK signaling pathway during inositol starvation leads to the up-regulation of genes involved in cell wall organization and biogenesis controlled by the Rlm1p transcription factor (Nunez et al., 2008), which may relieve plasma membrane stress when inositol sphingolipid synthesis is reduced. In addition, the triggering of PKC-MAPK signaling by inositol starvation, or by interrupting inositol sphingolipid synthesis, results in increased Sir2p-dependent telomeric silencing (Lee et al., 2013), presumably through phosphorylation of Sir3p, a known target of Slt2p (Ai et al., 2002; Ray et al., 2003) and component of the highly conserved Sir complex (Hecht et al., 1996; Strahl-Bolsinger et al., 1997). Thus, inositol sphingolipid metabolism controls multiple downstream stress responses.
8. Conclusions and reflections
The yeast, Saccharomyces cerevisiae, has proven to be a powerful model system in which to dissect the biochemical pathways and regulatory mechanisms controlling lipid metabolism and lipid mediated signaling pathways that are broadly applicable to eukaryotic organisms. The studies described in this review, involving the work of dozens of laboratories over a span of more than four decades; a period during which the genetic, biochemical, molecular methodologies available in yeast underwent an enormous evolution. Many of the gene enzyme relationships in yeast lipid metabolism, described in this review, led to discoveries of the functions of related homologous genes and proteins in other eukaryotic organisms, including humans, including INO1, encoding inositol 3-phosphate synthase and CDS1, CDP-diacylglycerol synthase. The discovery that human LIPIN is an enzyme with PA-phosphatase activity came from the discovery that it is a homolog of the yeast gene Pah1p gene. Many other examples of the insights derived from yeast that have proven pivotal to progress in biomedical research in the broadest sense could be cited. However, the primary focus of this review is the enormous progress derived from the application of genetics, biochemical and molecular technologies, applied to the dissection of regulatory mechanisms governing lipid biosynthesis, metabolism and signaling in yeast. Most importantly, the approach of tracing the genetics of the biosynthesis and metabolism of inositol, a molecule essential to eukaryotic signaling and lipid metabolism, has enabled the discovery of intricate regulatory mechanisms that control both phospholipid biosynthesis and lipid mediated signaling. Looking to the future, many questions remain to be answered. For example, understanding the mechanism of activation and attenuation of stress response signaling by lipid metabolism as well as the interconnectedness of these pathways remain areas of great importance that are as yet poorly understood. Yeast will continue to be an important model system in which to probe these questions, using its powerful molecular genetics to build on insights discussed in this review.
Acknowledgments
We are indebted to Yu-Fang Chang and Manuel Aregullin for critical reading of the manuscript and many helpful discussions. We also acknowledge all members of the Henry laboratory, both past and present, who have contributed to the research discussed in this review. Work in the Henry laboratory is supported by National Institutes of Health (NIH) grant GM-19629.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF. Regulation of subtelomeric silencing during stress response. Mol Cell. 2002;10:1295–1305. doi: 10.1016/s1097-2765(02)00695-0. [DOI] [PubMed] [Google Scholar]
- Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433:425–430. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Current opinion in genetics & development. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- Anderson MS, Lopes JM. Carbon source regulation of PIS1 gene expression in Saccharomyces cerevisiae involves the MCM1 gene and the twocomponent regulatory gene, SLN1. J Biol Chem. 1996;271:26596–26601. doi: 10.1074/jbc.271.43.26596. [DOI] [PubMed] [Google Scholar]
- Ashburner BP, Lopes JM. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol Cell Biol. 1995a;15:1709–1715. doi: 10.1128/mcb.15.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner BP, Lopes JM. Regulation of yeast phospholipid biosynthetic gene expression in response to inositol involves two superimposed mechanisms. Proc Natl Acad Sci U S A. 1995b;92:9722–9726. doi: 10.1073/pnas.92.21.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280:37301–37309. doi: 10.1074/jbc.M507261200. [DOI] [PubMed] [Google Scholar]
- Atkinson K, Fogel S, Henry SA. Yeast mutant defective in phosphatidylserine synthesis. J Biol Chem. 1980a;255:6653–6661. [PubMed] [Google Scholar]
- Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S. Yeast mutants auxotrophic for choline or ethanolamine. J Bacteriol. 1980b;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson KD, Kolat AI, Henry SA. Osmotic imbalance in inositolstarved spheroplasts of Saccharomyces cerevisiae. J Bacteriol. 1977;132:806–817. doi: 10.1128/jb.132.3.806-817.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Developmental cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Bachhawat N, Ouyang Q, Henry SA. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- Bailis AM, Lopes JM, Kohlwein SD, Henry SA. Cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae. Nucleic acids research. 1992;20:1411–1418. doi: 10.1093/nar/20.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis AM, Poole MA, Carman GM, Henry SA. The membrane-associated enzyme phosphatidylserine synthase is regulated at the level of mRNA abundance. Mol Cell Biol. 1987;7:167–176. doi: 10.1128/mcb.7.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Phillips S, Yanagisawa L, Li X, Routt S, Xie Z. Phosphatidylinositol transfer protein function in the yeast Saccharomyces cerevisiae. Advances in enzyme regulation. 2005;45:155–170. doi: 10.1016/j.advenzreg.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Becker GW, Lester RL. Changes in phospholipids of Saccharomyces cerevisiae associated with inositol-less death. J Biol Chem. 1977;252:8684–8691. [PubMed] [Google Scholar]
