Abstract
Intracerebral hemorrhage (ICH) is the stroke subtype with the worst prognosis and has no established acute treatment. ICH is classified as lobar or nonlobar based on the location of ruptured blood vessels within the brain. These different locations also signal different underlying vascular pathologies. Heritability estimates indicate a substantial genetic contribution to risk of ICH in both locations. We report a genome-wide association study of this condition that meta-analyzed data from six studies that enrolled individuals of European ancestry. Case subjects were ascertained by neurologists blinded to genotype data and classified as lobar or nonlobar based on brain computed tomography. ICH-free control subjects were sampled from ambulatory clinics or random digit dialing. Replication of signals identified in the discovery cohort with p < 1 × 10−6 was pursued in an independent multiethnic sample utilizing both direct and genome-wide genotyping. The discovery phase included a case cohort of 1,545 individuals (664 lobar and 881 nonlobar cases) and a control cohort of 1,481 individuals and identified two susceptibility loci: for lobar ICH, chromosomal region 12q21.1 (rs11179580, odds ratio [OR] = 1.56, p = 7.0 × 10−8); and for nonlobar ICH, chromosomal region 1q22 (rs2984613, OR = 1.44, p = 1.6 × 10−8). The replication included a case cohort of 1,681 individuals (484 lobar and 1,194 nonlobar cases) and a control cohort of 2,261 individuals and corroborated the association for 1q22 (p = 6.5 × 10−4; meta-analysis p = 2.2 × 10−10) but not for 12q21.1 (p = 0.55; meta-analysis p = 2.6 × 10−5). These results demonstrate biological heterogeneity across ICH subtypes and highlight the importance of ascertaining ICH cases accordingly.
Introduction
Stroke is the most devastating manifestation of cerebrovascular disease and the second leading cause of death and acquired disability worldwide.1,2 Strokes are classified as ischemic or hemorrhagic (85% and 15% in people of European descent, respectively), and spontaneous intracerebral hemorrhage (ICH [MIM 614519]) is by far the most common type of hemorrhagic stroke.3 Largely a disease of the elderly, ICH occurs when cerebral vessels rupture and is associated with 40%–50% 3-month mortality as well as sustained disability in more than half of survivors.4,5 The incidence of ICH is expected to rise in coming years because of increases in life expectancy and widespread use of antithrombotic therapy in the elderly.6,7 Although treatment of hypertension partially reduces the risk of initial or recurrent ICH,8 there are no established acute treatments for this condition. Therefore, identification of biological pathways that could eventually be targeted by novel therapeutic strategies is vital to reducing the health care burden associated with this disease.
Histopathological observations demonstrate that the underlying cerebral small vessel disease differs according to the location of the ICH within the brain. Lobar ICH originates in the cerebral cortex or cortical-subcortical junction and is most commonly associated with cerebral amyloid angiopathy.9 Nonlobar ICH originates in deep structures of cerebral hemispheres, brainstem, and cerebellum and tends to be associated with what is typically called hypertensive vasculopathy.10 This heterogeneity in underlying biological processes leading to different ICH subtypes has been corroborated by epidemiologic,11 neuroimaging,12 and genetic studies.13,14 A large multicenter candidate gene study undertaken by the International Stroke Genetics Consortium in the same populations utilized in the present study established that the epsilon variants of APOE (MIM 107741), known to be risk factors for sporadic cerebral amyloid angiopathy, are associated at genome-wide significance levels specifically with lobar ICH.13 Likewise, it was subsequently found that the burden of risk alleles for high blood pressure associates specifically with nonlobar ICH.14
Heritability estimates indicate that common genetic variation plays a substantial role in risk of both ICH subtypes beyond APOE and blood-pressure-related variants.15 Identification of these genetic contributors would have a significant impact on the field of stroke, because it could help uncover specific biological pathways hitherto unsuspected to play a role in this condition that could be targeted by novel therapeutic strategies. In this study we meta-analyzed data from six previously unpublished genome-wide association studies of ICH that enrolled subjects of European ancestry in the United States and Europe under the auspices of the International Stroke Genetics Consortium, with subsequent replication of identified susceptibility loci in an independent study of ICH.
Subjects and Methods
Participating Studies
Case and control subjects included in the discovery phase were subjects of European ancestry aged >55 years in the Genetics of Cerebral Hemorrhage with Anticoagulation13 (GOCHA) study (multicenter study in the US) and aged >18 years in the Genetic and Environmental Risk Factors for Hemorrhagic Stroke16 (GERFHS) studies I and II in Cincinnati, OH; Hospital del Mar Intracerebral Hemorrhage17 study and Vall d’Hebron Hospital ICH18 study in Barcelona, Spain; Jagiellonian University Hemorrhagic Stroke Study19 in Krakow, Poland; and the Lund Stroke Register20 study in Lund, Sweden. Because of their limited sample sizes, data from the four European studies (ESs) were analyzed together for the purposes of quality control, imputation, and association testing.
Subjects
Cases were ascertained across participating studies according to predefined standardized criteria. Spontaneous ICH was defined as a new and acute neurological deficit with compatible brain imaging (computed tomography or magnetic resonance imaging) showing the presence of intraparenchymal bleeding. According to standard research and clinical practice in the field,3 ICH location was assigned based on admission images by neurologists who were blinded to genotype data. ICH originating at the cerebral cortex or cortical-subcortical junction (with or without involvement of subcortical white matter) was defined as lobar, and ICH originating at the thalamus, internal capsule, basal ganglia, deep periventricular white matter, cerebellum, or brain stem was defined as nonlobar. Exclusion criteria included trauma, brain tumor, hemorrhagic transformation of ischemic stroke, vascular malformation, and any other cause of secondary ICH.
Control subjects were ICH-free individuals enrolled from the same population that gave rise to the case subjects at each participating study site, aged >55 years (GOCHA) and >18 years (GERFHS and ESs). Control subjects were sampled by random digit dialing in GERFHS and from ambulatory clinics in the remainder of the studies.
All studies were approved by the Institutional Review Board or ethics committee at each participating site. Participants provided informed consent; when subjects were not able to communicate, consent was obtained from their legal proxies.
