Abstract
Genital condyloma-like lesions were observed on male and female cynomolgus macaque monkeys (Macaca fascicularis) originating from the island of Mauritius. Cytobrush and/or biopsy samples were obtained from lesions of 57 affected macaques. Primary histologic features included eosinophilic, neutrophilic, and lymphoplasmacytic penile and vulvar inflammation, epidermal hyperplasia with acanthosis, and increased collagenous stroma. Polymerase chain reaction–based assays to amplify viral DNA revealed the presence of macaque lymphocryptovirus (LCV) DNA but not papillomavirus or poxvirus DNA. Subsequent DNA analyses of 3 genomic regions of LCV identified isolates associated with lesions in 19/25 (76%) biopsies and 19/57 (33%) cytology samples. Variable immunolabeling for proteins related to the human LCV Epstein Barr Virus was observed within intralesional plasma cells, stromal cells, and epithelial cells. Further work is needed to characterize the epidemiologic features of these lesions and their association with LCV infection in Mauritian-origin macaques.
Keywords: genital, condyloma, macaque, primate, gammaherpesvirus, lymphocryptovirus, papillomavirus, Mauritius
Introduction
Cynomolgus macaques were introduced to the island of Mauritius by Portuguese explorers between 300 and 500 years ago (Sussman and Tattersall 1986). Descendants of this original population are now widely used in research, particularly for the study of simian/human immunodeficiency virus (S/HIV) pathogenesis and vaccine efficacy (Florese et al. 2008; Mee et al. 2009). These animals are an immunologically unique population due in part to low major histocompatibility complex (MHC) diversity (Blancher et al. 2008) and distinct MHC-dependent responses to S/HIV challenge (Florese et al. 2008), making them a valuable research resource.
Recently, veterinarians working with Mauritian-origin macaques identified distinct genital lesions ranging in appearance from plaques to papillomatous growths. These observations led to potential concerns related to colony health, reproductive function, and suitability for research studies. Increasing numbers of observed cases in recent years also suggested a potential infectious etiology, originally suspected to be papillomavirus (PV). Genital PVs are highly prevalent in some nonhuman primate (NHP) populations and associated with both warty (condylomatous) and invasive types of genital neoplasia, similar to human PVs (Ostrow et al. 1990; Wood et al. 2004, 2007, 2011). Other candidate agents included types of herpesvirus (HV) and poxvirus (PXV), which have also been associated with proliferative skin and/or mucosal lesions across numerous mammalian hosts (e.g., King et al. 2002; Lackovich et al. 1999; Marr-Belvin et al. 2008; Brown, Nalley, and Kraus 1981; Banks et al. 2002; Goldstein et al. 2006; Sakulwira et al. 2004). The primary goal of this study was to provide an initial characterization of these genital lesions and investigate associated viral agents.
Method
Animal Background
Samples for this case series were collected from 57 cynomolgus macaques (Macaca fascicularis) with genital condylomatous growths observed grossly by the attending veterinarians. All animals were housed in Mauritius except for 1 female macaque housed at Wake Forest University (WFU), which originated from Mauritius. Animals in Mauritius are housed socially, in outdoor enclosures, in Assessment and Accreditation of Laboratory Animal Care (AAALAC)/International Organization for Standardization (ISO)-accredited facilities (http://www.aaalac.org) at three sites located on the West Coast of the Republic of Mauritius off the southeast coast of Africa. Animals are provided a formulated, commercially prepared monkey chow (Meaders Feed Ltd or LFL Ltd), a variety of fresh fruits and vegetables, and fresh water ad libitum via an automatic system. The program for animal care and welfare here meets AAALAC standards, including daily health observations, routine husbandry practices, disease surveillance and prevention programs, emergency and routine veterinary care, and a behavioral management program provided by veterinarians, veterinary technicians, behavioral management staff, and animal care staff. The facilities and laboratory animal programs of WFU are also fully accredited by AAALAC. Approval for sample collection was obtained for all specimens used in the current study by respective Institutional Animal Care and Use Committees (IACUCs) in accordance with committee guidelines and AAALAC accreditation. All procedures were conducted in compliance with State and Federal laws and standards of the U.S. Department of Health and Human Services.