- Berroteran RW, Ware DE, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Molecular genetics and genomics : MGG. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci U S A. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumann HA, Gubbens J, Koorengevel MC, Oh CS, Martin CE, Heck AJ, Patton-Vogt J, Henry SA, de Kruijff B, de Kroon AI. Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol Biol Cell. 2006;17:1006–1017. doi: 10.1091/mbc.E05-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J, Greenberg DM. Mono- and dimethylethanolamine isolated from rat-liver phospholipids. Biochim Biophys Acta. 1959;35:287–288. doi: 10.1016/0006-3002(59)90375-0. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Weissman JS. Membranes in balance: mechanisms of sphingolipid homeostasis. Mol Cell. 2010;40:267–279. doi: 10.1016/j.molcel.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends in biochemical sciences. 2006;31:694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim Biophys Acta. 2007;1771:322–330. doi: 10.1016/j.bbalip.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J Biol Chem. 2009;284:2593–2597. doi: 10.1074/jbc.R800059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annual review of biochemistry. 2011;80:859–883. doi: 10.1146/annurev-biochem-060409-092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282:37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson MA, Atkinson KD, Waechter CJ. Properties of particulate and solubilized phosphatidylserine synthase activity from Saccharomyces cerevisiae. Inhibitory effect of choline in the growth medium. J Biol Chem. 1982;257:8115–8121. [PubMed] [Google Scholar]
- Carson MA, Emala M, Hogsten P, Waechter CJ. Coordinate regulation of phosphatidylserine decarboxylase activity and phospholipid N-methylation in yeast. J Biol Chem. 1984;259:6267–6273. [PubMed] [Google Scholar]
- Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muniz M. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol Biol Cell. 2011;22:2924–2936. doi: 10.1091/mbc.E11-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae M, Han GS, Carman GM. The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme. J Biol Chem. 2012;287:40186–40196. doi: 10.1074/jbc.M112.421776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HJ, Jesch SA, Gaspar ML, Henry SA. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics. 2004;168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HJ, Jones EW, Henry SA. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics. 2002;162:29–43. doi: 10.1093/genetics/162.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Hancock LC, Lopes JM. Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim Biophys Acta. 2007;1771:310–321. doi: 10.1016/j.bbalip.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Chirala SS. Coordinated regulation and inositol-mediated and fatty acidmediated repression of fatty acid synthase genes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992;89:10232–10236. doi: 10.1073/pnas.89.21.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala SS, Kuziora MA, Spector DM, Wakil SJ. Complementation of mutations and nucleotide sequence of FAS1 gene encoding beta subunit of yeast fatty acid synthase. J Biol Chem. 1987;262:4231–4240. [PubMed] [Google Scholar]
- Chirala SS, Zhong Q, Huang W, al-Feel W. Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression. Nucleic acids research. 1994;22:412–418. doi: 10.1093/nar/22.3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Su WM, Han GS, Plote D, Xu Z, Carman GM. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J Biol Chem. 2012;287:11290–11301. doi: 10.1074/jbc.M112.346023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Su WM, Morgan JM, Han GS, Xu Z, Karanasios E, Siniossoglou S, Carman GM. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER602), THR723), AND SER744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 2011;286:1486–1498. doi: 10.1074/jbc.M110.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Martin WE, Murphy RC, Voelker DR. Phosphatidylcholine and N-methylated phospholipids are nonessential in Saccharomyces cerevisiae. J Biol Chem. 2004;279:42321–42330. doi: 10.1074/jbc.M405074200. [DOI] [PubMed] [Google Scholar]
- Clancey CJ, Chang SC, Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem. 1993;268:24580–24590. [PubMed] [Google Scholar]
- Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Mallory MJ, Strich R, Yao TP. Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot Cell. 2008;7:1191–1199. doi: 10.1128/EC.00059-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Donahue TF, Henry SA. Control of inositol biosynthesis in Saccharomyces cerevisiae; inositol-phosphate synthetase mutants. J Bacteriol. 1976;126:243–250. doi: 10.1128/jb.126.1.243-250.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Henry SA. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975;80:23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Johnson M, Henry SA. Biosynthesis of inositol in yeast. Primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J Biol Chem. 1989;264:1274–1283. [PubMed] [Google Scholar]
- Deranieh RM, Greenberg ML. Cellular consequences of inositol depletion. Biochem Soc Trans. 2009;37:1099–1103. doi: 10.1042/BST0371099. [DOI] [PubMed] [Google Scholar]