Genome-wide Genotyping and Quality Control
Case and control subjects were genotyped with Affymetrix 6.0 in GERFHS and with Illumina HumanHap610-Quad in GOCHA and ESs. Case and control subjects from each study were genotyped side-by-side on the same plates with the exception of the replication controls from the Cincinnati Control Cohort (CCC) and Genomic Control Cohort (GCC), which were genotyped from separate studies. Plate-to-plate variability was assessed by comparison of SNP call and error rates. Standardized prespecified quality-control procedures21 were implemented separately in GOCHA, GERFHS, and ESs. These filters excluded SNPs with genotype call rate <0.95, significant differential missingness between case and control subjects (p < 0.05), deviation from Hardy-Weinberg equilibrium (p < 1 × 10−6), or minor allele frequency (MAF) <0.01. At the subject level, quality control excluded individuals with genotype call rate of <95%; inconsistency between self-reported and genotypic gender; an inferred first- or second-degree relative in the sample identified on the basis of pairwise allele sharing estimates (estimated genome proportion shared identical by descent; π > 0.1875); and extreme genome-wide heterozygosity f statistic, defined as >5 times its standard deviation.
Population Stratification
After quality-control procedures, principal-components analysis22 was implemented separately in GOCHA, GERFHS, and ESs, incorporating genotype data from 1000 Genomes23 populations. Population outliers were identified and removed by visual inspection of principal component plots, and the first four principal components were subsequently included in regression models fitted for association testing. Principal-component and identity-by-descent analyses were performed via a pruned subset of independent SNPs (61,325 SNPs in GOCHA, 95,013 in GERFHS, and 64,728 in ESs) to account for potential biases introduced by LD structure.
Imputation
After quality-control procedures and principal component analysis, imputation was performed separately in GOCHA, GERFHS, and ESs via IMPUTE2 v.2.224 and 1000 Genomes23 integrated reference panels (Phase I interim release in NCBI build 37). The number of SNPs that entered the imputation process were 525,752, 795,240, and 532,149 for GOCHA, GERFHS, and ESs, respectively. Postimputation filters excluded imputed SNPs with MAF <0.01, IMPUTE2 information score <0.7, confidence score <0.9, and missing estimates in association testing for 1 or more studies.
Genome-wide Association Testing
Given the biological differences in ICH subtypes outlined in the introduction and after a prespecified analysis plan, genome-wide association analyses were computed separately for all ICH (lobar and nonlobar combined), for lobar ICH, and for nonlobar ICH. These analyses were completed separately in GOCHA, GERFHS, and ESs via logistic regression, assuming additive genetic effects (1-degree-of-freedom additive trend test) and adjusting for age, gender, and principal components. In secondary analysis, association testing was carried out separately in each European study. Association p values obtained in GOCHA, GERFHS, and ESs were meta-analyzed via the inverse normal method weighting by sample size as implemented in METAL,25 and heterogeneity of pooled estimates was quantified by computing Cochrane’s Q and corresponding p and I2. After recent GWAS meta-analysis,26 only SNPs with available estimates in all three data sets are reported. Quantile-quantile plots were utilized to assess systematic inflation in association results resulting from population stratification or other systematic causes of bias.
Further analyses were undertaken to evaluate the presence of additional independent signals at each locus. Additional independent signals at each locus were evaluated by conditional testing completed by adding the dosages of the top SNP at each locus to logistic regression models. Pairwise linkage disequilibrium (LD) between SNPs was assessed and visualized with 1000 Genomes Ensemble-based genome browser.27 Regional association plots were constructed with LocusZoom software.28
Replication
Replication of associations with p < 1 × 10−6 in the discovery meta-analysis was pursued in case and control groups from the Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH)29 and GERFHS III16 studies, in adult control group from the CCC,16 and GCC.16 Enrolled ICH cases and controls from ERICH were non-Hispanic whites, African Americans, and Hispanic subjects (based on self-reported race and ethnicity) aged >18 years from multiple study centers across the US. ICH case subjects were ascertained with the same criteria utilized in the discovery phase. ICH-free control subjects were sampled by random digit dialing from the population that gave rise to the case cohort. The CCC and GCC cohorts are population-based cohorts of ICH-free individuals from the Greater Cincinnati area. The CCC cohort was specifically enrolled to approximate the age, sex, and race distribution of ICH subjects, and the GCC was enrolled to match the population and geographic distribution of the metropolitan area. Genotyping was completed with TaqMan assays (ERICH Study) or Affymetrix 6.0 (GERFHS III and CCC-GCC). For the latter, preimputation quality-control procedures, imputation, and postimputation filters were implemented as described for the discovery phase. Association testing was carried out by fitting logistic regression models and implementing a 1-degree-of-freedom trend test, assuming additive effects and including age and gender in all models. Meta-analyses across discovery and replication proceeded as described above for the discovery phase. Replication results were considered significant at p < 0.05 and genome-wide significance was defined as p < 5 × 10−8.30
Overlap with Common Variants Related to Blood Pressure
Given the well-established role of hypertension in causing ICH, especially of nonlobar type, we specifically assessed the role in ICH of common genetic variants known to play a role in determining blood pressure. SNPs reported to be related to blood pressure at p < 1 × 10−5 were identified in the GWAS Catalog.31 Queried traits included blood pressure; hypertension; and diastolic, systolic, mean, and pulse blood pressures. Association results for these SNPs were identified for all, for lobar, and for nonlobar ICH.
Overlap with DNase I Hypersensitivity Sites
The positions of SNPs in ICH-associated loci were overlapped with DNase I hotspot regions from the Encyclopedia of DNA Elements (ENCODE) Project that mark generalized chromatin accessibility, mapped for each of 125 diverse cell lines and tissues.32 The genomic region of interest for each identified loci was defined based on the genomic variants that were in linkage disequilibrium with the top variant at each locus (defined as r2 > 0.5). In addition, ICH-associated SNPs were analyzed for other overlap with ENCODE data, including transcription factor motifs, via RegulomeDB.33
eQTL Analyses
Variants within each identified susceptibility locus for ICH were evaluated for gene expression in cis via publicly available resources. SNPs with p < 1 × 10−5 were assessed in four publicly available eQTL databases: SCAN (SNP and CNV Annotation Database), the NCBI and Broad Institute GTEx (Genotype-Tissue Expression) eQTL Browsers,34 the Pritchard laboratory UChicago eQTL browser,35 and mRNA by SNP Browser v.1.0.1.36 Gene expression was assessed in a range of tissue and cell types, including liver, brain, lymphoblastoid cell lines, monocytes, fibroblasts, and T cells. As in previous reports,37 we defined potential cis eQTLs as candidate SNPs associated with gene expression mapping to a 1 Mb window around each locus with p < 1 × 10−3.
