Abstract
KRIT1 is an 84kDa protein that lacks any relevant catalytic domains, associated with the cerebral cavernous malformation disease. We have investigated by means of ultrastructural immunocytochemistry the nuclear distribution of KRIT1 in different cell lines, revealing its unexpected localization on actively transcribing nuclear domains such as the perichromatin fibrils and the nucleolar dense fibrillar component. These preliminary data indicate a still undescribed and unknown role for KRIT1 inside the nucleus.
Key words: cell nucleus, cerebral cavernous malformations, KRIT1, transcription, electron microscopy, immunocytochemistry
Introduction
KRIT1 (Krev1 interaction trapped gene 1) is a gene associated with the cerebral cavernous malformation (CCM) disease, characterized by vascular abnormalities that cause risk of hemorrhages in the brain with consequent headache, seizures or even death.1 Along with KRIT1, two other genes, CCM2 (cerebral cavernous malformation 2) and PDCD10 (programmed cell death 10), were linked to CCM disease,2,3 all the three genes coding for scaffold proteins lacking catalytic domains but presenting several domains for protein-protein interaction.4 In the last years, several studies have provided new information on the signaling pathway in which KRIT1 takes part, suggesting a role in vascular development,5 cellcell junctions,6 and cell-extracellular matrix adhesion.7 However, none of these studies was focused on the nuclear compartment where it is known KRIT1 can be physiologically present,8,9 as seen by fluorescence resonance energy transfer (FRET) experiments and immunofluorescence. Indeed KRIT1, together with its protein interacting partner, ICAP1 (integrin beta-1-binding protein 1), shuttles between the nucleus and the cytoplasm bearing a putative NLS in its N-terminal amino acid sequence.
Although the shuttling dynamics involving KRIT1, ICAP1 and CCM2 is only partially understood,9 there is no evidence in literature of a possible active function of KRIT1 inside the nuclear compartment. One of the current hypotheses is that KRIT1 and ICAP1 regulate each other by sequestering the partner inside the nucleus, thus preventing interaction with cytoplasmic proteins, such as beta-1 integrin or the p-38-MAPK associated complex.8 By means of ultrastructural immunocytochemistry we detected KRIT1 inside the nucleus with the aim to gain insight into other possible roles of this protein, as suggested by its localization on nuclear structures of known function. The nuclear location of KRIT1 was investigated by transmission electron microscope in HeLa cells under normal growth conditions or after heat shock when the nuclear distribution of ribonucleoproteins (RNPs) is modified,10 as well as in rat hepatocytes and in Chironomus thummi salivary glands (the latter being an excellent model for transcription due to the presence of actively transcribed polytenic chromosomes).
The results obtained suggest that KRIT1 is involved in the maturation of nascent RNA transcripts, due to its prevalent localization in nuclear sites where active transcription takes place.
Materials and Methods
Cells and treatments
HeLa cells were grown in DMEM medium supplemented with 10% fetal bovine serum, 2mM glutamine and 100 units each of streptomycin and penicillin, in 25 cm2 plastic flasks or on 4 cm2 glass coverslips in multi-well dishes on air atmosphere containing 5% of CO2 at 37°C (all reagents and plastics were from Euroclone, Milan, Italy). For the heat shock treatment, the cells were incubated at 42°C for 1 h, and then kept at 37°C for additional 3 h before fixation.
Transmission electron microscopy and ultrastructural immunocytochemistry
The cells were harvested by mild trypsinization (0.25% trypsin in PBS containing 0.5% EDTA) and immediately fixed with 4% paraformaldehyde (2 h at 4°C) in the culture medium. The cell pellets were embedded in 2% agar in water, thoroughly rinsed with Sörensen buffer (pH 7.2) and dehydrated in ethanol. Finally, the cells were embedded in LR White resin and polymerized at 60°C for 24 h. Thin sections were collected on nickel grids covered with a Formvar carbon film. For immunolabeling, the grids were incubated on normal goat serum (NGS) 1:50 for 3 min and then floated for 17 h at room temperature onto a solution of 0.1% bovine serum albumine and 0.05% Tween 20 in PBS containing the primary anti-KRIT1 antibody (rabbit-polyclonal against human epitope of KRIT1, GeneTex, GTX109560) diluted 1:5, and eventually one of the following primary mouse-monoclonal antibody to detect co-localization: anti-Pol II (against human heptapeptide epitope of RNA Polymerase II, and recognizing both the active and inactive forms; Biodesign, M55701M) diluted 1:5; anti H20 (recognizing the 3mG cap of snRNAs, kindly provided by Dr. R. Lührmann) diluted 1:5. After rinsing with PBS-Tween and PBS, grids were incubated with NGS as above, and then reacted with a goat-anti rabbit and a goat anti-mouse secondary antibodies conjugated with 12 nm or 6 nm colloidal gold particles (The Jackson Lab., Bar Harbor, ME, USA) for 30 min at room temperature, rinsed and finally stained. To reduce chromatin contrast and selectively reveal nuclear ribonucleoprotein constituents, the sections were bleached by the EDTA method11 or selectively stained for RNA with terbium citrate.12
The samples were observed on a Zeiss 900 EM operating at 80kV. In some of the micrographs, the gold grains have been digitally contrasted with the Photoshop software.