Cell and Biopsy Collection
Brush cytology samples of gross genital lesions were collected from 22 female and 35 male adult cynomolgus monkeys (M. fascicularis) of Mauritian origin and stored in SurePath preservative (Tripath Imaging Inc., Burlington, NC). For this procedure, animals were briefly anesthetized with ketamine, and a soft nylon cytobrush (Surgipath Medical Industries, Richmond, IL)~0.3 cm in diameter was used to collect exfoliated keratinocytes from cervical, vaginal, and vulvar areas in females or directly from the exophytic penile lesions in males. After collection, the brush was placed in a preservative solution (SurePath, Burlington, NC) and coded so that the laboratory was blinded to information about animal identification and disease status. In addition, 25 biopsies from suspected lesions were collected from a total of 24 individuals. For biopsies, animals were sedated with ketamine, the genital area was cleaned with betadyne solution, and a small (<50 mg) pinch biopsy sample was taken from the lesion site.
Histopathology
Biopsies collected from the 16 animals listed in Table 1 were fixed in 10% neutral buffered formalin and processed using standard histologic procedures. Fixed tissues were embedded in paraffin and sectioned to 5 μm in thickness for H&E staining. H&E-stained slides were evaluated qualitatively for histologic changes by 2 board-certified pathologists. Biopsies from the remaining 8 animals (in the initial cohort of samples) were placed directly in cytology preservative and were thus not available for histologic evaluation.
Table 1.
Histologic characteristics of condylomatous genital lesions in Mauritian cynomolgus macaques.
| Case no. | Animal ID | Sex | Lesion location | Epithelial hyperplasiaa | Inflammatory cell infiltrateb | Eosinophilic componentc | PCR for LCV DNAd |
|---|---|---|---|---|---|---|---|
| 1 | 6974 | F | Vulva | 3+ | 2+ | 5+ | +(gB, MDBP) |
| 2 | BA457J | M | Penis | 3+ | 1+ | 1+ | +(gB) |
| 3 | BB433B | M | Penis | 2+ | 1+ | 1+ | – |
| 4 | BB606D | M | Penis | 3+ | 1+ | 1+ | – |
| 5 | BC200D | M | Penis | 2+ | 2+ | 3+ | – |
| 6 | BC232B | M | Penis | 3+ | 1+ | 3+ | – |
| 7 | BC530 | F | Vulva | 3+ | 3+ | 4+ | – |
| 8 | BC754B | M | Penis | 2+ | 2+ | 1+ | – |
| 9 | BC759A | M | Penis | 3+ | 2+ | 2+ | – |
| 10 | BC810B | M | Penis | 2+ | 2+ | 1+ | +(gB) |
| 11 | BC992B | M | Penis | 2+ | 2+ | 4+ | +(gB) |
| 12 | CA142A | M | Penis | 2+ | 1+ | 1+ | +(gB) |
| 13 | CA514B | M | Penis | 1+ | 1+ | 1+ | +(DPOL) |
| 14 | CA644B | M | Penis | 3+ | 1+ | 1+ | – |
| 15 | CB626A | M | Penis | 3+ | 2+ | 4+ | – |
| 16 | J541P | M | Penis | 3+ | 1+ | 5+ | +(gB) |
Note: LCV = lymphocryptovirus.
Epithelial hyperplasia within condyloma-like lesions; graded as absent (0), mild (1+), moderate (2+), or marked (3+).
Inflammatory cell infiltrate within condyloma-like genital lesions; graded as absent (0), mild (1+), moderate (2+), or severe (3+).
Eosinophilic component of the inflammatory cell population; graded as 0% to 10% of total inflammatory cells (1+), 11% to 25% (2+), 26% to 50% (3+), 51% to 75% (4+), and 76% to 100% (5+).
DNA obtained from cytology and/or biopsy samples, LCV region amplified is noted.