Results
After excluding subjects based on quality-control procedures (n = 23) and principal component analysis (n = 93), a case cohort of 1,545 subjects and a control cohort of 1,481 subjects were available for association testing in the discovery analysis (mean age 67 [SD 10], female sex 45%, Table 1). After preimputation quality-control procedures, imputation to 1000 Genomes reference panels, and postimputation quality-control filters, a total of 5,258,103 SNPs were available for association testing across all data sets included in the discovery sample.
Table 1.
Descriptive Characteristics of Participating Studies
| Covariate |
Discovery |
Replication |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Multicenter, US |
European Studies |
Cincinnati, US |
Cincinnati, US |
|||||||||
|
GOCHA |
HM-ICH + VVH-ICH |
JUHSS |
LSR |
GERFHS I & II |
ERICH/GERFHS III/CCC-GCC |
|||||||
| Case Cohort | Control Cohort | Case Cohort | Control Cohort | Case Cohort | Control Cohort | Case Cohort | Control Cohort | Case Cohort | Control Cohort | Case Cohort | Control Cohort | |
| Subjects, n | 298 | 457 | 212 | 169 | 122 | 163 | 116 | 153 | 797 | 539 | 1,681 | 2,261 |
| Age, mean (SD) | 74 (10) | 72 (8) | 74 (11) | 71 (9) | 67 (12) | 65 (13) | 75 (10) | 75 (10) | 67 (15) | 66 (15) | 62 (15) | 43a (26) |
| Female, n (%) | 134 (45) | 231 (51) | 103 (49) | 77 (46) | 69 (57) | 93 (57) | 49 (42) | 69 (45) | 383 (48) | 243 (45) | 701 (42) | 1,113 (49) |
| Hypertension, n (%) | 217 (73) | 280 (61) | 126 (60) | 99 (64) | 96 (81) | 74 (45) | 76 (67) | 65 (43) | 494 (62) | 280 (52) | 1,009 (60) | 738b (51) |
| Lobar ICH, n (%) | 184 (58) | – | 88 (40) | – | 51 (39) | – | 36 (28) | – | 327 (41) | – | 484 (29) | – |
| Nonlobar ICH, n (%) | 132 (42) | – | 133 (60) | – | 80 (61) | – | 94 (72) | – | 470 (59) | – | 1,197 (71) | – |
Abbreviations are as follows: ICH, intracerebral hemorrhage; GOCHA, Genetics of Cerebral Hemorrhage on Anticoagulation Study; HM-ICH, Hospital del Mar Intracerebral Hemorrhage Study; VVH-ICH, Vall d’Hebron Hospital ICH Study; JUHSS, Jagiellonian University Hemorrhagic Stroke Study; LSR, Lund Stroke Register; GERFHS, Genetic and Environmental Risk Factors for Hemorrhagic Stroke Study; ERICH, Ethnic/Racial Variations of Intracerebral Hemorrhage Study; CCC-GCC, Cincinnati Control Cohort - Genomic Control Cohort.
Includes the 819 GCC pediatric control subjects with ages ≤18.
Excludes the 819 GCC pediatric control subjects with no hypertension history.
Susceptibility loci were identified for lobar and nonlobar ICH, but not for all ICH (lobar and nonlobar combined) (Figures 1A–1C, Tables S1–S3 available online). The estimated inflation factors (λ of 1.039, 1.016, and 1.038 for all, lobar, and nonlobar ICH, respectively) and quantile-quantile plots (Figure S1) indicated absence of inflation resulting from systematic bias caused by population substructure or other artifacts. Several SNPs on chromosomal region 12q21.1, an intergenic region near TRHDE (MIM 606950), were associated with lobar ICH, with peak association detected at rs11179580 (MAF = 0.24, per additional major allele [C], odds ratio [OR] 1.56, 95% confidence interval [CI] 1.33–1.84; p = 7.0 × 10−8; Q = 0.43, I2 = 0%; Figure 1B and Table 2). Similar results were obtained when analyzing each European study separately (Figure S2). A similar effect was observed for all ICH (per additional C allele, OR 1.36, 95% CI 1.21–1.54; p = 5.4 × 10−8; Q = 0.07, I2 = 62%; Table 2 and Figure 1A), but the combination of similar effect magnitudes (despite double the sample size) and significant increase in heterogeneity suggests that the observed association is driven by lobar ICH. rs11179580 also showed some effect in nonlobar ICH (OR 1.25, 95% CI 1.09–1.42, p = 0.002; Table 2), indicating that the affected biological pathway could impact both types of cerebral hemorrhage. Neither adjustment for the most significant SNPs at this locus (Figure S3A) nor haplotype testing identified additional associations of interest.
Figure 1.
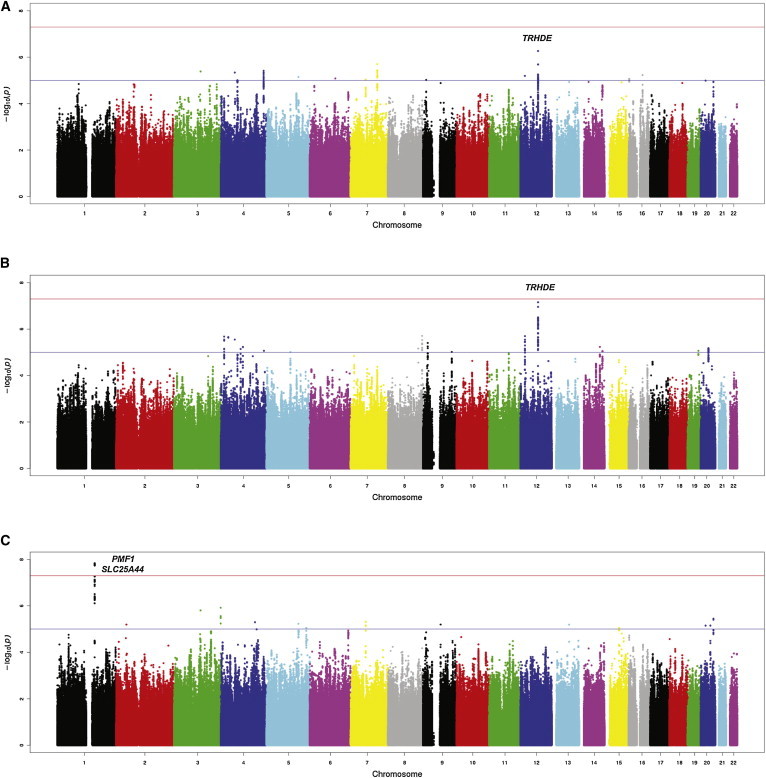
Genome-wide Association Study Results
Genome-wide association study results of autosomal SNPs: (A) all (lobar ICH and nonlobar ICH combined), (B) lobar ICH, and (C) nonlobar ICH. The plots show –log10-transformed p values for genotyped and imputed SNPs with respect to their physical positions. The threshold for association at genome-wide significance (p = 5 × 10−8) is shown by the upper dashed line, and the lower dashed line corresponds to p = 1 × 10−5. Landmark genes are indicated for loci that reached the threshold to pursue replication.