- Deranieh RM, He Q, Caruso JA, Greenberg ML. Phosphorylation regulates myo-inositol-3-phosphate synthase: A novel regulatory mechanism of inositol biosynthesis. J Biol Chem. 2013 doi: 10.1074/jbc.M113.479121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL, Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. Inositol Mutants of SACCHAROMYCES CEREVISIAE: Mapping the ino1 Locus and Characterizing Alleles of the ino1, ino2 and ino4 Loci. Genetics. 1981a;98:491–503. doi: 10.1093/genetics/98.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981b;256:7077–7085. [PubMed] [Google Scholar]
- Dowd SR, Bier ME, Patton-Vogt JL. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Molecular microbiology. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- Elkhaimi M, Kaadige MR, Kamath D, Jackson JC, Biliran H, Jr, Lopes JM. Combinatorial regulation of phospholipid biosynthetic gene expression by the UME6, SIN3 and RPD3 genes. Nucleic acids research. 2000;28:3160–3167. doi: 10.1093/nar/28.16.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakas S, Konstantinou C, Carman GM. DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J Biol Chem. 2011;286:1464–1474. doi: 10.1074/jbc.M110.194308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner A, Chen X, Rush J, Horazdovsky B, Waechter CJ, Carman GM, Sternweis PC. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J Biol Chem. 1999;274:14831–14837. doi: 10.1074/jbc.274.21.14831. [DOI] [PubMed] [Google Scholar]
- Fernandez-Murray JP, Gaspard GJ, Jesch SA, McMaster CR. NTE1-encoded phosphatidylcholine phospholipase b regulates transcription of phospholipid biosynthetic genes. J Biol Chem. 2009;284:36034–36046. doi: 10.1074/jbc.M109.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Murray JP, McMaster CR. Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J Biol Chem. 2005;280:38290–38296. doi: 10.1074/jbc.M507700200. [DOI] [PubMed] [Google Scholar]
- Fujita M, Kinoshita T. GPI-anchor remodeling: potential functions of GPIanchors in intracellular trafficking and membrane dynamics. Biochim Biophys Acta. 2012;1821:1050–1058. doi: 10.1016/j.bbalip.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Galdieri L, Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J Biol Chem. 2012;287:23865–23876. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansheroff LJ, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harbor perspectives in biology. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardocki ME, Jani N, Lopes JM. Phosphatidylinositol biosynthesis: biochemistry and regulation. Biochim Biophys Acta. 2005;1735:89–100. doi: 10.1016/j.bbalip.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Gardocki ME, Lopes JM. Expression of the yeast PIS1 gene requires multiple regulatory elements including a Rox1p binding site. J Biol Chem. 2003;278:38646–38652. doi: 10.1074/jbc.M305251200. [DOI] [PubMed] [Google Scholar]
- Gaspar ML, Aregullin MA, Jesch SA, Henry SA. Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J Biol Chem. 2006;281:22773–22785. doi: 10.1074/jbc.M603548200. [DOI] [PubMed] [Google Scholar]
- Gaspar ML, Aregullin MA, Jesch SA, Nunez LR, Villa-Garcia M, Henry SA. The emergence of yeast lipidomics. Biochim Biophys Acta. 2007;1771:241–254. doi: 10.1016/j.bbalip.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Gaspar ML, Hofbauer HF, Kohlwein SD, Henry SA. Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J Biol Chem. 2011;286:1696–1708. doi: 10.1074/jbc.M110.172296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar ML, Jesch SA, Viswanatha R, Antosh AL, Brown WJ, Kohlwein SD, Henry SA. A block in endoplasmic reticulum-to-Golgi trafficking inhibits phospholipid synthesis and induces neutral lipid accumulation. J Biol Chem. 2008;283:25735–25751. doi: 10.1074/jbc.M802685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor PM, Gill T, Toutenhoofd S, Summers EF, McGraw P, Homann MJ, Henry SA, Carman GM. Regulation of phosphatidylethanolamine methyltransferase and phospholipid methyltransferase by phospholipid precursors in Saccharomyces cerevisiae. Biochim Biophys Acta. 1991;1090:326–332. doi: 10.1016/0167-4781(91)90197-t. [DOI] [PubMed] [Google Scholar]
- Geiger JH, Jin X. The structure and mechanism of myo-inositol-1-phosphate synthase. Sub-cellular biochemistry. 2006;39:157–180. doi: 10.1007/0-387-27600-9_7. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Bankaitis VA. Phosphatidylinositol transfer proteins: negotiating the regulatory interface between lipid metabolism and lipid signaling in diverse cellular processes. Biofactors. 2011;37:290–308. doi: 10.1002/biof.180. [DOI] [PubMed] [Google Scholar]
- Gibson KD, Wilson JD, Udenfriend S. The enzymatic conversion of phospholipid ethanolamine to phospholipid choline in rat liver. J Biol Chem. 1961;236:673–679. [PubMed] [Google Scholar]
- Gilstring CF, Melin-Larsson M, Ljungdahl PO. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol Biol Cell. 1999;10:3549–3565. doi: 10.1091/mbc.10.11.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA, Henry SA. Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics. 2000;154:1485–1495. doi: 10.1093/genetics/154.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. "Sleeping beauty": quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ML, Goldwasser P, Henry SA. Characterization of a yeast regulatory mutant constitutive for synthesis of inositol-1-phosphate synthase. Molecular & general genetics : MGG. 1982a;186:157–163. doi: 10.1007/BF00331845. [DOI] [PubMed] [Google Scholar]
