Abstract
Ultrastructural studies conducted in situ using conventional transmission electron microscopy have had relatively little impact on defining the structural organization of chromatin. This is due to the fact that in routine transmission electron microscopy, together with the deoxyribonucleoprotein, many different intermingled substances are contrasted, masking the ultrastructure of chromatin. By selective staining of DNA in thin sections, using the Feulgen-like osmium-ammine reaction, these drawbacks have been overcome and worthwhile data have been obtained both on the gross morphology and the ultrastructural-functional organization of chromatin in situ. In the present study these results are reviewed and discussed in light of recent achievements in both interphase nuclear chromatin compartmentalization in interphase nuclei and in the structural organization of chromatin fibers in transcriptionally active and inactive chromatin.
Key words: chromatin structure in situ, DNA cytochemistry, transmission electron microscopy, chromatin fibers, nucleosomes, osmium-ammine
Introduction
The investigation of the structural-functional organization of chromatin in situ using conventional transmission electron microscopy has been hindered by a number of technical limitations. The major handicap derives from the fact that in most electron microscopy preparations, the agents employed for thin-section contrasting render electron-opaque, together with the deoxyribonucleoprotein, many other different intermingled substances, which masks the morphology of the chromatin structures.1,2 Therefore, very little in situ data have been produced on chromatin structural organization and its functionally related changes. However, valuable information has been obtained from studies applying a highly selective and specific staining method for DNA, the Feulgen-like osmium ammine technique. This staining procedure, introduced by Cogliati and Gautier,3 contributed to electron microscopy by exclusively visualizing the DNA-containing structures, thus allowing a study of the chromatin organization down to the molecular level in thin sections of samples fixed in situ.4-6 For this reason, it is of interest to review these in situ studies, even though obtained some years ago, and to discuss them in the light of recent results on the chromatin structural-functional organization, coming from investigations conducted using different technical approaches.7 In fact, comparisons of these data may help to shed light on topics which are still debated, such as: the relationship between chromosome territories and the interchromatin space within the interphase cell nucleus; the structure of chromatin fibres (i.e., whether with a diameter of 10 or 30 nm); the structure of transcriptionally-active and inactive chromatin. A brief description of the characteristics of the Feulgen-like technique for the visualization of DNA-containing structures at the electron microscope level and its potential for studying both gross and fine chromatin structure in situ will be discussed first.
The Feulgen-like osmium-ammine staining reaction
In 1973 Cogliati and Gautier proposed a staining procedure for the visualization of the DNA-containing structures at the electron microscope level.3 This method was based on a Feulgen-like reaction which utilizes an electron-opaque osmium-ammine complex as a Schiff-like reagent. The classic Schiff reagent is a solution of pararosaniline chloride (red-purple coloured) rendered colourless by SO2 exposure, which after reacting with aldehyde groups, restored to the red-purple colour of pararosaniline. This reagent is used for cytochemical detection of substances in which aldehyde groups are generated either by mild hydrochloric acid hydrolysis (e.g., DNA in the Feulgen reaction) or by periodic acid treatment (e.g., glycoconjugates in the PAS reaction).8 The osmium-ammine complex prepared by Cogliati and Gautier, and in a more reproducible protocol established by Olins et al.,9 behaves as a Schiff reagent in the presence of SO2 by reacting with aldehyde groups produced in the Feulgen and in the periodic acid Schiff (PAS) reaction, rendering electron opaque with high selectivity and specificity the DNA- and the glycoconjugate-containing structures in thin sections. For the selective staining of the DNA or the glycoconjugate-containing structures, formaldehyde fixation is mandatory.4 The staining reaction works on thin sections derived from routinely dehydrated and resin-embedded samples. The high selectivity for DNA and glyconjugates of the osmium-ammine-SO2 staining in the Feulgen and in the PAS reaction, respectively, has been demonstrated.10 In thin section of mammalian cells, only the DNA-containing structures are rendered electron-opaque in the Feulgen-like reaction whereas the cytoplasmic organelles exhibit very low electron opacity. In the nucleus, all the ribonu-cleoprotein components remain unstained (Supplementary Figure 1A). After periodic acid pre-treatment the staining reaction allows selective visualization of substances containing a high proportion of carbohydrate macromolecules such as glycogen, glycoprotein and proteoglycans; whereas DNA-containing structures are not rendered electron-opaque (Supplementary Figure 1B). For more complete information on the osmium-ammine complex as a Schiff-like reagent in electron microscopy, the reader is directed to.,6,8,9,11
Figure 1.
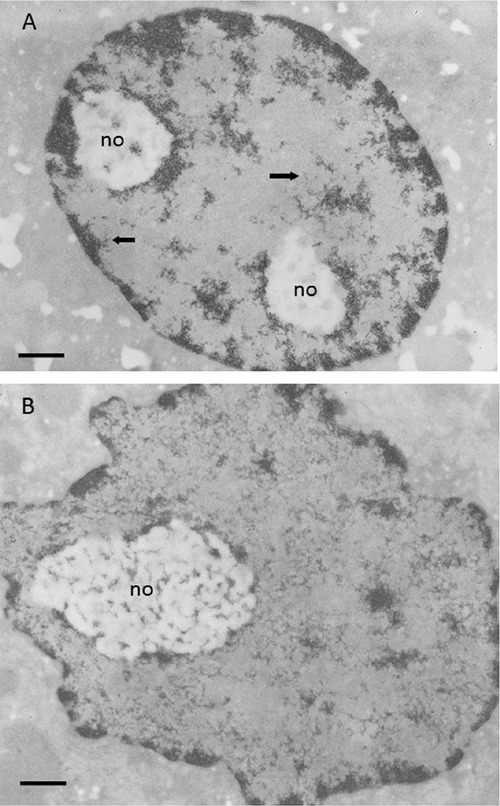
A) Resting rat hepatocyte stained with the Feulgen-like osmium-ammine reaction; the condensed chromatin is visible either closely associated to the inner nuclear membrane, scattered within the nucleoplasmic space, or surrounding the unstained nucleolar bodies (no); large portions of the nucleoplasm are devoid of chromatin structures; arrows indicate loosened chromatin structures branching out at the periphery of the condensed chromatin. B) Regenerating rat hepatocyte 24 h after partial hepatectomy; Feulgen-like osmium-ammine staining; the amount of condensed chromatin is strongly reduced; the loosened chromatin structures are uniformly distributed throughout the nucleoplasmic space, where they constitute a close-mesh network. Scale bar: 0.5 μm.