Table 2.
Loci with p < 1 × 10−6 in at least One Subtype of Intracerebral Hemorrhage
|
SNP Characteristics |
Meta-analysis Discovery Phase |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | Locus | Major/Minor Allele | Tested Allelea | MAF | Type | Gene Landmarks | Trait | OR | p | Q | I |
| rs11179580 | 12 | 73586579 | 12q21.1 | C/T | C | 0.24 | intergenic | TRHDE | all ICH | 1.36 | 7.1 × 10−7 | 0.07 | 61% |
| lobar ICH | 1.56 | 7.0 × 10−8 | 0.43 | 0% | |||||||||
| nonlobar ICH | 1.25 | 0.002 | 0.03 | 72% | |||||||||
| rs2984613 | 1 | 156197380 | 1q22 | C/T | C | 0.32 | intronic | PMF1 SLC25A44 | all ICH | 1.21 | 6.0 × 10−4 | 0.15 | 48% |
| lobar ICH | 0.97 | 0.62 | 0.7 | 1% | |||||||||
| nonlobar ICH | 1.44 | 1.6 × 10−8 | 0.05 | 66% | |||||||||
Abbreviations are as follows: Chr, chromosome; MAF, minor allele frequency; OR, odds ratio; ICH, intracerebral hemorrhage.
Showing effect estimates when testing the major allele to depict genetic variation associated with increased risk of ICH.
For nonlobar ICH, a susceptibility locus was identified on chromosomal region 1q22, a region that contains PMF1 (MIM 609176) and SLC25A44 (MIM 610824). The top-associated variant within this locus was the intronic SNP rs2984613 (MAF 0.31; per additional major allele [C] OR 1.44, 95% CI 1.27–1.64; p = 1.6 × 10−8; Q = 0.05, I2 = 66%; Table 2 and Figure 1C). Although some heterogeneity was observed for this specific variant, several SNPs within this locus achieved genome-wide significance with substantially lower heterogeneity (as showed by Q > 0.05, Table S5). Comparable results were obtained when analyzing each European study separately (Figure S2). rs2984613 also had some effect in all ICH (OR 1.21, 95% CI 1.12–1.38, p = 6.0 × 10−4), probably driven by nonlobar cases, and had no effect on lobar ICH. Neither analysis adjusting for the most significant SNPs (Figure S3B) nor haplotype testing within these loci identified additional associations of interest.
Replication of identified associations was pursued in 1,681 case subjects (513 non-Hispanic whites, 634 African Americans, and 534 Hispanics) and 2,261 control subjects (1,552 non-Hispanic whites, 449 African Americans, and 260 Hispanics) (Tables 1 and S4). Genotyping at this stage included both direct genotyping (ERICH study) and genome-wide genotyping (GERFHS III and CCC-GCC studies). Direct genotyping in ERICH included the top SNP at each locus: rs11179580 for 12q21.1 in lobar ICH and rs2758605 for 1q22 in nonlobar ICH. For technical reasons, rs2758605 (second top SNP) was genotyped in lieu of rs2984613 (r2 = 0.99). The association of chromosomal region 1q22 with nonlobar ICH replicated but that of 12q22.1 with lobar ICH did not. No association was found between rs11179580, the top SNP at 12q21.1, and lobar ICH (OR 1.05, 95% CI 0.89–1.24; p = 0.55; meta-analysis p = 2.6 × 10−5; Figure 2A). For rs2758605 (1q22) in nonlobar ICH, each additional C allele was associated with a 24% increase in risk of hemorrhage (95% CI 1.09–1.40; p = 6.5 × 10−4; meta-analysis p = 2.2 × 10−10; Figure 2B). When considering ethnic-specific analysis (Table S6), the association was significant for non-Hispanic whites (p = 0.04) and African Americans (p = 1 × 10−4) but not for Hispanics (p = 0.90).
Figure 2.
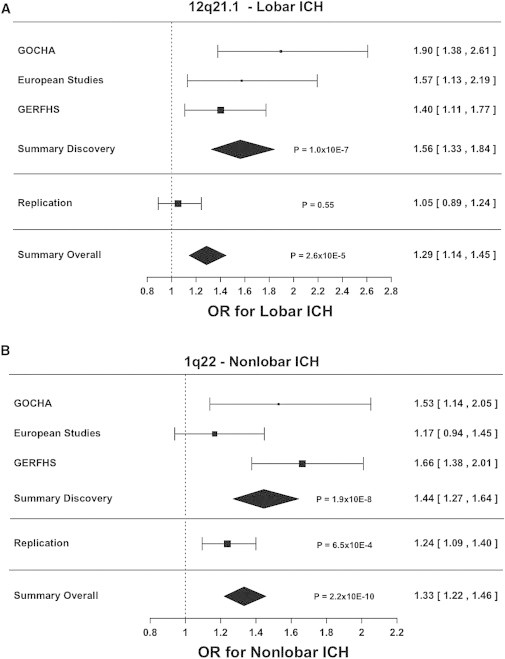
Meta-analysis Results
Forest plots describing effect estimates for participating studies, as well as for the replication effort. Pooled estimates for odds ratios and 95% confidence intervals were calculated by fixed effects, inverse variance weighting meta-analysis.
(A) Association results for rs11179580 at chromosomal region 12q21.1 in lobar ICH.
(B) Association results for rs2984613 at chromosomal region 1q22 in nonlobar ICH.
Results correspond to effect estimates when testing the major allele to depict genetic variation associated with increased risk of ICH.