- Greenberg ML, Klig LS, Letts VA, Loewy BS, Henry SA. Yeast mutant defective in phosphatidylcholine synthesis. J Bacteriol. 1983;153:791–799. doi: 10.1128/jb.153.2.791-799.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ML, Lopes JM. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ML, Reiner B, Henry SA. Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol-excreting mutants. Genetics. 1982b;100:19–33. doi: 10.1093/genetics/100.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P. Regulation of yeast phospholipid biosynthetic genes in phosphatidylserine decarboxylase mutants. J Bacteriol. 1997;179:5843–5848. doi: 10.1128/jb.179.18.5843-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P. Sec14 related proteins in yeast. Biochim Biophys Acta. 2007;1771:737–745. doi: 10.1016/j.bbalip.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Griac P, Henry SA. The yeast inositol-sensitive upstream activating sequence, UASINO, responds to nitrogen availability. Nucleic acids research. 1999;27:2043–2050. doi: 10.1093/nar/27.9.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P, Swede MJ, Henry SA. The role of phosphatidylcholine biosynthesis in the regulation of the INO1 gene of yeast. J Biol Chem. 1996;271:25692–25698. doi: 10.1074/jbc.271.41.25692. [DOI] [PubMed] [Google Scholar]
- Grigat M, Jaschke Y, Kliewe F, Pfeifer M, Walz S, Schuller HJ. Multiple histone deacetylases are recruited by corepressor Sin3 and contribute to gene repression mediated by Opi1 regulator of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Molecular genetics and genomics : MGG. 2012;287:461–472. doi: 10.1007/s00438-012-0692-x. [DOI] [PubMed] [Google Scholar]
- Gururaj C, Federman R, Chang A. Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J Biol Chem. 2013;288:20453–20463. doi: 10.1074/jbc.M113.472860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Han GS, Carman GM. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 2010;285:14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Johnston CN, Chen X, Athenstaedt K, Daum G, Carman GM. Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J Biol Chem. 2001;276:10126–10133. doi: 10.1074/jbc.M011421200. [DOI] [PubMed] [Google Scholar]
- Han GS, O'Hara L, Carman GM, Siniossoglou S. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem. 2008a;283:20433–20442. doi: 10.1074/jbc.M802903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, O'Hara L, Siniossoglou S, Carman GM. Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J Biol Chem. 2008b;283:20443–20453. doi: 10.1074/jbc.M802866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282:37026–37035. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci U S A. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Han GS, Iwanyshyn WM, Carman GM. Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc. J Biol Chem. 2005;280:29017–29024. doi: 10.1074/jbc.M505881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanscho M, Ruckerbauer DE, Chauhan N, Hofbauer HF, Krahulec S, Nidetzky B, Kohlwein SD, Zanghellini J, Natter K. Nutritional requirements of the BY series of Saccharomyces cerevisiae strains for optimum growth. FEMS yeast research. 2012;12:796–808. doi: 10.1111/j.1567-1364.2012.00830.x. [DOI] [PubMed] [Google Scholar]
- Hanson BA, Lester RL. Effects of inositol starvation on phospholipid and glycan syntheses in Saccharomyces cerevisiae. J Bacteriol. 1980;142:79–89. doi: 10.1128/jb.142.1.79-89.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Henneberry AL, Sturley SL. Sterol homeostasis in the budding yeast, Saccharomyces cerevisiae. Semin Cell Dev Biol. 2005;16:155–161. doi: 10.1016/j.semcdb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Henry SA. Death resulting from fatty acid starvation in yeast. J Bacteriol. 1973;116:1293–1303. doi: 10.1128/jb.116.3.1293-1303.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SA, Atkinson KD, Kolat AI, Culbertson MR. Growth and metabolism of inositol-starved Saccharomyces cerevisiae. J Bacteriol. 1977;130:472–484. doi: 10.1128/jb.130.1.472-484.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SA, Donahue TF, Culbertson MR. Selection of spontaneous mutants by inositol starvation in yeast. Molecular & general genetics : MGG. 1975;143:5–11. doi: 10.1007/BF00269415. [DOI] [PubMed] [Google Scholar]
- Henry SA, Horowitz B. A new method for mutant selection in Saccharomyces cerevisiae. Genetics. 1975;79:175–186. doi: 10.1093/genetics/79.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SA, Patton-Vogt JL. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Petti AA, Ho-Shing O, Silverman SJ, McIsaac RS, Lee TA, Botstein D. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol Biol Cell. 2011;22:4192–4204. doi: 10.1091/mbc.E11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann MJ, Bailis AM, Henry SA, Carman GM. Coordinate regulation of phospholipid biosynthesis by serine in Saccharomyces cerevisiae. J Bacteriol. 1987;169:3276–3280. doi: 10.1128/jb.169.7.3276-3280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann MJ, Henry SA, Carman GM. Regulation of CDP-diacylglycerol synthase activity in Saccharomyces cerevisiae. J Bacteriol. 1985;163:1265–1266. doi: 10.1128/jb.163.3.1265-1266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K, Kodaki T. [Regulation of expression of phospholipids-biosynthetic genes in yeast: coordinate repression by inositol and choline]. Seikagaku. The Journal of Japanese Biochemical Society. 1990;62:451–456. [PubMed] [Google Scholar]
- Hosaka K, Murakami T, Kodaki T, Nikawa J, Yamashita S. Repression of choline kinase by inositol and choline in Saccharomyces cerevisiae. J Bacteriol. 1990;172:2005–2012. doi: 10.1128/jb.172.4.2005-2012.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshizaki DK, Hill JE, Henry SA. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- Howe AG, McMaster CR. Regulation of phosphatidylcholine homeostasis by Sec14. Canadian journal of physiology and pharmacology. 2006;84:29–38. doi: 10.1139/Y05-138. [DOI] [PubMed] [Google Scholar]