The ultrastructure of chromatin in situ
Chromatin architecture in the interphase nucleus
The first studies conducted using osmium-ammine staining in the Feulgen-like reaction allowed easy distinguishing of condensed heterochromatin from dispersed euchromatin of mammalian cell nuclei in situ and clearly established the gross ultrastructural morphology of transcriptionally-active chromatin.12,13 It has been shown that chromatin organization depends on the metabolic state on the cell, more precisely on RNA polymerase II transcriptional activity.14-16 In cells with a moderate RNA polymerase II-mediated transcriptional activity, such as resting rat hepatocytes, a significant portion of chromatin is condensed in electron-dense masses, well separated from the completely electron-translucent nucleoplasmic space, the interchromatin compartment (Figure 1A). Thinner chromatin threads appear to branch out for a short length from the periphery of the chromatin masses. On the contrary, in highly transcriptionally-active cells, such as regenerating rat hepatocytes, chromatin appears mainly in a very dispersed form, constituting a network of interwoven threads uniformly distributed through the nucleoplasmic space. Very small clumps of condensed chromatin are associated with the inner nuclear membrane and sparsely distributed in the nucleoplasm (Figure 1B). Selective inhibition of RNA polymerase II activity by α-amanitin causes highly dispersed chromatin to compact into electron dense masses, thus increasing the interchromatin space completely devoid of chromatin structures (Figure 2). The dispersion of the chromatin clumps, with the increased occupancy of the interchromatin space that accompanies the progressive increase in RNA polymerase II-mediated transcriptional activity, is particularly striking in human circulating lymphocytes stimulated to proliferate by phytohemagglutinin (Figure 3A-C). These observations indicate that chromosomes during interphase are highly plastic structures and that the relationship between chromatin and the interchromatin space is highly variable depending upon RNA transcription and cell cycle phases.17 The chromatin and the inter-chromatin compartment appear to be clearly segregated only in the case of low nuclear transcriptional activity, whereas in cells with up-regulated synthesis of mRNAs a clear compartmentalization is not detected. These findings may ameliorate the differences between models proposed for the relationship between the chromatin and interchromatin compartment in interphase cell nucleus.18-20
Figure 2.
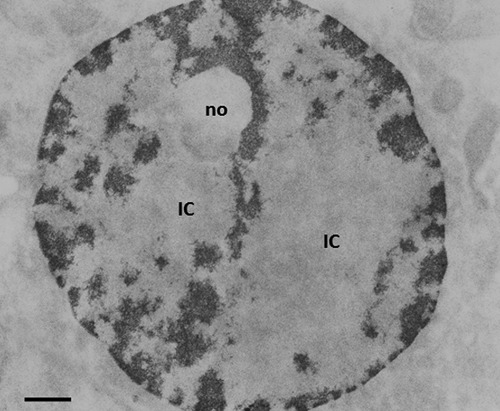
Feulgen-like osmium-ammine staining. Regenerating hepatocyte at 24 h after partial hepatectomy, from a rat treated with α-amanitin for 4 h. As a consequence of the inhibition of RNA polymerase II activity, all the dispersed chromatin was condensed into compact chromatin masses. no: nucleolar body. IC, interchromatin space. Scale bar: 0.5 μm.
Figure 3.
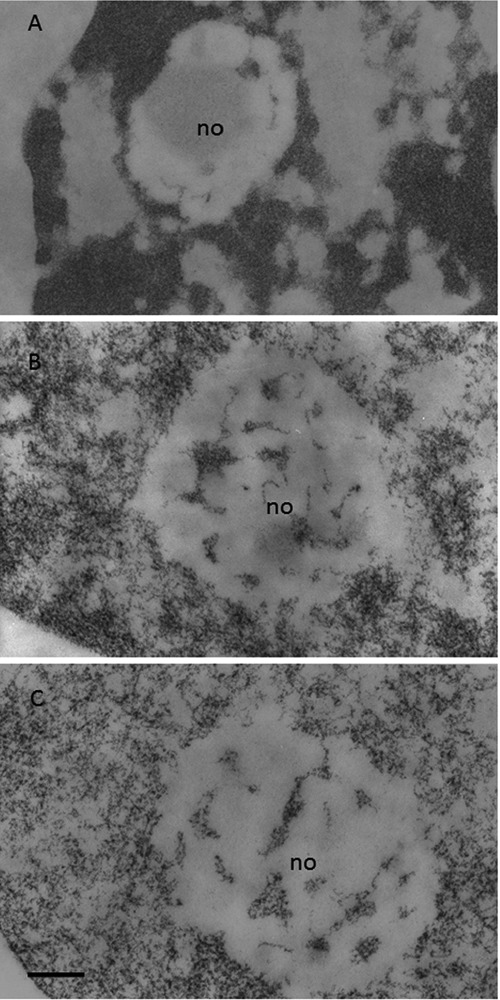
Circulating human lymphocytes. Feulgen-like osmium-ammine staining; no, nucleolus. A) Resting lymphocyte; almost all the chromatin is in a highly condensed state; a clear separation between chromatin and the interchromatin space is present. B) Lymphocyte stimulated to proliferate by a 24-hour phytohemagglutinin exposure; the compactness of the chromatin masses is reduced; the loosened chromatin structures begin to occupy the interchromatin space. C) Lymphocyte stimulated to proliferate by a 48-hour phytohemagglutinin exposure; chromatin is in a dispersed configuration and is uniformly distributed throughout the nucleoplasm. Scale bar: 0.2 μm.