DNA Extraction
All samples were processed in an ultraviolet-light sterilized hood in a room physically separated from the laboratory where polymerase chain reaction (PCR) amplification and analyses of products were performed. DNA was extracted from cytologic specimens and biopsies by digestion in a solution containing 100 mM Tris, 2 mM EDTA, 2% Laureth-12, and 400 μg/ml Proteinase K at pH 8.0 at 55°C for 18 hr, heated at 95°C for 10 min, and precipitated with ammonium acetate and ethanol. Pelleted DNA was resuspended in Tris-EDTA buffer for amplification by PCR.
Viral Screening
Macaque DNA isolates were screened for PV, PXV, and HV sequences. Previously described primer sets included the following: PV primers (FAP59/64 and MY11/09) targeting conserved sites within the L1 gene (Burk et al. 1996; Castle et al. 2002; Chen et al. 2009); PXV primers targeting either the p43K or the MC080R genes of the molluscum contagiosum virus (MCV; Trama, Adelson, and Mordechai 2007); and HV primers targeting a conserved region of amino acid motifs within the coding region of the DNA-dependent DNA Polymerase (DPOL) gene (VanDevanter et al. 1996). Amplification of HV DNA utilized a multiplex, nested PCR method. The initial reaction used 2 sense primers, DFA and ILK, and a single antisense primer, KG1, in a final reaction volume of 10 μl. The second PCR round used 5 μl of first round PCR product as template in a final reaction of 50 μl, and amplification used the sense TGV primer and the antisense IYG primer, as described (VanDevanter et al. 1996). Degenerate PCR primers targeting either the Major DNA Binding Protein (MDBP) gene or the glycoprotein B (gB) gene of known simian lymphocrypto-viruses (LCVs) were used to further characterize HV-positive samples (Ehlers et al. 2010).
An equal mixture of Ampli Taq Gold DPOL (Applied Biosystems, Foster City, CA) and Platinum Taq DPOL High Fidelity (Invitrogen, Carlsbad, CA) was used as previously described (Chen et al. 2009). Thermocycler conditions for nested PCR included initial denaturing for 5 min at 94°C followed by 40 cycles as follows: 30 sec denaturing at 94°C, 60 sec annealing at 46°C, and 60 sec extension at 68°C followed by a final 7 min extension at 68°C. PCR products were separated by electrophoresis in 1% agarose gels, and the amplified DNA was purified using the Quickstep 2 PCR Purification Kit (EdgeBio, Gaithersburg, MD) and sequenced at the Einstein Genomics Facility (Bronx, NY).
Viral Sequence Analysis
Sequences were assembled using Geneious Pro version 5.5.2 (Biomatters LTD, Auckland, New Zealand) from bidirectional Sanger sequencing of PCR products. The compiled sequences from each sample were used as a nucleotide query in an individual National Center for Biotechnology Information (NCBI) Blastn search for comparison to known viral sequences in GenBank (NCBI, Bethesda, MD). Nucleotide sequences identified by Blast as gamma-HV (gHV) DNA were aligned using ClustalW, according to gene region, in Geneious Pro. The identified query sequences were subsequently aligned by codon, starting with the first ATG, using the FFT-NS-2 algorithm in MAFFT v6.814b to generate multiple sequence alignments (Katoh et al. 2002).