- Hudak KA, Lopes JM, Henry SA. A pleiotropic phospholipid biosynthetic regulatory mutation in Saccharomyces cerevisiae is allelic to sin3 (sdi1, ume4, rpd1) Genetics. 1994;136:475–483. doi: 10.1093/genetics/136.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JC, Lopes JM. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic acids research. 1996;24:1322–1329. doi: 10.1093/nar/24.7.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani NM, Lopes JM. Transcription regulation of the Saccharomyces cerevisiae PIS1 gene by inositol and the pleiotropic regulator, Ume6p. Molecular microbiology. 2008;70:1529–1539. doi: 10.1111/j.1365-2958.2008.06506.x. [DOI] [PubMed] [Google Scholar]
- Jani NM, Lopes JM. Regulated transcription of the Saccharomyces cerevisiae phosphatidylinositol biosynthetic gene, PIS1, yields pleiotropic effects on phospholipid synthesis. FEMS yeast research. 2009;9:552–564. doi: 10.1111/j.1567-1364.2009.00514.x. [DOI] [PubMed] [Google Scholar]
- Jaschke Y, Schwarz J, Clausnitzer D, Muller C, Schuller HJ. Pleiotropic corepressors Sin3 and Ssn6 interact with repressor Opi1 and negatively regulate transcription of genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Molecular genetics and genomics : MGG. 2011;285:91–100. doi: 10.1007/s00438-010-0589-5. [DOI] [PubMed] [Google Scholar]
- Jensen-Pergakes K, Guo Z, Giattina M, Sturley SL, Bard M. Transcriptional regulation of the two sterol esterification genes in the yeast Saccharomyces cerevisiae. J Bacteriol. 2001;183:4950–4957. doi: 10.1128/JB.183.17.4950-4957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch SA, Gaspar ML, Stefan CJ, Aregullin MA, Henry SA. Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J Biol Chem. 2010;285:41947–41960. doi: 10.1074/jbc.M110.188607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch SA, Liu P, Zhao X, Wells MT, Henry SA. Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J Biol Chem. 2006;281:24070–24083. doi: 10.1074/jbc.M604541200. [DOI] [PubMed] [Google Scholar]
- Jesch SA, Zhao X, Wells MT, Henry SA. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch, S.A.a.H SA. Yeast inositol phospholipids: Synthesis, regulation and involvement in membrane trafficking and lipid signaling. In: Daum G, editor. Cell biology and dynamics of yeast lipids. Kerala, India: Research Signpost; 2005. pp. 105–131. [Google Scholar]
- Jin X, Geiger JH. Structures of NAD(+)- and NADH-bound 1-l-myoinositol 1-phosphate synthase. Acta crystallographica. Section D Biological crystallography. 2003;59:1154–1164. doi: 10.1107/s0907444903008205. [DOI] [PubMed] [Google Scholar]
- Jiranek V, Graves JA, Henry SA. Pleiotropic effects of the opi1 regulatory mutation of yeast: its effects on growth and on phospholipid and inositol metabolism. Microbiology. 1998;144(Pt 10):2739–2748. doi: 10.1099/00221287-144-10-2739. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S, Shaltiel G, Shamir A, Agam G, Greenberg ML. Human 1-Dmyo-inositol-3-phosphate synthase is functional in yeast. J Biol Chem. 2004;279:21759–21765. doi: 10.1074/jbc.M312078200. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanipes MI, Henry SA. The phospholipid methyltransferases in yeast. Biochim Biophys Acta. 1997;1348:134–141. doi: 10.1016/s0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- Karanasios E, Barbosa AD, Sembongi H, Mari M, Han GS, Reggiori F, Carman GM, Siniossoglou S. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol Biol Cell. 2013;24:2124–2133. doi: 10.1091/mbc.E13-01-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E, Han GS, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci U S A. 2010;107:17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MJ, Bailis AM, Henry SA, Carman GM. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J Biol Chem. 1988;263:18078–18085. [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim KH, Storey MK, Voelker DR, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase. J Biol Chem. 1999;274:14857–14866. doi: 10.1074/jbc.274.21.14857. [DOI] [PubMed] [Google Scholar]
- Klig LS, Henry SA. Isolation of the yeast INO1 gene: located on an autonomously replicating plasmid, the gene is fully regulated. Proc Natl Acad Sci U S A. 1984;81:3816–3820. doi: 10.1073/pnas.81.12.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig LS, Homann MJ, Carman GM, Henry SA. Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: pleiotropically constitutive opi1 mutant. J Bacteriol. 1985;162:1135–1141. doi: 10.1128/jb.162.3.1135-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig LS, Homann MJ, Kohlwein SD, Kelley MJ, Henry SA, Carman GM. Saccharomyces cerevisiae mutant with a partial defect in the synthesis of CDP-diacylglycerol and altered regulation of phospholipid biosynthesis. J Bacteriol. 1988a;170:1878–1886. doi: 10.1128/jb.170.4.1878-1886.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig LS, Hoshizaki DK, Henry SA. Isolation of the yeast INO4 gene, a positive regulator of phospholipid biosynthesis. Curr Genet. 1988b;13:7–14. doi: 10.1007/BF00365749. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Hosaka K, Nikawa J, Yamashita S. Identification of the upstream activation sequences responsible for the expression and regulation of the PEM1 and PEM2 genes encoding the enzymes of the phosphatidylethanolamine methylation pathway in Saccharomyces cerevisiae. J Biochem. 1991a;109:276–287. [PubMed] [Google Scholar]
- Kodaki T, Nikawa J, Hosaka K, Yamashita S. Functional analysis of the regulatory region of the yeast phosphatidylserine synthase gene, PSS. J Bacteriol. 1991b;173:7992–7995. doi: 10.1128/jb.173.24.7992-7995.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Tsuji S, Otani N, Yamamoto D, Rao KS, Watanabe S, Tsukatsune M, Makino K. Differential transcriptional regulation of two distinct Sadenosylmethionine synthetase genes (SAM1 and SAM2) of Saccharomyces cerevisiae. Nucleic Acids Res Suppl. 2003:303–304. doi: 10.1093/nass/3.1.303. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 1987;262:15428–15435. [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur J Biochem. 1989;185:243–251. doi: 10.1111/j.1432-1033.1989.tb15109.x. [DOI] [PubMed] [Google Scholar]