Evidence supports that after mitosis the compact chromosomes, even though they undergo marked decondensation, still occupy distinct regions, the chromosomes territories, within the interphase nucleus.21-24 The inter-chromatin domain contains proteins and ribonucleoproteins which constitute the nuclear machinery necessary for nucleic acid synthesis and processing. From an ultrastructural point of view, two major models for chromosome territory and interchromatin domain compartmentalization have been proposed. One model, proposed by Cremer and Cremer,18 suggests that the chromosomes territories are separated by the interchromatin domain, which are almost devoid of DNA, and that transcriptionally-active genes are located at the periphery of a given chromosome territory, where they constitute a thin layer, the perichromatin region, in which extended chromatin fibers and RNA transcripts intermingle.25 The other model, proposed by Branco and Pombo,19,20 argues that chromatin from different chromosomes is not separated by the interchromatin domain. Rather, the chromatin structures form a lattice-like network of fibers which intermingle in an uniform way within individual chromosome territory and between different chromosome territories. A defined perichromatin region is excluded, RNA transcription taking place everywhere within the chromatin domains. Thus, this model does not support the compartmentalization in chromosomes territories, interchromatin domain and perichromatin region. Indeed, the data obtained using the osmium-ammine Feulgen-like reaction suggest that the model of the chromosome territories, interchromatin domain and perichromatin region compartmentalization may be more valid for nuclei characterized by a low RNA transcription rate in which a clear compartmentalization in chromatin domains and the interchromatin space is present. This appears not to be the case for cells characterized by an enhanced transcription rate. In such cells, chromatin constitutes a diffuse network uniformly distributed throughout the nucleoplasm, with all the nucleoplasmic components intermingled together and the consequent loss of any obvious compartmentalization. Also, ultrastructural data coming from the preferential visualization of the ribonucleoprotein components using the uranyl-EDTA-lead staining procedure11,26 were consistent with the apparent presence of three different compartments in the nuclei of resting cells and with the loss of this compartmentalization in cells with an enhanced Pol II transcriptional activity. This is clearly visible in Supplementary Figure 2 A,B showing a resting and a regenerating rat hepatocyte, respectively. It appears that, depending on the transcriptional activity of the cells, the organization of the nuclear compartments can be consistent with one or the other model. However it is worth noting that, whereas the Cremer and Cremer model18 can be consistent also with a modulation of chromatin architecture from resting cells (characterized by a low RNA transcription rate) to proliferating cells (characterized by a high RNA transcription rate), just by expanding the perichromatin region, the Branco and Pombo model19,20 cannot justify the chromatin compartmentalization in cells with a low RNA transcription rate, being in fact applicable only to highly transcriptionally active cells.
Chromatin structure: nucleosome visualization
In the late 20th century there were major achievements in the study of chromatin structure and function derived from investigations conducted using isolated chromatin that was subjected to treatment with detergents, different ionic strength solutions and metal cation concentrations, and to nuclease digestion. Many years ago these studies led to the discovery that chromatin was constituted by repeating units,27 the nucleosomes, made up of histones and DNA,28 thus opening new scenarios for the understanding of the structural organization of DNA in chromatin and of the mechanisms controlling gene expression.29
Visualization of nucleosomes in situ was obtained using the osmium-ammine staining in the Feulgen-like reaction. In fact, this staining procedure was found to possess high resolution of the stained structures in thin sections, thus allowing a clear visualization of DNA filaments (as thin as the 2-3 nm DNA double helix molecule)4 and visualization of nucleosomes in thin sections of mammalian cells routinely processed for electron microscopy.30 The nucleosome is composed by a histone octamer, two each of histones H2A, H2B, H3 and H4, which constitute a protein core around which 147 base pairs of DNA are wrapped in 1.7 left-handed super-helical turns; histone H1 is attached to the complex.31 Ultrastructurally, nucleosomes are flat cylinders with a diameter of 11 nm and with a height of 5.5 nm. Therefore, after the Feulgen-like osmium-ammine reaction, since only DNA is stained, the particles appear as small rings, with a diameter of about 11 nm, constituted by electron-opaque thin filament, about 3 nm thick (corresponding to the thickness of double helix DNA filament), delineating an unstained inner core. The observation of these ring-shaped structures is obviously limited to those portions of chromatin where the electron beam is perpendicular to the diameter of the nucleosomes. Moreover, nucleosomes in thin sections of chromatin fixed in situ do not lie in the same plane, but in fact (given the thickness of sections that range from 50-80 nm) they may be distributed on overlapping planes. Thus, section thickness and varying orientations of the chromatin reduces a generalized visualization of the ring-shaped particles. Nevertheless, they can be frequently recognized both in the extended fibres of the dispersed chromatin (compare Figure 4A, in which nucleosomes are shown in a spread chromatin preparation, with Figure 4B, showing a dispersed in situ chromatin fibre) and within the small clumps and the masses of highly condensed chromatin (Figure 4C,D). Measurements of the diameter of clearly recognizable ring-shaped particles (performed on micrographs with an original magnification of 50,000 and photographically enlarged to 175,000) resulted in a mean value of 11±0.8 nm (S.D.) with a range from 9.5 to 13.0 nm,30 strongly suggesting their identification with nucleosomes.