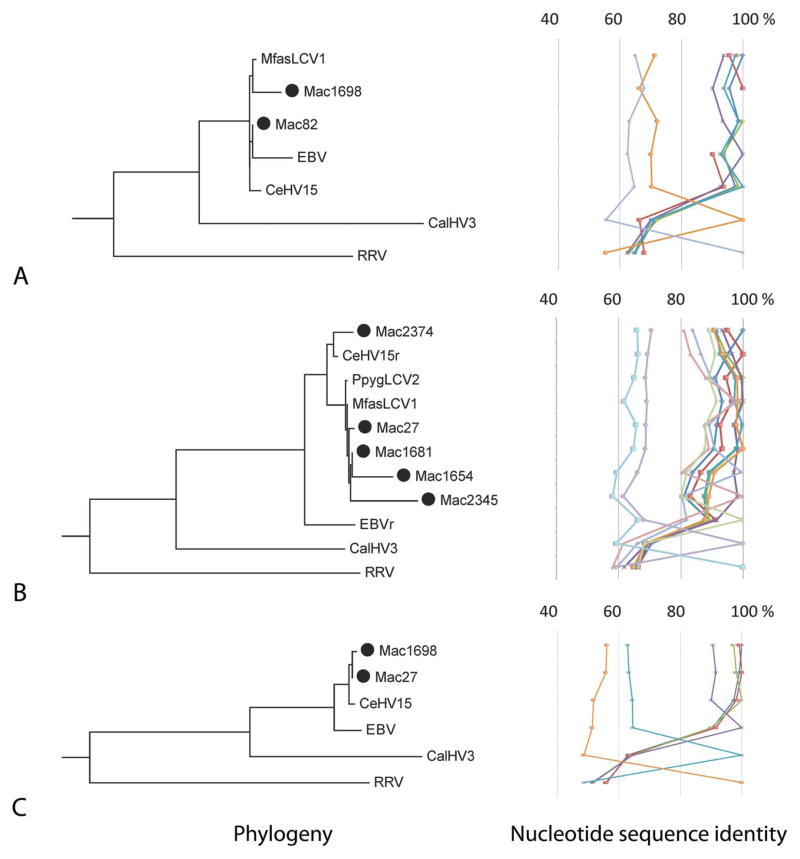
HV sequences, as determined by blast searches, were extracted and aligned for each of the 3 genomic regions sequenced (i.e., DPOL, gB, and MDBP). Blastn results generated the top 100 maximum identity hits from the NCBI database. Of these, we selected reference sequences with maximum identity to the query sequences, selecting those blast hits that aligned to at least 80% of the query sequence and had greater than 95% identity, harbored no more than 2 gaps, and had the most significant E-values. All sequences from isolates were aligned to at least 4 common gHV reference genomes: CeHV-15 (Rhesus LCV, accession # AY037858), CalHV-3 (Marmoset LCV, accession # AF319782), EBV (Human HV 4, accession # DQ279927), and RRV (Rhesus rhadinovirus, accession # AF083501). Where available, nucleotide sequences from orangutan LCV (PpygLCV2, accession # GQ921926) and cynomolgus macaque LCV (MfasLCV1, accession # AF534221) were included. The available reference sequences for each of the assayed LCV genes varied with region. The number of homologous sequences in the gB and DPOL region exceeded those for the MDBP region.
Phylogenetic Trees
Nucleotide sequences for the complete open reading frames (ORFs) from homologous simian LCV genomes identified by NCBI Blastn searches were used in phylogenetic analysis using RRV as the gamma-1 outgroup. Multiple sequence alignments generated by MAFFT and ClustalW were used to build a Maximum-Likelihood (ML) tree using PhyML (Guindon et al. 2010) within SeaView v4.3.0 (Gouy, Guindon, and Gascuel 2010). ModelTestv3.7 (Posada and Crandall 1998) was used to identify the best parameters for phylogenetic tree construction. The general time reversible (GTR) model was applied to our data set and used as the model for the rate of nucleotide substitution; data were bootstrapped 1,000 times with a fixed Ts/Tv ratio of 4.0 and optimized for across-site variability. Tree topology was generated using the nearest neighbor interchange to identify the most likely tree. Nucleotide polymorphisms were identified from multiple sequence alignments using MEGA5 (Tamura et al. 2011; data not shown).
Immunohistochemistry (IHC)
IHC to detect the EBV-associated proteins EBV nuclear antigen 2 (EBNA2) and EBV latent membrane protein 1 (LMP1) was performed on tissue sections from the 16 animals in Table 1. Lymphoma tissue from a previously reported simian EBV-positive lymphoma from a SIV-infected rhesus macaque (Macaca mulatta; Blaschke et al. 2001) was kindly provided by Dr. Sherry Klump for use as positive control tissue. Briefly, heat-induced epitope retrieval was performed in a citrate buffer (pH 6.0) using a pressure cooker at high pressure for 1 min. Specimens were incubated overnight with primary antibodies for EBNA2 (mouse monoclonal, clone PE2: 1:25 dilution at 4°C; Dako, Carpinteria, CA) and LMP1 (mouse monoclonal, clone CS.1–4: 1:100 dilution at 37°C; Dako, Carpinteria, CA). Antibody labeling was detected using a biotinylated anti-mouse secondary antibody and a streptavidin–alkaline phosphatase enzyme conjugate.