- Kohlwein SD. Triacylglycerol homeostasis: insights from yeast. J Biol Chem. 2010;285:15663–15667. doi: 10.1074/jbc.R110.118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koipally J, Ashburner BP, Bachhawat N, Gill T, Hung G, Henry SA, Lopes JM. Functional characterization of the repeated UASINO element in the promoters of the INO1 and CHO2 genes of yeast. Yeast. 1996;12:653–665. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C653::AID-YEA953%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Buratowski S, Nonet M, Young RA. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Korennykh A, Walter P. Structural basis of the unfolded protein response. Annual review of cell and developmental biology. 2012;28:251–277. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- Kovac L, Gbelska I, Poliachova V, Subik J, Kovacova V. Membrane mutants: a yeast mutant with a lesion in phosphatidylserine biosynthesis. Eur J Biochem. 1980;111:491–501. doi: 10.1111/j.1432-1033.1980.tb04965.x. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nature genetics. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Lai K, McGraw P. Dual control of inositol transport in Saccharomyces cerevisiae by irreversible inactivation of permease and regulation of permease synthesis by INO2, INO4, and OPI1. J Biol Chem. 1994;269:2245–2251. [PubMed] [Google Scholar]
- Lamping E, Kohlwein SD, Henry SA, Paltauf F. Coordinate regulation of phosphatidylserine decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:6432–6437. doi: 10.1128/jb.173.20.6432-6437.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E, Luckl J, Paltauf F, Henry SA, Kohlwein SD. Isolation and characterization of a mutant of Saccharomyces cerevisiae with pleiotropic deficiencies in transcriptional activation and repression. Genetics. 1994;137:55–65. doi: 10.1093/genetics/137.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc MA, McMaster CR. Surprising roles for phospholipid binding proteins revealed by high throughput genetics. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2010;88:565–574. doi: 10.1139/O09-171. [DOI] [PubMed] [Google Scholar]
- Lee S, Gaspar ML, Aregullin MA, Jesch SA, Henry SA. Activation of Protein Kinase C-Mitogen-activated Protein Kinase Signaling in Response to Inositol Starvation Triggers Sir2p-dependent Telomeric Silencing in Yeast. J Biol Chem. 2013;288:27861–27871. doi: 10.1074/jbc.M113.493072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester HE, Gross SR. Efficient method for selection of auxotrophic mutants of Neurospora. Science. 1959;129:572. doi: 10.1126/science.129.3348.572. [DOI] [PubMed] [Google Scholar]
- Letts VA, Henry SA. Regulation of phospholipid synthesis in phosphatidylserine synthase-deficient (chol) mutants of Saccharomyces cerevisiae. J Bacteriol. 1985;163:560–567. doi: 10.1128/jb.163.2.560-567.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts VA, Klig LS, Bae-Lee M, Carman GM, Henry SA. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc Natl Acad Sci U S A. 1983;80:7279–7283. doi: 10.1073/pnas.80.23.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Haase E, Brendel M. Hyper-resistance to nitrogen mustard in Saccharomyces cerevisiae is caused by defective choline transport. Curr Genet. 1991;19:423–427. doi: 10.1007/BF00312732. [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy BS, Henry SA. The INO2 and INO4 loci of Saccharomyces cerevisiae are pleiotropic regulatory genes. Mol Cell Biol. 1984;4:2479–2485. doi: 10.1128/mcb.4.11.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JM, Henry SA. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic acids research. 1991;19:3987–3994. doi: 10.1093/nar/19.14.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JM, Hirsch JP, Chorgo PA, Schulze KL, Henry SA. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic acids research. 1991;19:1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JM, Schulze KL, Yates JW, Hirsch JP, Henry SA. The INO1 promoter of Saccharomyces cerevisiae includes an upstream repressor sequence (URS1) common to a diverse set of yeast genes. J Bacteriol. 1993;175:4235–4238. doi: 10.1128/jb.175.13.4235-4238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, York JD. Phosphoinositide phosphatases and disease. J Lipid Res. 2009;50(Suppl):S249–S254. doi: 10.1194/jlr.R800072-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder AL, Chatterjee A, Ghosh Dastidar K, Majee M. Diversification and evolution of L-myo-inositol 1-phosphate synthase. FEBS Lett. 2003;553:3–10. doi: 10.1016/s0014-5793(03)00974-8. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Johnson MD, Henry SA. 1L-myo-inositol-1-phosphate synthase. Biochim Biophys Acta. 1997;1348:245–256. doi: 10.1016/s0005-2760(97)00122-7. [DOI] [PubMed] [Google Scholar]
- Malanovic N, Streith I, Wolinski H, Rechberger G, Kohlwein SD, Tehlivets O. S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem. 2008;283:23989–23999. doi: 10.1074/jbc.M800830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P, Henry SA. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics. 1989;122:317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster CR, Bell RM. Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases. J Biol Chem. 1994a;269:28010–28016. [PubMed] [Google Scholar]
- McMaster CR, Bell RM. Phosphatidylcholine biosynthesis via the CDPcholine pathway in Saccharomyces cerevisiae. Multiple mechanisms of regulation. J Biol Chem. 1994b;269:14776–14783. [PubMed] [Google Scholar]
- Michell RH. Evolution of the diverse biological roles of inositols. Biochemical Society symposium. 2007:223–246. doi: 10.1042/BSS0740223. [DOI] [PubMed] [Google Scholar]