Results
After EDTA staining to reveal RNPs,11 KRIT1 labeling in Hela cells was localized on the perichromatin region, where transcription takes place and, in particular, on the perichromatin fibrils (PF, the in situ morphological expression of the extranucleolar RNA transcription13) (Figure 1a; Supplementary Figure 1). This pattern was also observed after heat shock, suggesting that the nuclear localization of KRIT1 is not influenced by this type of stress. More in detail, staining with terbium citrate12 for RNA allowed us to better visualize the nascent PFs labeled with the KRIT1 antibody (Figure 1b).
Figure 1.
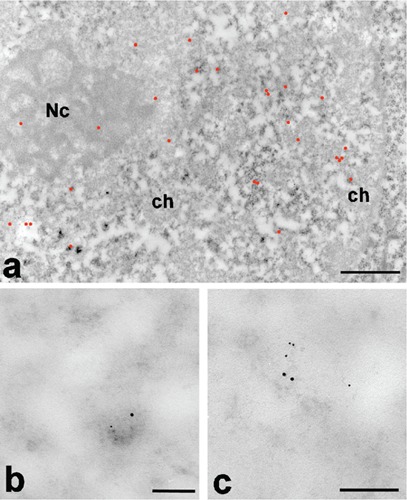
a) KRIT1 labeling pattern at low magnification; EDTA staining; the gold particles (highlighted in red) are present on the PF at the periphery of condensed chromatin areas (ch) and on the nucleolus (Nc); scale bar: 500 nm. b) A nascent single PF stained with terbium for RNA in a HeLa cell nucleus; the large gold grain represents RNA Pol II while the small grain indicates KRIT1); scale bar: 100 nm. c) The same labeling pattern as in (b) was observed in a Chironomus nascent fibril; scale bar: 100 nm.
We were able to observe the same pattern in salivary glands of Chironomus thummi: Fig 1c shows a PF where the labelings for RNA Pol II and KRIT1 are present at the opposite extremities. Since the antibody does not discriminate between the active and inactive form of RNA Pol II, we cannot determine if that PF is being transcribed, although it is tempting to assume so. In this model system, KRIT1 is also present in the large (perichromatin-granules like) Balbiani granules, where the mRNA transcribed is stored for the export to the cytoplasm (Figure 2a). We rarely found KRIT1 co-localizing with RNA Pol II, whereas we observed a co-localization with the methyl-cap of snRNA. In Figure 2b an example of double labeling in a Hela cell after heat shock is presented. In all the cell types used in these experiments, we have observed focal electron dense areas of the nucleus that show a higher labeling for KRIT1 suggesting the presence of nuclear bodies of unknown-function (Figure 2 c,d). These labeled nuclear domains were observed in all immunolabeling experiments in condition of minimum background, which leads us to exclude an artifactual labeling. Labeling for Pol II was never found on these nuclear domains.
Figure 2.
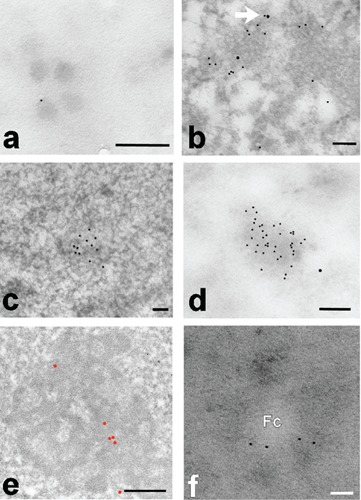
a) Four Balbiani granules are shown, one of which is labeled for KRIT1 at the periphery; terbium staining; scale bar: 50 nm. b) HeLa cell double labeled for m3Gcap (small grains) and KRIT1 (large grains); in some cases (arrow) we can observe a colocalization; EDTA staining; scale bar: 100 nm. c) Rat liver after EDTA staining; a nuclear domain is shown with a strong labeling for KRIT1; scale bar: 100 nm. d) A similar nuclear domain, terbium stained, is present in a HeLa cell; also in this case a heavy labeling for KRIT1 is present; scale bar: 100 nm. e) Labeling for KRIT1 in the nucleolus of a HeLa cells; the grains are present on the dense fibrillar component; scale bar: 200 nm. f) Part of a nucleolus of a Hela cell nucleus; KRIT1 is present in the dense fibrillar component surrounding the fibrillar center (Fc); terbium staining; scale bar: 50 nm.
Finally, we have documented KRIT1 presence in the dense fibrillar component (DFC) of the nucleolus in numerous Hela samples (Figure 2 e,f; Supplementary Figure 2). The same labeling distribution is present on the nucleolus in hepatocytes (not shown).