The 1q22 locus contains PMF1 and SLC25A44 (Figure 3A), two genes that code polyamine-modulated factor 1 and solute carrier family 25-member 44, respectively. A total of 49 SNPs in this locus achieved p < 1 × 10−5 (Table S7) and at least one of these was directly genotyped in all participating studies. Although most of these SNPs are intronic, rs1052053 has transcriptional impact, resulting in substitution of glutamine by arginine in position 75 of exon 2 of PMF1. The resulting transcript variant (isoform 3) is shorter than isoform 1 and has a distinct C terminus. This variation has benign functional impact according to PolyPhen-2 (score 0) and is well tolerated according to SIFT (score 1). In addition, rs2241107 is located in a promoter region in front of PMF1, and this region also corresponds to a DNase I hypersensitivity region in all ENCODE cell types.
Figure 3.
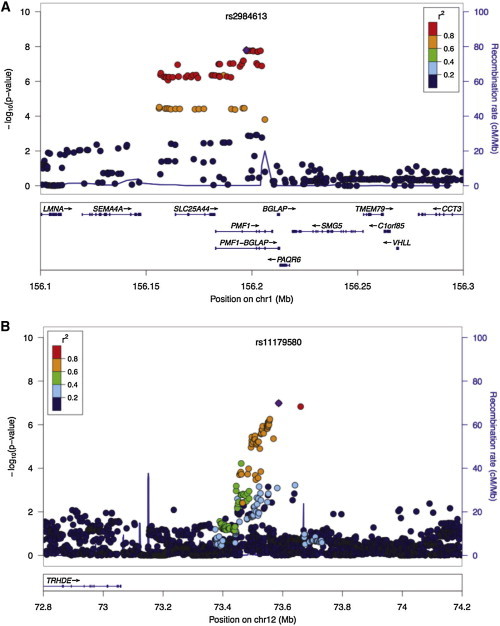
Zoom Plots
Regional association results.
(A) Chromosomal region 1q22 in nonlobar ICH.
(B) Chromosomal region 12q21.1 in lobar ICH.
The index-associated SNP is labeled with violet color.
Expression data from GTEx show that gene expression in neural tissues is high for SLC25A44 (Figure S4) and moderate for PMF1 (Figure S5). Moreover, eQTL data from this same source raises the possibility of 1q22 exerting its effect by regulating nearby genes, with 27 of the 49 SNPs mentioned above being associated with expression of SEMA4A (MIM 607292) in tibial nerve tissue (most significant p = 5.3 × 10−6 for rs6427304, Table S8). Axons, the main component of peripheral nerves, are extensions of neurons that, together with the blood vessel wall, extracellular matrix, and glia, form the neurovascular unit, a biological compartment suggested to play an important role in cerebrovascular disease.38 SEMA4A is located upstream of SLC25A44 and codes for semaphorin-4A, a member of the semaphorin family of soluble and transmembrane proteins that is involved in numerous functions, including axon guidance, morphogenesis, carcinogenesis, and immunomodulation. SEMA4A has moderate-to-high expression in neural tissues (Figure S6).
Variants known to influence blood pressure did not significantly affect ICH risk. A total of 172 entries related to blood pressure were identified in the GWAS catalog. These corresponded to 72 unique SNPs. None of these SNPs was significantly associated with all, lobar, or nonlobar ICH after adjusting for multiple testing (Bonferroni-corrected threshold p < 0.0007, Table S9).
Discussion
In this GWAS of ICH, we meta-analyzed data from 3,223 case subjects and 3,725 control subjects and identified and replicated a susceptibility locus for this condition located at 1q22 (nonlobar ICH). Importantly, the replication spanned European- and African-derived ethnicities. These results confirm previous heritability studies that indicated that common genetic variation may play an important role in the occurrence of this condition.15 The subtype specificity of the identified susceptibility locus is consistent with the findings of histologic, epidemiologic, and candidate-gene studies that different mechanisms underlie each ICH subtype. Given the absence of an effective treatment and the heavy burden imposed by ICH to society, the present findings inaugurate the possibility of identifying novel biological mechanisms involved in causing this disease that could eventually be targeted by new therapeutic interventions.
Providing important supportive evidence for a role of 1q22 in cerebral small vessel disease, a recent GWAS of cerebral white matter lesions in healthy adults identified 1q22 as a suggestive locus. Cerebral white matter lesions constitute a radiological manifestation of cerebral small vessel disease,39 and several studies have shown that nonlobar ICH is the most devastating clinical manifestation of this vasculopathy.40,41 Importantly, several SNPs identified in our study were also listed in the mentioned GWAS (Table 3).42 This previous study of white matter hyperintensities informs our GWAS of ICH because the former was undertaken in individuals of European ancestry, thus providing crucial support to our findings by confirming the association between 1q22 and cerebral small disease in whites. Likewise, our study informs the GWAS of white matter hyperintensities, because in the latter 1q22 achieved only sub-genome-wide significance (p = 5 × 10−6).42 In this setting, our findings represent an important contribution to definitively establishing the role of this locus in cerebrovascular disease by taking the association signal beyond the genome-wide threshold and extending the effect of this genomic region beyond the realm of imaging endophenotypes to meaningful clinical impact.
Table 3.