- Morash SC, McMaster CR, Hjelmstad RH, Bell RM. Studies employing Saccharomyces cerevisiae cpt1 and ept1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J Biol Chem. 1994;269:28769–28776. [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner H, Daum G. Dynamics of neutral lipid storage in yeast. Acta Biochim Pol. 2004;51:323–347. [PubMed] [Google Scholar]
- Murray M, Greenberg ML. Regulation of inositol monophosphatase in Saccharomyces cerevisiae. Molecular microbiology. 1997;25:541–546. doi: 10.1046/j.1365-2958.1997.4881840.x. [DOI] [PubMed] [Google Scholar]
- Murray M, Greenberg ML. Expression of yeast INM1 encoding inositol monophosphatase is regulated by inositol, carbon source and growth stage and is decreased by lithium and valproate. Molecular microbiology. 2000;36:651–661. doi: 10.1046/j.1365-2958.2000.01886.x. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Akiyoshi M, Hirata S, Fukuda T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic acids research. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Kodaki T, Yamashita S. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J Biol Chem. 1987a;262:4876–4881. [PubMed] [Google Scholar]
- Nikawa J, Tsukagoshi Y, Kodaki T, Yamashita S. Nucleotide sequence and characterization of the yeast PSS gene encoding phosphatidylserine synthase. Eur J Biochem. 1987b;167:7–12. doi: 10.1111/j.1432-1033.1987.tb13297.x. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Tsukagoshi Y, Yamashita S. Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J Bacteriol. 1986;166:328–330. doi: 10.1128/jb.166.1.328-330.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Molecular microbiology. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Nikoloff DM, Henry SA. Functional characterization of the INO2 gene of Saccharomyces cerevisiae. A positive regulator of phospholipid biosynthesis. J Biol Chem. 1994;269:7402–7411. [PubMed] [Google Scholar]
- Nikoloff DM, McGraw P, Henry SA. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid synthesis. Nucleic acids research. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez LR, Jesch SA, Gaspar ML, Almaguer C, Villa-Garcia M, Ruiz-Noriega M, Patton-Vogt J, Henry SA. Cell wall integrity MAPK pathway is essential for lipid homeostasis. J Biol Chem. 2008;283:34204–34217. doi: 10.1074/jbc.M806391200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Oshiro J, Rangaswamy S, Chen X, Han GS, Quinn JE, Carman GM. Regulation of the DPP1-encoded diacylglycerol pyrophosphate (DGPP) phosphatase by inositol and growth phase. Inhibition of DGPP phosphatase activity by CDP-diacylglyceron and activation of phosphatidylserine synthase activity by DGPP. J Biol Chem. 2000;275:40887–40896. doi: 10.1074/jbc.M008144200. [DOI] [PubMed] [Google Scholar]
- Paltauf FK, SD, Henry SA. Regulation and Compartimentalization of Lipid Synthesis in Yeast. In: EWJ Broach EW, Pringle JR, editors. The Molecular Biology of the Yeast Saccharomyces: Gene Expression. Plainview, NY: Cold Spring Harbor Press; 1992. pp. 415–500. [Google Scholar]
- Pascual F, Carman GM. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim Biophys Acta. 2013;1831:514–522. doi: 10.1016/j.bbalip.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual F, Soto-Cardalda A, Carman GM. PAH1-encoded Phosphatidate Phosphatase Plays a Role in the Growth Phase- and Inositol-mediated Regulation of Lipid Synthesis in Saccharomyces cerevisiae. J Biol Chem. 2013;288:35781–35792. doi: 10.1074/jbc.M113.525766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nature genetics. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Petti AA, Crutchfield CA, Rabinowitz JD, Botstein D. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc Natl Acad Sci U S A. 2011;108:E1089–E1098. doi: 10.1073/pnas.1101494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti AA, McIsaac RS, Ho-Shing O, Bussemaker HJ, Botstein D. Combinatorial control of diverse metabolic and physiological functions by transcriptional regulators of the yeast sulfur assimilation pathway. Mol Biol Cell. 2012;23:3008–3024. doi: 10.1091/mbc.E12-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS biology. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Poole MA, Homann MJ, Bae-Lee MS, Carman GM. Regulation of phosphatidylserine synthase from Saccharomyces cerevisiae by phospholipid precursors. J Bacteriol. 1986;168:668–672. doi: 10.1128/jb.168.2.668-672.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Hector RE, Roy N, Song JH, Berkner KL, Runge KW. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nature genetics. 2003;33:522–526. doi: 10.1038/ng1132. [DOI] [PubMed] [Google Scholar]
- Ridgway GJ, Douglas HC. Unbalanced growth of yeast due to inositol deficiency. J Bacteriol. 1958;76:163–166. doi: 10.1128/jb.76.2.163-166.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KA, Lopes JM. SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic acids research. 2000;28:1499–1505. doi: 10.1093/nar/28.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Yeung T, Schekman RW. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandager L, Dahlqvist A, Banas A, Stahl U, Lenman M, Gustavsson M, Stymne S. An acyl-CoA:cholesterol acyltransferase (ACAT)-related gene is involved in the accumulation of triacylglycerols in Saccharomyces cerevisiae. Biochem Soc Trans. 2000;28:700–702. [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- Santiago TC, Mamoun CB. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- Scafe C, Chao D, Lopes J, Hirsch JP, Henry S, Young RA. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Schuller HJ, Hahn A, Troster F, Schutz A, Schweizer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992a;11:107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ, Schorr R, Hoffmann B, Schweizer E. Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a transactivator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae. Nucleic acids research. 1992b;20:5955–5961. doi: 10.1093/nar/20.22.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller HJ. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic acids research. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Dowhan W. Regulation of phospholipid biosynthetic enzymes by the level of CDP-diacylglycerol synthase activity. J Biol Chem. 1997;272:11215–11220. doi: 10.1074/jbc.272.17.11215. [DOI] [PubMed] [Google Scholar]