Results
Gross and Histologic Features
Exophytic genital lesions were initially brought to our attention from reference samples (not included here) sent by independent animal facilities that had acquired affected cynomolgus macaques from colonies in Mauritius. Subsequent correspondence with veterinarians and managers associated with one of these colonies revealed that the lesions had been observed more commonly in recent years, primarily among male macaques, prompting this case survey. Grossly, the penile lesions ranged from raised focal or multifocal verrucous papillomas up to approximately 2.5 × 2.0 × 1.0 cm in size to more widespread plaques on both the prepuce and the glans penis (Figure 1A). Lesions showed variable loss of pigmentation with areas of erythema and ulceration. Vulvar/vaginal lesions also presented as multifocal to coalescing papillomas, often with a domed appearance (Figure 1B). The vulvar lesion in Figure 1B was originally removed in 2002, gradually recurred, and was removed again in 2010.
Figure 1.
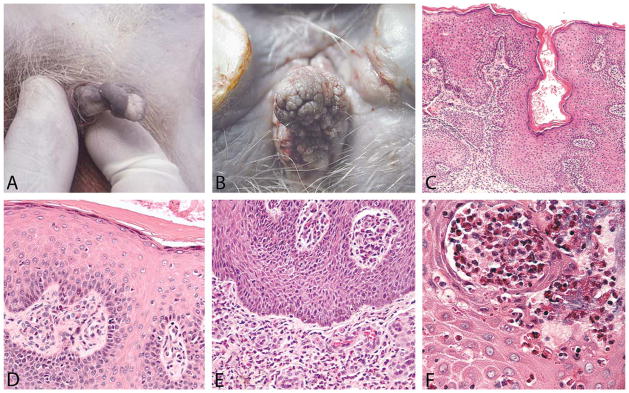
Morphologic features of genital lesions in Mauritian-origin cynomolgus macaques. Representative gross images show exophytic penile (A) and vulvar (B) masses. Condyloma-like lesions were composed of a collagenous stromal core lined by a hyperplastic keratinized stratified squamous epithelium. Histologic features of penile (C, E) and vulvar (D, F) papillomas included epidermal hyperplasia (C–E); mild-to-severe eosinophilic and lymphoplasmacytic inflammation, primarily within the superficial dermis (C–E); fibrovascular stroma often with plump endothelial cells (E); intraepidermal eosinophilic pustules (F); and bacterial cocci (F). Images were taken at an objective magnification of 4× (C), 20× (E), and 40× (D, F).
Histologic findings in the reference penile cases and the initial vulvar biopsy from a Mauritian-origin female animal at WFU (Figure 1B, Case 1 in Table 1) were similar. Primary changes included epidermal hyperplasia with acanthosis and hypergranulosis (Figure 1C) and a pleocellular eosinophilic, neutrophilic, and lymphoplasmacytic balanoposthitis or vulvitis, often with dermal and intraepidermal pustules (Figure 1D). Other changes included orthokeratotic hyperkeratosis, dermal edema, and stromal proliferation. In some cases, aggregates of cellular debris, degenerate eosinophils or neutrophils, and bacterial colonies (cocci) were present within the superficial keratin layers.