Overlapping Results: GWAS of Nonlobar ICH and White Matter Hyperintensities
| SNP | Minor Allele | Other Allele | MAF | Landmark Gene | p Value Nonlobar ICH | p Value White Matter Hyperintensities |
|---|---|---|---|---|---|---|
| rs2758605 | c | g | 0.36 | PMF1;BGLAP | 1.5 × 10−8 | 9.4 × 10−6 |
| rs2251847 | a | g | 0.36 | PMF1;BGLAP | 1.6 × 10−8 | 1.0 × 10−5 |
| rs887953 | c | t | 0.34 | PMF1;BGLAP | 7.5 × 10−8 | 8.2 × 10−5 |
| rs2244144 | a | g | 0.34 | PMF1;BGLAP | 8.2 × 10−8 | 8.2 × 10−5 |
| rs1052053 | g | a | 0.38 | PMF1;BGLAP | 9.2 × 10−8 | 5.0 × 10−6 |
| rs2842873 | t | c | 0.38 | PMF1;BGLAP | 1.4 × 10−7 | 6.4 × 10−6 |
| rs6427304 | g | a | 0.36 | SLC25A44 | 4.0 × 10−7 | 6.8 × 10−5 |
| rs2853641 | g | a | 0.35 | SLC25A44 | 4.2 × 10−7 | 5.3 × 10−5 |
| rs2241108 | g | c | 0.35 | SLC25A44 | 4.2 × 10−7 | 5.2 × 10−5 |
| rs2241107 | c | t | 0.35 | PMF1;BGLAP | 4.2 × 10−7 | 5.4 × 10−5 |
Abbreviations are as follows: PMF1, Polyamine Modulating Factor 1; BGLAP, bone gamma-carboxyglutamate protein; SLC25A44, solute carrier family 25, member 44. See also Fornage et al.42
Variants within 1q22 are located in an LD block of 48 kb that contains PMF1 and SLC25A44 (Figure 3A). PMF1 codes for polyamine-modulated factor 1, a nuclear protein regulated by polyamines that is required for normal chromosome alignment and kinetochore formation during mitosis.43 In conjunction with the transcription factor NFE2L2 (nuclear factor, erythroid 2-like 2; also known as NRF2), PMF1 also mediates the transcriptional induction of SAT1, which codes for an acetyltransferase responsible for the rate-limiting enzyme in the catabolic pathway of polyamine metabolism.44 Polyamines are proteins with multiple amine groups that have been linked to cerebrovascular disease by several studies. They have been found to be elevated in stroke subjects and to be implicated in breakdown of the blood-brain barrier45,46 and regulation of the receptor for the excitatory neurotransmitter NMDA through a polyamine-specific binding site.47 SLC25A44 encodes solute carrier family 25-member 44, a member of the SLC25 family of mitochondrial carrier proteins,48 and no pathological consequences have been reported so far for genetic variation within this gene.
Although the lack of replication of 12q21.1 raises the possibility that the result of the discovery phase could be spurious, the consistency in effect estimates across the discovery cohorts suggests that follow-up studies should explore this locus further. Identified SNPs at this locus are located within an intergenic region that lies 400 kb upstream of TRHDE (Figure 3B), a gene that codes for thyrotropin-releasing hormone-degrading enzyme. However, this gene is located beyond the adjacent recombination hotspot. Four different publicly available databases on eQTLs were queried with no significant findings. Likewise, data from the ENCODE project were also utilized to evaluate whether this genomic region contains histone modification patterns observed in regulatory regions, also with no significant findings. This locus has not been linked to other clinical traits.
Several additional complementary analyses could enrich the results presented here. In particular, evaluation of gene- and pathway-based association with ICH could allow identification of both genes and relevant biologic pathways involved in causing ICH but that had only sub-GW results of the present analysis. Additionally, now that GW data become available for ICH, joint genetic contribution to ischemic and hemorrhagic stroke can be assessed, as well as joint genetic contribution to ICH and white matter hyperintensities, a widely studied endophenotype for cerebral small vessel disease.
Our study has limitations. Although we strived to reduce possible misclassification of cases when assigning lobar and nonlobar categories, some residual misclassification may still have occurred. However, it is reasonable to assume that, if present, this misclassification was independent and nondifferential, thus biasing our results toward the null. A second limitation is sample size: although the discovery cohort was large, additional samples will be critical to further depict the genetic architecture of ICH and its subtypes and to better understand how genetic effects vary by race and ethnicity. Although the null result obtained in replication for 1q22 in Hispanics could be truly negative—reflecting heterogeneity of effects across ethnic groups—power limitations could also account for it. Finally, because direct genotyping was performed in the majority of samples included in replication, only self-reported ancestry could be utilized to account for population structure at that stage.
Importantly, other complex genetic disorders show that there are probably numerous susceptibility loci with small or modest effects that require significant increases in sample size to be discovered.49,50 In this regard, the overlap of chromosomal region 1q22 with another cerebral-small-vessel-disease-related trait (white matter hyperintensities42) points to the interesting opportunity of collaborating with consortia that focus on phenotypes like cognitive decline, late-life depression, and non-parkinsonian gait disturbances, all believed to be mediated to some degree by small vessel diseases of the brain.
We report a GWAS of ICH, the most devastating stroke type that currently has no effective treatment. As has been the case for ischemic stroke,37,51 careful phenotyping of ICH based on known differences in cerebrovascular histopathology proved essential for successful discovery of susceptibility loci. We identified one susceptibility locus for nonlobar ICH (chromosomal region 1q22), confirmed through independent replication. The results of a prior GWAS report of white matter hyperintensities, an established risk factor for ICH, provide complementary evidence for a role of 1q22 in cerebral small vessel disease. The specific biological mechanisms underlying the described associations remain to be elucidated.
Acknowledgments
We thank the Biorepository and Center for Genome Technology (University of Miami) (specifically Sandra West, Ioanna Konidari, and Susan Slifer) and Miguel Hernán for providing expert advice. Computing support, in part, provided by the Wake Forest Center for Public Health Genomics. Funding entities had no direct involvement in study design; data collection, analysis, and interpretation; writing of the manuscript; or the decision to submit for publication. Funding provided as follows: GERFHS, NIH grants NS36695 and NS30678; GOCHA, NIH grant R01NS059727, the Keane Stroke Genetics Research Fund, the Edward and Maybeth Sonn Research Fund, and the University of Michigan General Clinical Research Center M01 RR000042; ERICH, NIH grant NS069763; HM-ICH, Instituto de Salud Carlos III with the grants “Registro BASICMAR” Funding for Research in Health (PI051737), “GWALA project” from Fondos de Investigación Sanitaria ISC III (PI10/02064), and Fondos FEDER/EDRF Red de Investigación Cardiovascular (RD12/0042); JUHSS, Polish Ministry of Education grant N402 083934; LSR, Lund University, Region Skåne, the Swedish Research Council (K2010-61X-20378-04-3), the Swedish Stroke Association, the Freemasons Lodge of Instruction EOS in Lund, and the King Gustaf V and Queen Victoria’s foundations; G.J.F. and H.B.B., NIH SPOTRIAS fellowship P50NS061343; C.D.D., fellowship from the American Brain Foundation; J.N.G., NIH grant 5K23NS059774; P.M.R., awards from the NIHR and the Wellcome Trust; M.S., NIH grant U01 NS074425; and D.L.B., NIH grants R01 NS062675, R01 HL098065, R01 NS070941, and R18 HS017690, the Blue Cross Blue Shield of Michigan Foundation, Michigan Department of Community Health, and the University of Michigan.