- Shen H, Dowhan W. Regulation of phosphatidylglycerophosphate synthase levels in Saccharomyces cerevisiae. J Biol Chem. 1998;273:11638–11642. doi: 10.1074/jbc.273.19.11638. [DOI] [PubMed] [Google Scholar]
- Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003a;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATPdependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003b;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shetty A, Lopes JM. Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot Cell. 2010;9:1845–1855. doi: 10.1128/EC.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, Henry SA, Arndt KM. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Skinner HB, McGee TP, McMaster CR, Fry MR, Bell RM, Bankaitis VA. The Saccharomyces cerevisiae phosphatidylinositol-transfer protein effects a ligand-dependent inhibition of choline-phosphate cytidylyltransferase activity. Proc Natl Acad Sci U S A. 1995;92:112–116. doi: 10.1073/pnas.92.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D, Daum G. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J Bacteriol. 2002;184:519–524. doi: 10.1128/JB.184.2.519-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & development. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Summers EF, Letts VA, McGraw P, Henry SA. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swede MJ, Hudak KA, Lopes JM, Henry SA. Strategies for generating phospholipid synthesis mutants in yeast. Methods in enzymology. 1992;209:21–34. doi: 10.1016/0076-6879(92)09005-n. [DOI] [PubMed] [Google Scholar]
- Tehlivets O, Hasslacher M, Kohlwein SD. S-adenosyl-L-homocysteine hydrolase in yeast: key enzyme of methylation metabolism and coordinated regulation with phospholipid synthesis. FEBS Lett. 2004;577:501–506. doi: 10.1016/j.febslet.2004.10.057. [DOI] [PubMed] [Google Scholar]
- Thomas D, Rothstein R, Rosenberg N, Surdin-Kerjan Y. SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol. 1988;8:5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. SAM1, the structural gene for one of the Sadenosylmethionine synthetases in Saccharomyces cerevisiae. Sequence and expression. J Biol Chem. 1987;262:16704–16709. [PubMed] [Google Scholar]
- Toke DA, Bennett WL, Dillon DA, Wu WI, Chen X, Ostrander DB, Oshiro J, Cremesti A, Voelker DR, Fischl AS, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding diacylglycerol pyrophosphate phosphatase. J Biol Chem. 1998a;273:3278–3284. doi: 10.1074/jbc.273.6.3278. [DOI] [PubMed] [Google Scholar]
- Toke DA, Bennett WL, Oshiro J, Wu WI, Voelker DR, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J Biol Chem. 1998b;273:14331–14338. doi: 10.1074/jbc.273.23.14331. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Voelker DR. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem. 1993;268:21416–21424. [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Yates R, Voelker DR. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J Biol Chem. 1995;270:6071–6080. doi: 10.1074/jbc.270.11.6071. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Tsui MM, York JD. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Advances in enzyme regulation. 2010;50:324–337. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Garcia MJ, Choi MS, Hinz FI, Gaspar ML, Jesch SA, Henry SA. Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Molecular genetics and genomics : MGG. 2011;285:125–149. doi: 10.1007/s00438-010-0592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci U S A. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waechter CJ, Lester RL. Regulation of phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1971;105:837–843. doi: 10.1128/jb.105.3.837-843.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waechter CJ, Lester RL. Differential regulation of the N-methyl transferases responsible for phosphatidylcholine synthesis in Saccharomyces cerevisiae. Archives of biochemistry and biophysics. 1973;158:401–410. doi: 10.1016/0003-9861(73)90637-1. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wilson MS, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. The Biochemical journal. 2013;452:369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- Wu WI, Liu Y, Riedel B, Wissing JB, Fischl AS, Carman GM. Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:1868–1876. doi: 10.1074/jbc.271.4.1868. [DOI] [PubMed] [Google Scholar]
- Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht JA, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci U S A. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Oshima A. Regulation of phosphatidylethanolamine methyltransferase level by myo-inositol in Saccaromyces cerevisiae. Eur J Biochem. 1980;104:611–616. doi: 10.1111/j.1432-1033.1980.tb04465.x. [DOI] [PubMed] [Google Scholar]
- Ye C, Bandara WM, Greenberg ML. Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J Biol Chem. 2013;288:24898–24908. doi: 10.1074/jbc.M113.493353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, Smits GJ, Loewen CJ. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- Zaccheo O, Dinsdale D, Meacock PA, Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem. 2004;279:24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]