Figure 4.
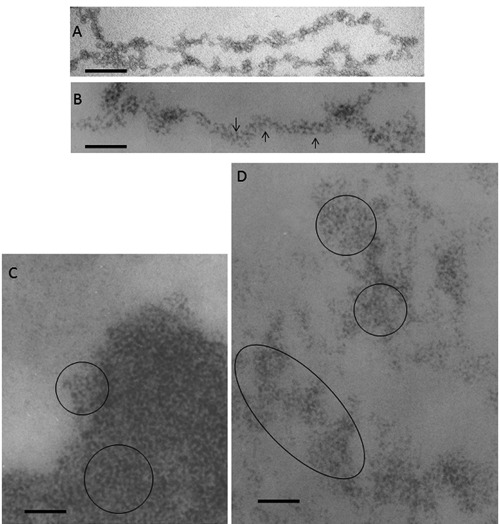
A) Chromatin fibers spilling out of a ruptured chicken erythrocyte nucleus, negative staining with 0.5% ammonium molybdate adjusted to pH 7.4 to 8.0 with NH4OH. Scale bar: 50 nm. B,C,D) Thin sections from samples fixed in situ, resin embedded and stained with the Feulgen-like osmium-ammine reaction. B) regenerating rat hepatocytes, 24 h after partial hepatectomy; a long chromatin fiber is visible, 20-25 nm thick, which appears to be composed by a series of round particles with a diameter of about 11 nm, exhibiting an unstained inner core encircled by a DNA ring with a thickness of 2-3 nm (arrows). C) human resting lymphocyte; in the highly compact chromatin, particles similar to those described in (B) can be detected (encircled). D) regenerating rat hepatocyte; a particulate organization is present in the loosened chromatin structures (encircled). Scale bar: 50 nm
Chromatin structure: the chromatin fibers
After the discovery of the nucleosomal organization of chromatin in 1974, a series of investigations were conducted using different methological approaches in order to determine whether the nucleosomal filament (in the remainder of this article the nucleosomal filament will be described as an 11-nm fiber) gives rise to higher order structures for DNA compaction within interphase and metaphase chromosomes. These studies, conducted in vitro, lead to the conviction that the nucleosome filament, the first level of DNA compaction, was further folded into a 30 nm chromatin fiber, that was considered to represent the basic unit of interphase chromatin.32 Furthermore, 30 nm chromatin fibres were visualised in isolated mitotic chromosomes swollen in low salt solution,33,34 in starfish sperm intact nuclei35 and in polytene chromosome puffs after inhibition of transcription.36 This firm belief of the existence of 30 nm chromatin fibers was questioned by a series of studies conducted in recent years using different approaches to visualize chromatin in situ.7 Using cryo-electron microscopy and image processing of vitreous sections of human mitotic chromosomes, it was possible to demonstrate that mitotic chromatin exhibited a homogeneous grainy texture, in which there were no visible 30-nm fibers, but the bulk of compact chromatin existed in a highly disordered and interdigitated state, comparable with a polymer melt.37,38 Furthermore, the application of the electron spectroscopic imaging (ESI) technique to the study of chromatin structure39 allowed the visualization in situ of 10 nm thick chromatin fibres40 even in compact chromatin domains known as condensed heterochromatin.41 These ESI data indicated that a large portion of chromatin in mammalian cell nuclei was organized in 10 nm thick fibers.42 Indeed, very recently using cryo-electron microscopy and synchrotron X-ray scattering, it was shown that human mitotic HeLa chromosomes were composed mainly of irregularly folded nucleosome fibers no thicker than 11 nm, thus indicating the near absence of regular 30-nm chromatin fibers within chromosomes.43
In this context, has the study of the structural organization of chromatin in situ using the Feulgen-like osmium-ammine technique added to our knowledge? The major questions explored were the following: i) Are the 11-nm and the 30-nm chromatin fibers present in situ? ii) Which of these two kinds of fibers represents the basic unit of chromatin in situ? The studies conducted in situ were mainly carried out using mammalian cells characterized by a physiological dispersion of chromatin, such as regenerating hepatocytes, which greatly facilitated the observations of chromatin structural organization. Furthermore, these cells have very large nucleolar bodies characterized by a greater electron-translucency than the nucleoplasmic space, a consequence of rRNA extraction due to the HCl hydrolysis. In the nucleolar bodies most of the intranucleolar chromatin structures consist of very small clumps and of thin fibers which are well separated from each other and can be followed for a very long tract, unlike the fibers in the extra-nucleolar compartment (Figure 5A). These characteristics rendered the regenerating rat hepatocyte nuclei, and especially the intranucleolar chromatin, particularly suitable for the high resolution study of the chromatin structure in situ. The studies conducted on this type of cells revealed the presence of both 11 and 20-25 nm chromatin fibers.30,44 Regarding the 11-nm fibers, they were easily recognized within the nucleolar body and frequently appear to be loosely interwoven to each other (Figure 5B). Visualization of the 11-nm fibers was improved by the use of stereo-pair micrographs, which demonstrated that many of the chromatin structures were composed of these kind of fibers (Figure 6A). The 11-nm fibers were also visualized in the extranucleolar nucleoplasmic space. The analysis of stereopair micrographs allowed these fibers to be detected giving rise to rings of different diameter, frequently intermingled together (Figure 6B). Regarding the thicker chromatin fibers, the analysis of chromatin structure in situ did not reveal the presence of fibers with an actual thickness of 30 nm, but, in fact showed fibers with a thickness ranging from 20 to 25 nm. This kind of fibers were mainly observed within the nucleolar body (Supplementary Figure 3), always as solitary fibers, and never being clearly observed to constitute more complex chromatin structures, in contrast to the 11 nm thick fibers. The 20-25 nm thick fibers did not show either a solenoid or a beaded organization. Other than the 11 and the 20-25 nm fibers, fibers with intermediate thickness were also observed. Analysis of these fibers using high magnification stereopair micrographs indicated that these fibers appeared to be frequently composed of two 11 nm thick nucleofilaments wound around one-another (Figure 7A), thus suggesting that the 20-25 nm thick fibers might result from the association of two 11 nm fibers. Regarding the structure of the chromatin in metaphase chromosomes, in situ analysis after the Feulgen-like osmium ammine reaction, also carried out using stereo-pair micrographs, did not allow detection of a clear structural organization in basic units, constituted by either 11 or 25-30 nm thick chromatin fibers (Figure 7B).