A subsequent batch of 15 biopsies was collected from macaques in Mauritius exhibiting grossly exophytic genital lesions (cases 2–16 in Table 1). Microscopically, these condyloma-like masses were polypoid and composed of a vascular collagenous core lined by irregularly hyperplastic, keratinizing stratified squamous epithelium with marked acanthosis, occasional thickening of the stratum granulosum, and diffuse predominantly orthokeratotic hyperkeratosis. The hyperplastic epithelium formed broad anastomosing rete ridges that projected into the stroma. The stroma contained congested, thin-walled vessels lined by plump endothelial cells, and mild to heavy lymphoplasmacytic and eosinophilic inflammation. Inflammatory cells were frequently limited to the stroma immediately subjacent to the epithelium. The quantity of the eosinophilic inflammatory cell component varied among specimens, although eosinophils were present in all lesions and predominated in some specimens (Figure 1, Table 1). Secondary changes observed across multiple samples included randomly scattered keratinocyte apoptosis, perinuclear clearing within keratinocytes (intracellular edema), intercellular edema, ulceration, and intracorneal bacterial cocci. One animal displayed segmented interface dermatitis, mild epithelial disorganization, and basal cell proliferation. Intranuclear or intracytoplasmic inclusions were not observed, and there was no evidence of cytologic atypia, including karyomegaly or koilocytosis.
Virus Sequencing and Analysis
Given the exophytic appearance of these wart-like lesions, it was initially suspected that they would be associated with PV infection. However, despite exhaustive efforts to amplify PV fragments using multiple primer sets, no PV DNA was identified in any of these samples. Subsequently, we pursued other viral agents associated with proliferative cutaneous or mucosal growth. The first alternative was MCV, a human PXV associated with proliferative genital lesions. All samples were tested for PXV DNA using a broad-spectrum degenerate primer PCR system, but no MCV DNA was detected. We next looked for evidence of HV infection using a nested PCR method previously reported to amplify a wide range of gHVs in primates (Ehlers et al. 2010). Consensus primers generated PCR products that upon sequencing revealed the presence of gHV DNA. Using degenerate PCR primers designed to amplify a range of known simian gHVs (Ehlers et al. 2010), we amplified DNA from 3 distinct regions of the gHV genome: DPOL (DNA-dependent Polymerase), gB, and MDBP. In total, we detected gHV DNA by PCR in 33% of cytologic samples and 76 % of biopsies, as summarized in Table 2.
Table 2.
Summary of lymphocryptovirus detection by PCR assay.
| Sample type | PCR assay | Male | Female | All | % Positive |
|---|---|---|---|---|---|
| Cytology Preps | N = 35 | N = 22 | N = 57 | ||
| BALF5/DPOL | 1 | – | 1 | ||
| BALF4/gB | 8 | 6 | 14 | ||
| BALF2/MDBP | 2 | 2 | 4 | ||
| Total positive | 11 | 8 | 19 | 33 | |
| Biopsy | N = 22 | N = 3a | N =25 | ||
| BALF5/DPOL | 4 | 1 | 5 | ||
| BALF4/gB | 8 | 1 | 9 | ||
| BALF2/MDBP | 4 | 1 | 5 | ||
| Total positive | 16 | 3 | 19 | 76 |
Note: PCR = polymerase chain reaction.
Includes repeat biopsies of 1 animal.
The DPOL region was amplified from 6 cynomolgus macaque samples and characterized by sequence analyses (see Table 2, Figure 2A). This gene is highly conserved across HV genomes and often targeted by consensus primer PCR to discriminate among subfamilies of HVs for identification of viral isolates (Ehlers et al. 1999; VanDevanter et al. 1996). To distinguish LCV species, pairwise DNA sequence identity of less than 95% similarity within the DPOL region has been used by some investigators (Ehlers et al. 2003). To assess genomic heterogeneity within the conserved DPOL region, nucleotide identity derived from our study was compared to available DPOL (partial) sequences from Old World primate LCVs: rhesus LCV (CeHV-15), orangutan LCV (PpygLCV-2), and a previously described partial isolate from a captive Japanese macaque, MfasLCV-1 (Ehlers et al. 2003).
Figure 2.
Phylogenetic trees and nucleotide identity of viral isolates. Phylogenetic tree topologies and percent nucleotide identity between isolates and known gHV types in a given genomic region are shown on the left and right side of each panel, respectively. Panels show maximum-likelihood (ML) tree and percent nucleotide sequence identity derived from partial sequences of the DNA Polymerase (DPOL) region from indicated samples (A); ML tree and percent nucleotide sequence identity for isolates and known gHV types in the gB region (B); and ML tree and percent nucleotide sequence identity for isolates and known gHV types in the major DNA binding protein (MDBP) region (C). Representative sequences for isolates determined in this study are denoted with a black circle in front of the sample name.