Contributor Information
Daniel Woo, Email: daniel.woo@uc.edu.
Jonathan Rosand, Email: jrosand@partners.org.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
eqtl.uchicago.edu, http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl
IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
LocusZoom, http://csg.sph.umich.edu/locuszoom/
mRNA by SNP Browser, http://www.sph.umich.edu/csg/liang/asthma/
NCBI GTEx (Genotype-Tissue Expression) eQTL Browser, http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
SCAN: SNP and CNV Annotation Database, http://www.scandb.org/newinterface/about.html
SNP Annotation and Proxy Search (SNAP), http://www.broadinstitute.org/mpg/snap/
SNPDoc, https://www.phs.wakehealth.edu/public/bios/gene/downloads.cfm
SNPLASH, https://www.phs.wakehealth.edu/public/bios/gene/downloads.cfm
UNPHASED, https://sites.google.com/site/fdudbridge/software/
Accession Numbers
The dbGaP accession number for data reported in this paper is phs000416.v1.p1.
References
- 1.Feigin V.L., Lawes C.M.M., Bennett D.A., Barker-Collo S.L., Parag V. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: a systematic review. Lancet Neurol. 2009;8:355–369. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- 2.Strong K., Mathers C., Bonita R. Preventing stroke: saving lives around the world. Lancet Neurol. 2007;6:182–187. doi: 10.1016/S1474-4422(07)70031-5. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi A.I., Mendelow A.D., Hanley D.F. Intracerebral haemorrhage. Lancet. 2009;373:1632–1644. doi: 10.1016/S0140-6736(09)60371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kissela B., Schneider A., Kleindorfer D., Khoury J., Miller R., Alwell K., Woo D., Szaflarski J., Gebel J., Moomaw C. Stroke in a biracial population: the excess burden of stroke among blacks. Stroke. 2004;35:426–431. doi: 10.1161/01.STR.0000110982.74967.39. [DOI] [PubMed] [Google Scholar]
- 5.Taylor C.L., Yuan Z., Selman W.R., Ratcheson R.A., Rimm A.A. Mortality rates, hospital length of stay, and the cost of treating subarachnoid hemorrhage in older patients: institutional and geographical differences. J. Neurosurg. 1997;86:583–588. doi: 10.3171/jns.1997.86.4.0583. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty M.L., Kissela B., Woo D., Kleindorfer D., Alwell K., Sekar P., Moomaw C.J., Haverbusch M., Broderick J.P. The increasing incidence of anticoagulant-associated intracerebral hemorrhage. Neurology. 2007;68:116–121. doi: 10.1212/01.wnl.0000250340.05202.8b. [DOI] [PubMed] [Google Scholar]
- 7.Mayo N.E., Nadeau L., Daskalopoulou S.S., Côté R. The evolution of stroke in Quebec: a 15-year perspective. Neurology. 2007;68:1122–1127. doi: 10.1212/01.wnl.0000258664.12423.4c. [DOI] [PubMed] [Google Scholar]
- 8.Collins R., Peto R., MacMahon S., Hebert P., Fiebach N.H., Eberlein K.A., Godwin J., Qizilbash N., Taylor J.O., Hennekens C.H. Blood pressure, stroke, and coronary heart disease. Part 2, Short-term reductions in blood pressure: overview of randomised drug trials in their epidemiological context. Lancet. 1990;335:827–838. doi: 10.1016/0140-6736(90)90944-z. [DOI] [PubMed] [Google Scholar]
- 9.Vinters H.V. Cerebral amyloid angiopathy. A critical review. Stroke. 1987;18:311–324. doi: 10.1161/01.str.18.2.311. [DOI] [PubMed] [Google Scholar]
- 10.Fisher C.M. Pathological observations in hypertensive cerebral hemorrhage. J. Neuropathol. Exp. Neurol. 1971;30:536–550. doi: 10.1097/00005072-197107000-00015. [DOI] [PubMed] [Google Scholar]
- 11.Martini S.R., Flaherty M.L., Brown W.M., Haverbusch M., Comeau M.E., Sauerbeck L.R., Kissela B.M., Deka R., Kleindorfer D.O., Moomaw C.J. Risk factors for intracerebral hemorrhage differ according to hemorrhage location. Neurology. 2012;79:2275–2282. doi: 10.1212/WNL.0b013e318276896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falcone G.J., Biffi A., Brouwers H.B., Anderson C.D., Battey T.W.K., Ayres A.M., Vashkevich A., Schwab K., Rost N.S., Goldstein J.N. Predictors of hematoma volume in deep and lobar supratentorial intracerebral hemorrhage. JAMA Neurol. 2013;70:988–994. doi: 10.1001/jamaneurol.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biffi A., Sonni A., Anderson C.D., Kissela B., Jagiella J.M., Schmidt H., Jimenez-Conde J., Hansen B.M., Fernandez-Cadenas I., Cortellini L., International Stroke Genetics Consortium Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann. Neurol. 2010;68:934–943. doi: 10.1002/ana.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falcone G.J., Biffi A., Devan W.J., Jagiella J.M., Schmidt H., Kissela B., Hansen B.M., Jimenez-Conde J., Giralt-Steinhauer E., Elosua R., International Stroke Genetics Consortium Burden of risk alleles for hypertension increases risk of intracerebral hemorrhage. Stroke. 2012;43:2877–2883. doi: 10.1161/STROKEAHA.112.659755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devan W.J., Falcone G.J., Anderson C.D., Jagiella J.M., Schmidt H., Hansen B.M., Jimenez-Conde J., Giralt-Steinhauer E., Cuadrado-Godia E., Soriano C., International Stroke Genetics Consortium Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke. 2013;44:1578–1583. doi: 10.1161/STROKEAHA.111.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo D., Sauerbeck L.R., Kissela B.M., Khoury J.C., Szaflarski J.P., Gebel J., Shukla R., Pancioli A.M., Jauch E.C., Menon A.G. Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke. 2002;33:1190–1195. doi: 10.1161/01.str.0000014774.88027.22. [DOI] [PubMed] [Google Scholar]
- 17.Gomis M., Ois A., Rodríguez-Campello A., Cuadrado-Godia E., Jiménez-Conde J., Subirana I., Dávalos A., Roquer J. Outcome of intracerebral haemorrhage patients pre-treated with statins. Eur. J. Neurol. 2010;17:443–448. doi: 10.1111/j.1468-1331.2009.02838.x. [DOI] [PubMed] [Google Scholar]