Figure 5.
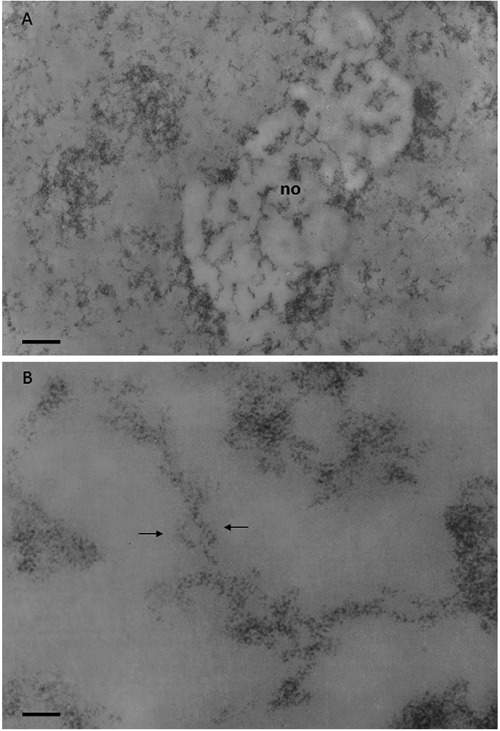
Regenerating rat hepatocyte at 24 h after partial hepatectomy; Feulgen-like osmium-ammine staining. A) Chromatin is mainly in a dispersed form, only small clumps of compact chromatin can be detected; the nucleolar body (no) appears to be more electron-translucent than the remaining nucleoplasm, very likely due to RNA extraction; within the nucleolar body many chromatin fibers, well separated from each other, can be followed for a considerable length; scale bar: 0.4 μm. B) Loosened organization of the intranucleolar chromatin which appears to be composed of fibers with a thickness ranging from 11 to 20 nm; arrows indicate 11 nm thick chromatin fibers intertwined with each other, as the serpent around the rod of Asclepius; scale bar: 50 nm.
Figure 6.
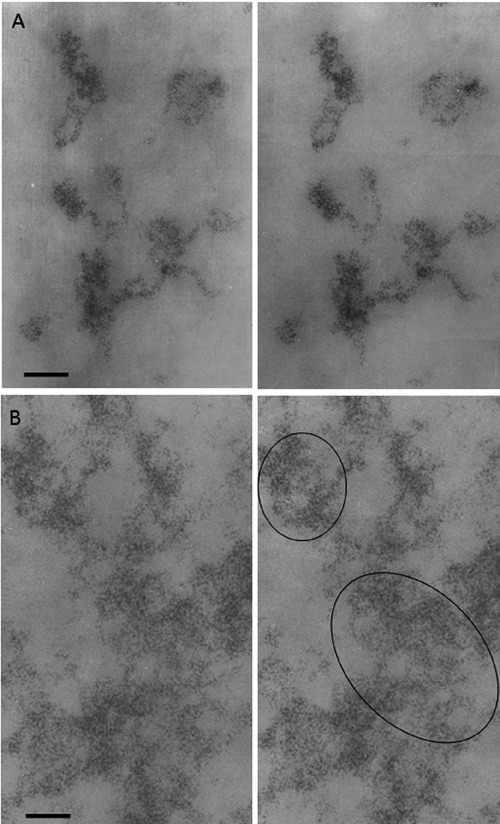
Regenerating rat hepatocyte at 24 h after partial hepatectomy; Feulgen-like osmium-ammine staining; stereo-pair micrographs, tilt angle ±5. A) Intranucleolar chromatin structures show their organization in 11-nm thick fibers. B) Extranucleolar loose chromatin masses appear to be composed of 11-nm chromatin fibers (in the circles). Scale bar: 50 nm.
Figure 7.
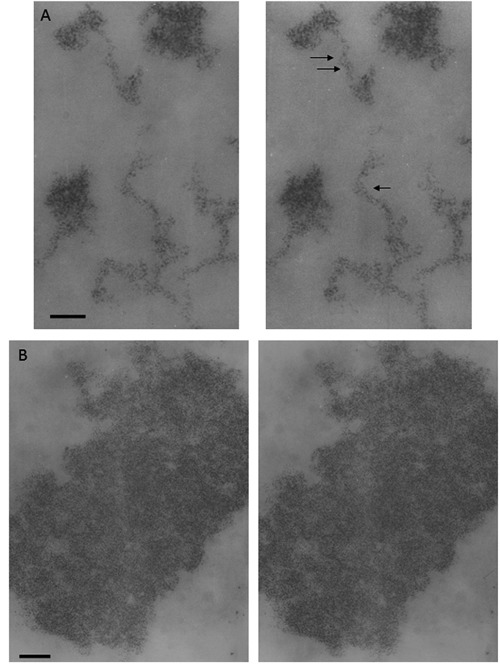
Regenerating rat hepatocyte at 24 h after partial hepatectomy; Feulgen-like osmium-ammine staining; stereo-pair micrographs, tilt angle ±5. A) Intranucleolar chromatin structures; arrows indicate two 20-nm chromatin fibers which appear to be composed of two 11-nm fibers wrapped around each other; scale bar: 50 nm. B) Metaphase chromosome; even though the 3D image allows visualization of a generalized particulate organization of chromatin, no clear evidence can be obtained demonstrating widespread presence of fibers either 11 or 25-30 nm thick; scale bar: 0.1 μm.