Discussion
Our results provide novel information about the fine nuclear distribution of the protein KRIT1 and its spatial relationships with the RNP components involved in RNA transcription and maturation, confirming and extending previous data on its nuclear localization.8
Despite its known role in cell-cell junction6 and cell-matrix anchorage,7 KRIT1 shows a peculiar nuclear labeling on the PFs and nucleolar DFC, where transcription actively takes place.13,14 The recurrent presence of this labeling pattern in most of the cells in the samples observed, from human Hela cells to Chironomus thummi salivary glands, suggests a possible conserved function of KRIT1 inside the nuclear compartment, reasonably involving RNA transcription. The presence of the labeling at the terminus of the newly transcribed PFs (i.e., far away from Pol II but associated with the m3G cap), together with the lack of DNA binding domains in the KRIT1 protein structure, leads us to hypothesize that KRIT1 could be involved in RNA maturation rather than be directly involved in transcription. Also the presence of KRIT1 labeling in the perichromatin-granules-like Balbiani granules could support this idea.
No marked difference was observed in Hela cells before and after heat shock, suggesting that KRIT1 nuclear localization and/or shuttling is not modified or increased by heat shock. Only few cells within the various samples studied showed the high labeled nuclear domains. As it is the case for other nuclear bodies,15 their function could be related to the nucleus-cytoplasmic shuttling of KRIT1. Being positive to Terbium and EDTA staining we know they are composed of RNPs, but further studies are needed to understand the nature of these nuclear domains.
These data lay the basis for a further characterization of KRIT1 function inside the nuclear compartment. It is not excluded that an impairment of this unknown KRIT1 nuclear function could contribute to the development of CCM in the individuals that carry mutations on this gene. Thus we expect an altered KRIT1 labeling pattern in CCM patients relative to healthy. Furthermore, these data can be implemented by observing the nuclear labeling pattern of the CCM2 and PDCD10 protein products, involved in the development of the disease. Being the pathogenetic mechanisms by which KRIT1 mutation cause familial CCM still unknown, we believe that adding new information on the complex molecular pathway of these genes could lead to a better understanding of the CCM pathology.
References
- 1.Marchuk DA, Srinivasan S, Squire TL, Zawistowski JS. Vascular morphogenesis: tales of two syndromes. Hum Mol Genet 2003;1:R97-112 [DOI] [PubMed] [Google Scholar]
- 2.Craig HD, Günel M, Cepeda O, Johnson EW, Ptacek L, Steinberg GK, et al. Multilocus linkage identifies two new loci for a mendelian form of stroke, cerebral cavernous malformation, at 7p15-13 and 3q25.2-27. Hum Mol Genet 1998;7:1851-8 [DOI] [PubMed] [Google Scholar]
- 3.Dubovsky J, Zabramski JM, Kurth J, Spetzler RF, Rich SS, Orr HT, et al. A gene responsible for cavernous malformations of the brain maps to chromosome 7q. Hum Mol Genet 1995;4:453-8 [DOI] [PubMed] [Google Scholar]
- 4.Faurobert E, Albiges-Rizo C. Recent insights into cerebral cavernous malformations: a complex jigsaw puzzle under construction. FEBS J 2010;277:1084-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer A, Zalvide J, Faurobert E, Albiges-Rizo C, Tournier-Lasserve E. Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol Med 2013;19:302-8 [DOI] [PubMed] [Google Scholar]
- 6.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol 2007;179:247-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum Mol Genet 2002;11:389-96 [DOI] [PubMed] [Google Scholar]
- 8.Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson qGL, et al. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet 2005;14:2521-31 [DOI] [PubMed] [Google Scholar]
- 9.Francalanci F, Avolio M, De Luca E, Longo D, Menchise V, Guazzi P, et al. Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp Cell Res 2009;315:285-303 [DOI] [PubMed] [Google Scholar]
- 10.Chiodi I, Biggiogera M, Denegri M, Corioni M, Weighardt F, Cobianchi F, et al. Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J Cell Sci. 2000;113:4043-53 [DOI] [PubMed] [Google Scholar]
- 11.Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res 1969;27:250-65 [DOI] [PubMed] [Google Scholar]
- 12.Biggiogera M, Fakan S. Fine structural specific visualization of RNA on ultrathin sections. J Histochem Cytochem 1998;46:389-95 [DOI] [PubMed] [Google Scholar]
- 13.Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol 1994;4:86–90 [DOI] [PubMed] [Google Scholar]
- 14.Cisterna B, Biggiogera M. Ribosome biogenesis: from structure to dynamics. Int Rev Cell Mol Biol 2010;284:67-111 [DOI] [PubMed] [Google Scholar]
- 15.Morimoto M, Boerkoel C F. The Role of nuclear bodies in gene expression and disease. Biology (Basel) 2013;2:976-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]