- 18.Domingues-Montanari S., Hernandez-Guillamon M., Fernandez-Cadenas I., Mendioroz M., Boada M., Munuera J., Rovira A., Maisterra O., Parés M., Gutierrez M., Stroke Project Cerebrovascular Diseases Study Group, Spanish Society of Neurology ACE variants and risk of intracerebral hemorrhage recurrence in amyloid angiopathy. Neurobiol. Aging. 2011;32:e13–e22. doi: 10.1016/j.neurobiolaging.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Pera J., Slowik A., Dziedzic T., Pulyk R., Wloch D., Szczudlik A. Glutathione peroxidase 1 C593T polymorphism is associated with lobar intracerebral hemorrhage. Cerebrovasc. Dis. 2008;25:445–449. doi: 10.1159/000126918. [DOI] [PubMed] [Google Scholar]
- 20.Hallström B., Jönsson A.-C., Nerbrand C., Norrving B., Lindgren A. Stroke incidence and survival in the beginning of the 21st century in southern Sweden: comparisons with the late 20th century and projections into the future. Stroke. 2008;39:10–15. doi: 10.1161/STROKEAHA.107.491779. [DOI] [PubMed] [Google Scholar]
- 21.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anttila V., Winsvold B.S., Gormley P., Kurth T., Bettella F., McMahon G., Kallela M., Malik R., de Vries B., Terwindt G., North American Brain Expression Consortium. UK Brain Expression Consortium. International Headache Genetics Consortium Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat. Genet. 2013;45:912–917. doi: 10.1038/ng.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo D., Rosand J., Kidwell C., McCauley J.L., Osborne J., Brown M.W., West S.E., Rademacher E.W., Waddy S., Roberts J.N. The Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) study protocol. Stroke. 2013;44:e120–e125. doi: 10.1161/STROKEAHA.113.002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.-B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De La Vega F.M., Isaac H.I., Scafe C.R. A tool for selecting SNPs for association studies based on observed linkage disequilibrium patterns. Pac. Symp. Biocomput. 2006;2006:487–498. [PubMed] [Google Scholar]
- 37.Holliday E.G., Maguire J.M., Evans T.-J., Koblar S.A., Jannes J., Sturm J.W., Hankey G.J., Baker R., Golledge J., Parsons M.W., Australian Stroke Genetics Collaborative. International Stroke Genetics Consortium. Wellcome Trust Case Control Consortium 2 Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat. Genet. 2012;44:1147–1151. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Zoppo G.J. Toward the neurovascular unit. A journey in clinical translation: 2012 Thomas Willis Lecture. Stroke. 2013;44:263–269. doi: 10.1161/STROKEAHA.112.653618. [DOI] [PubMed] [Google Scholar]
- 39.Wardlaw J.M., Smith C., Dichgans M. Mechanisms of sporadic cerebral small vessel disease: insights from neuroimaging. Lancet Neurol. 2013;12:483–497. doi: 10.1016/S1474-4422(13)70060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rost N.S., Rahman R.M., Biffi A., Smith E.E., Kanakis A., Fitzpatrick K., Lima F., Worrall B.B., Meschia J.F., Brown R.D., Jr. White matter hyperintensity volume is increased in small vessel stroke subtypes. Neurology. 2010;75:1670–1677. doi: 10.1212/WNL.0b013e3181fc279a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rost N.S., Fitzpatrick K., Biffi A., Kanakis A., Devan W., Anderson C.D., Cortellini L., Furie K.L., Rosand J. White matter hyperintensity burden and susceptibility to cerebral ischemia. Stroke. 2010;41:2807–2811. doi: 10.1161/STROKEAHA.110.595355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fornage M., Debette S., Bis J.C., Schmidt H., Ikram M.A., Dufouil C., Sigurdsson S., Lumley T., DeStefano A.L., Fazekas F. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann. Neurol. 2011;69:928–939. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Devereux W., Stewart T.M., Casero R.A., Jr. Cloning and characterization of human polyamine-modulated factor-1, a transcriptional cofactor that regulates the transcription of the spermidine/spermine N(1)-acetyltransferase gene. J. Biol. Chem. 1999;274:22095–22101. doi: 10.1074/jbc.274.31.22095. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Devereux W., Stewart T.M., Casero R.A., Jr. Characterization of the interaction between the transcription factors human polyamine modulated factor (PMF-1) and NF-E2-related factor 2 (Nrf-2) in the transcriptional regulation of the spermidine/spermine N1-acetyltransferase (SSAT) gene. Biochem. J. 2001;355:45–49. doi: 10.1042/0264-6021:3550045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dempsey R.J., Başkaya M.K., Doğan A. Attenuation of brain edema, blood-brain barrier breakdown, and injury volume by ifenprodil, a polyamine-site N-methyl-D-aspartate receptor antagonist, after experimental traumatic brain injury in rats. Neurosurgery. 2000;47:399–404. doi: 10.1097/00006123-200008000-00024. discussion 404–406. [DOI] [PubMed] [Google Scholar]
- 46.Koenig H., Goldstone A.D., Lu C.Y. Blood-brain barrier breakdown in cold-injured brain is linked to a biphasic stimulation of ornithine decarboxylase activity and polyamine synthesis: both are coordinately inhibited by verapamil, dexamethasone, and aspirin. J. Neurochem. 1989;52:101–109. doi: 10.1111/j.1471-4159.1989.tb10903.x. [DOI] [PubMed] [Google Scholar]
- 47.Georgiev D., Taniura H., Kambe Y., Takarada T., Yoneda Y. A critical importance of polyamine site in NMDA receptors for neurite outgrowth and fasciculation at early stages of P19 neuronal differentiation. Exp. Cell Res. 2008;314:2603–2617. doi: 10.1016/j.yexcr.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Haitina T., Lindblom J., Renström T., Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88:779–790. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 49.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott R.A., Lagou V., Welch R.P., Wheeler E., Montasser M.E., Luan J., Mägi R., Strawbridge R.J., Rehnberg E., Gustafsson S., DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bellenguez C., Bevan S., Gschwendtner A., Spencer C.C.A., Burgess A.I., Pirinen M., Jackson C.A., Traylor M., Strange A., Su Z., International Stroke Genetics Consortium (ISGC) Wellcome Trust Case Control Consortium 2 (WTCCC2) Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat. Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