Altogether these observations are consistent with the available data obtained from different methodological approaches indicating that, in situ, interphase chromatin is mainly organized in fibers with a thickness of about 11 nm; while the image data are not consistent with a structural organization of chromatin based upon units composed of 30 nm thick chromatin fibers.
The fine structure of transcriptionally-active chromatin
Another question that has been posed after the demonstration that DNA is organized into nucleosomes was to ascertain the relationship between the nucleosomal organization of DNA and gene transcription. In other words, whether DNA uncoils from the histone octamer as a pre-requisite for transcription and whether it must resume the nucleosome configuration for gene silencing. In this context, special attention has been paid to the structural-functional relationship of ribosomal genes. This is due to the fact that, in higher eukaryotes, the rRNA genes are present in multiple copies and, therefore, easily studied by different methodological approaches. For example, human diploid cells contain about 400 ribosomal genes. Data obtained from spread chromatin preparations indicated that the transcribing ribosomal genes are constituted by a central DNA axis connected with nascent growing RNA transcripts whose length progressively increases from the transcription initiation site to the termination site, the well-known Christmas tree structure.45 A series of in vitro studies showed that the DNA of the transcribing ribosomal genes is in a nonnucleosomal configuration, whereas that of the repressed transcriptionallyinactive ribosomal genes exhibits a nucleosome organization, suggesting that the activity of ribosomal genes could be regulated by DNA unravelling versus compaction into nucleosome.46 By contrast, in vivo psoralen cross-linking assays have demonstrated that the ratio between the non-nucleosomal and the nucleosomal structured chromatin did not change, even with a marked variation of transcriptional activity,46-52 questioning the assumption that ribosome gene transcription can be controlled by DNA unravelling and compaction. The study of chromatin in situ using the Feulgen-like osmium-ammine staining has greatly contributed to elucidation of the structural-functional organization of ribosomal chromatin. In mammalian cells, during interphase the ribosomal genes associated with transcription factors are located in the nucleolus, within the so-called fibrillar centers (Figure 8A). This precise localization has been established by the fact that some factors associated with ribosomal genes (i.e., nucleolin, upstream binding factor, and the largest RNA polymerase I subunit) are selectively stained by silver, which is located in the fibrillar centers42 (Figure 8B). It has been repeatedly demonstrated, using the Feulgen-like staining, that the ribosomal chromatin is present within the fibrillar centres (Figure 8C) and it is organized into very thin filaments with a thickness of 2-3 nm (the same thickness as that of the DNA filament) and are loosely interwoven (Figure 8D). Of note, the greater portion of the ribosomal chromatin within the fibrillar centres is transcriptionally inactive, despite being associated with a number of transcription factors, rRNA transcription taking place only at the periphery of the fibrillar centers.44 In addition, the DNA present in metaphase nucleolar organizer regions, which consist of transcriptionally-inactive ribosomal genes are also in an extended non-nucleosomal configuration. This was demonstrated by combining the silver staining procedure for the AgNOR proteins with the Feulgen-like osmium-ammine reaction53 (Figure 8E,F). Therefore, these in situ studies support the conclusion that, at least for ribosome genes, the extended, non-nucleosomal structure is a necessary, but not sufficient condition for transcription and that gene inactivation during interphase and metaphase does not always occur by DNA compaction into nucleosomes. Is this a general rule that can be extended to non-ribosomal genes? Unfortunately there are no available data on the structure-function relation of other genes in situ that might lead to similar or divergent conclusions. Indeed, extended DNA filaments similar to those observed in the nucleolus have never been clearly found in the nucleoplasmic space, even in high transcriptionally-active nuclei such as those of regenerating hepatocytes. However, the possibility cannot be ruled out that the lack of visualization of extended DNA filaments in the extra-nucleolar compartment may be due to the low number of copies of the transcriptionally active non-ribosomal genes.
Figure 8.
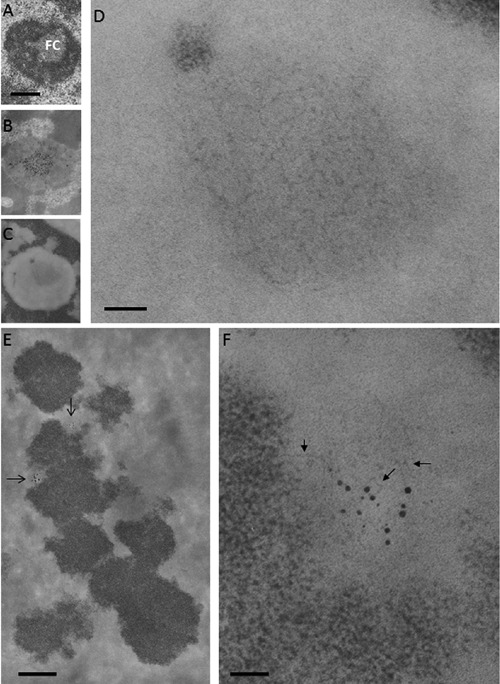
Ribosomal chromatin. A, B, C) Human circulating lymphocytes; scale bar: 0.4 μm. A) Aldehyde and osmium fixation, uranium and lead staining: a solitary, large fibrillar center (FC) is present in the central area of the nucleolus. B) En-bloc silver staining for the visualization of proteins associated to the ribosomal genes; thin section stained with uranium and lead: silver deposits are mainly located in the FC. C) Feulgen-like osmium ammine reaction: in the nucleolus, a light electron-opaque area is located in the same central zone occupied by the fibrillar center. D) Detail of (C) showing that the central area of the nucleolus is occupied by a large agglomerate of extended DNA filaments with a thickness of 2-3 nm; scale bar: 50 nm. E,F) En-bloc silver staining for the visualization of the proteins associated with the ribosomal genes in metaphase chromosomes; thin section stained with the Feulgen-like osmium-ammine reaction. E) Silver grain deposits identify two nucleolar organizer regions (arrows); scale bar: 10 μm. F) Detail of (E) showing that chromatin associated with the silver grains is composed of thin DNA filaments (arrows), similar to the fibrillar centers of interphase nuclei; scale bar: 50 nm.
Conclusions
The ultrastructural studies conducted in situ at the electron microscope level using the Feulgen-like osmium-ammine reaction for the selective staining of DNA have helped to define the structural-functional organization of interphase and metaphase chromatin in mammalian cells. The major contributions are the following: i) The gross chromatin morphology appears to depend upon RNA polymerase II transcriptional activity; a clear compartmentalization into chromatin territories, peri-chromatin region and inter-chromatin space is present only within nuclei with a low transcriptional activity; compartmentalization is lost during enhanced RNA synthesis. This demonstrated high plasticity of chromatin is consistent both with the chromosome territory-interchromatin model18,25 and with the interchromatin network model:19,20 the nuclear architecture model depending upon the transcriptional activity of the cell. ii) The structural organization of interphase chromatin appears to be mainly based on the about 11 nm thick fiber. Thicker chromatin fibers (20-25 nm thick) were also visualized, but only in special nuclear districts (i.e., the nucleolar body), where these fibers sometimes appeared to be composed of two interwoven 11 nm thick fibers. No evidence was obtained that the 20-25 nm thick chromatin fiber represents a basic unit of condensed interphase chromatin or of metaphase chromosomes. Therefore, these data are consistent with a model of structural organization of chromatin based on the 11 thick fibers, but not upon on higher-order (20-30 nm thick) fibers. iii) Not all transcriptionallyinactive chromatin is organized into nucleosomes, as observed for a portion of chromatin containing the ribosomal genes. This conclusion is consistent with the data obtained from the psoralen photocrosslinking assay of ribosomal chromatin structure,46-51 suggesting a model for control of ribosome gene activity that is not mediated simply by DNA unraveling from, or compaction into the nucleosome structure.
Acknowledgments
This work was supported by grants for orientated fundamental research from the Italian Ministry of Education, University and Research (MD). DEO and ALO thank the German Cancer Research Center, Heidelberg, Germany for Guest Scientist Awards and the University of New England, College of Pharmacy, Portland, ME, USA, for support.
References
- 1.Rapkin LM, Anchel DR, Li R, Bazett-Jones DP. A view of the chromatin landscape. Micron. 2012;43:150-8 [DOI] [PubMed] [Google Scholar]
- 2.Fakan S. Ultrastructural cytochemical analyses of nuclear functional architecture. Eur J Histochem. 2004; 48:5-14 [PubMed] [Google Scholar]
- 3.Cogliati R, Gautier A. Mise en évidence de l’ADN et des polysaccharides à l’aide d’un nouveau réactif “de type Schiff”. CR Acad Sci Hebd Seances Acad Sci D. 1973;276:3041-4 [PubMed] [Google Scholar]
- 4.Derenzini M, Viron A, Puvion-Dutilleul F. The Feulgen-like osmium-ammine reaction as a tool to investigate chromatin structure in thin sections. J Ultrastruct Res. 1982;80:133-47 [DOI] [PubMed] [Google Scholar]
- 5.Derenzini M. Ultrastructural cytochemistry, pp 69-93 Morel G. () Visualization of nucleic acids. CRC Press, 1995 [Google Scholar]
- 6.Biggiogera M, Courtens JL, Derenzini M, Fakan S, Hernandez-Verdun D, Risueno MC, et al. Osmium ammine: review of current applications to visualize DNA in electron microscopy. Biol Cell. 1996;87:121-32 [PubMed] [Google Scholar]
- 7.Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Curr Opin Cell Biol. 2012;24:359-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chieco P, Derenzini M. The Feulgen reaction 75 years on. Histochem Cell Biol. 1999;111:345-58 [DOI] [PubMed] [Google Scholar]
- 9.Olins AL, Moyer BA, Kim SH, Allison DP. Synthesis of a more stable osmium ammine electron-dense DNA stain. J Histochem Cytochem. 1989;37:395-8 [DOI] [PubMed] [Google Scholar]
- 10.von Schack ML, Fakan S. Retention of glycogen in cryosubstituted mouse liver. Histochemistry. 1994;102:451-5 [DOI] [PubMed] [Google Scholar]
- 11.Moyne G. Methods in ultrastructural cytochemistry of the cell nucleus. Prog Histochem Cytochem. 1980;13:1-72 [DOI] [PubMed] [Google Scholar]
- 12.Derenzini M, Lorenzoni E, Marinozzi V, Barsotti P. Ultrastructural cytochemistry of active chromatin in regenerating rat hepatocytes. J Ultrastruct Res. 1977;59:250-62 [DOI] [PubMed] [Google Scholar]
- 13.Derenzini M. Fine structure of chromatin as visualized in thin sections with the Gautier selective stain for DNA. J Ultrastruct Res. 1979;69:239-48 [DOI] [PubMed] [Google Scholar]
- 14.Derenzini M, Novello F, Pession-Brizzi A. Perichromatin fibrils and chromatin ultrastructural pattern. Exp Cell Res. 1978; 112:443-54 [DOI] [PubMed] [Google Scholar]
- 15.Derenzini M, Pession-Brizzi A, Bonetti E, Novello F. Relationship between ultrastructure and function of hepatocyte chromatin: a study with adrenalectomized rats after cortisol administration. J Ultrastruct Res. 1979;67:161-79 [DOI] [PubMed] [Google Scholar]
- 16.Derenzini M, Pession-Brizzi A, Betts-Eusebi C, Novello F. Relationship between the fine structural organization of chromatin and nucleic acid synthesis in regenerating rat hepatocytes. J Ultrastruct Res. 1981;75:229-42 [DOI] [PubMed] [Google Scholar]
- 17.Cremer M, Zinner R, Stein S, Albiez H, Wagler B, Cremer C, et al. Three dimensional analysis of histone methylation patterns in normal and tumor cell nuclei. Eur J Histochem. 2004;48:15-28 [PubMed] [Google Scholar]
- 18.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branco MR, Pombo A. Chromosome organization: new facts, new models. Trends Cell Biol. 2007;17:127-34 [DOI] [PubMed] [Google Scholar]
- 21.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292-301 [DOI] [PubMed] [Google Scholar]
- 22.Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541-67 [DOI] [PubMed] [Google Scholar]
- 23.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur J Histochem. 2006;50:161-176 [PubMed] [Google Scholar]
- 24.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem. 2006;50:223-272 [PubMed] [Google Scholar]
- 25.Fakan S, van Driel R. The perichromatin region: a functional compartment in the nucleus that determines large-scale chromatin folding. Semin Cell Dev Biol. 2007;18:676-81 [DOI] [PubMed] [Google Scholar]
- 26.Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250-65 [DOI] [PubMed] [Google Scholar]
- 27.Olins A L, Olins D E. Spheroid chromatin units (v bodies). Science. 1974;183:330:2. [DOI] [PubMed] [Google Scholar]
- 28.Kornberg R. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868-71 [DOI] [PubMed] [Google Scholar]
- 29.Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4:809-14 [DOI] [PubMed] [Google Scholar]
- 30.Derenzini M, Hernandez-Verdun D, Bouteille M. Visualization of a repeating subunit organization in rat hepatocyte chromatin fixed in situ. J Cell Sci. 1983;61:137-49 [DOI] [PubMed] [Google Scholar]
- 31.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285-94 [DOI] [PubMed] [Google Scholar]
- 32.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rattner JB, Hamkalo BA. Higher order structure in metaphase chromosomes. II. The relationship between the 250 A fiber, superbeads and beads-on-a-string. Chromosoma. 1978;69:373-9 [DOI] [PubMed] [Google Scholar]
- 34.Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849-58 [DOI] [PubMed] [Google Scholar]
- 35.Woodcock CL. Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length. J Cell Biol. 1994;125:11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson K, Mähr R, Björkroth B, Daneholt B. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma. 1982;87:33-48 [DOI] [PubMed] [Google Scholar]
- 37.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol. 2010;22:291-7 [DOI] [PubMed] [Google Scholar]
- 39.Bazett-Jones D P, Hendzel MJ. Electron spectroscopic imaging of chromatin. Methods. 1999;17:188-200 [DOI] [PubMed] [Google Scholar]
- 40.Ahmed K, Li R, Bazett-Jones DP. Electron spectroscopic imaging of the nuclear landscape. Methods Mol Biol. 2009;464:415-23 [DOI] [PubMed] [Google Scholar]
- 41.Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fussner E, Ching RW, Bazett-Jones D P. Living without 30nm chromatin fibers. Trends Biochem Sci. 2011;36:1-6 [DOI] [PubMed] [Google Scholar]
- 43.Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derenzini M, Pasquinelli G, O’Donohue MF, Ploton D, Thiry M. Structural and functional organization of ribosomal genes within the mammalian cell nucleolus. J Histochem Cytochem. 2006;54:131-45 [DOI] [PubMed] [Google Scholar]
- 45.Miller OL, Jr, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955-7 [DOI] [PubMed] [Google Scholar]
- 46.Toussaint M, Levasseur G, Tremblay M, Paquette M, Conconi A. Psoralen photocrosslinking, a tool to study the chromatin structure of RNA polymerase I--transcribed ribosomal genes. Biochem Cell Biol. 2005;83:449-59 [DOI] [PubMed] [Google Scholar]
- 47.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753-61 [DOI] [PubMed] [Google Scholar]
- 48.Conconi A, Sogo JM, Ryan CA. Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc Natl Acad Sci USA. 1992;89:5256-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dammann R, Lucchini R, Koller T, Sogo JM. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fahy D, Conconi A, Smerdon MJ. Rapid changes in transcription and chromatin structure of ribosomal genes in yeast during growth phase transitions. Exp Cell Res. 2005;305:365-73 [DOI] [PubMed] [Google Scholar]
- 52.Sirri V, Roussel P, Hernandez-Verdun D. The AgNOR proteins: qualitative and quantitative changes during the cell cycle. Micron. 2000;31:121-6 [DOI] [PubMed] [Google Scholar]
- 53.Hernandez-Verdun D, Derenzini M. Non-nucleosomal configuration of chromatin in nucleolar organizer regions of metaphase chromosomes in situ. Eur J Cell Biol. 1983;31:360-5 [PubMed] [Google Scholar]