DPOL nucleotide sequence alignments showed that our isolates sorted into 2 clades. The representative isolates, Mac82 and Mac1698, shown in Figure 2A, exemplify the range of sequence variability we detected from cynomolgus macaque isolates. Overall, DNA isolated from biopsies had higher sensitivity for amplification of the LCV DPOL region compared to the cytology prep samples (Table 2). The pairwise percent nucleotide identity of isolates was compared to previously identified simian LCV homologs and are displayed to the right of the tree (Figure 2A). All sequences from the isolates of this study were tightly grouped with the EBV and CeHV15 sequences, showing greater than 90% sequence identity. The DPOL sequence from the 2 representative isolates from this study had greater than 95% nucleotide identity to the previously reported isolate from a Japanese macaque (MfasLCV1; Figure 2A and Supplemental Table S2A). The isolate from Mac1698 was more frequently identified within our sampling population as compared to that from Mac82, as shown in the Supplemental Table S3.
Amplification of a 315 bp region of the gB gene by PCR from 23 cynomolgus macaques was confirmed by sequencing. Of these 23 animals, gB DNA was identified from 14 cytology samples and 9 biopsies (see Table 2), and the sequences were compared to homologous gB sequences from CeHV-15, EBV, PpygLCV-2, CalHV-3, and RRV. A partial sequence of 100 nucleotides in the gB region from MfasLCV-1 was previously identified from a cynomolgus macaque (Ehlers et al. 2003). Five isolates representative of partial gB sequences detected by PCR are included in the ML tree and sort to 2 subclades, one similar to rhesus LCV (CeHV15), and the other which includes the partial isolates from orangutan (PpygLCV2) and the cynomolgus macaque (MfasLCV1) viral homologs. The percent nucleotide sequence identity within the gB region is displayed to the right of the tree (see Figure 2B). Of the different gB sequence isolates, 3 exhibit greater than 90% identity to CeHV15 and 2 share more than 95% nucleotide identity to the partial 100 bp sequence available from MfasLCV1, as shown in Figure 2B. The most frequent isolate identified in our sampling population based on the gB region was isolate Mac1681, which when aligned to the CeHV15 reference genome contained 19 polymorphic sites (Supplemental Table S2B). This viral isolate was identified in both male and female macaques and from samples collected by both cytology and biopsy.
DNA from 9 cynomolgus macaques generated a PCR amplicon from the MDBP region (Table 2). Multiple sequence alignments revealed high conservation to the rhesus LCV (CeHV15) MDBP gene, represented by the 2 cynomolgus macaque isolates (Mac27 and Mac1698) included in the ML tree that exhibited greater than 95% pairwise nucleotide identity to CeHV15 (Figure 2C). The only references available for comparison in this region were from a limited set of LCV genomes (i.e., EBV and CalHV3). RRV was set as the out-group to root the tree. Of the 9 samples from which the MDBP sequence was determined, 2 representative viral isolates exhibited greater than 95% nucleotide sequenceidentitytoCeHV15(Figure 2C, Supplemental Table S2C).
Immunolabeling for EBV proteins
IHC for the EBV latency proteins EBNA2 and LMP1 was performed on tissue sections from 16 cases (Table 1). Nuclear immunolabeling for EBNA2 was present in lymphocytes from EBV-positive control tissue but absent in all 16 genital lesion cases (Figure 3A). In contrast, moderate-to-strong cytoplasmic LMP1 antibody labeling was observed in all tissue sections within intralesional plasma cells and fewer numbers of spindle-shaped stromal cells and epithelial cells (Figure 3B, Supplemental Figure 1). It should be noted, however, that non-specific labeling of plasma cells with LMP1 antibodies has been previously reported (Gulley et al. 2002), and some nuclei of epithelial cells labeled positive for LMP1 antibody, suggesting nonspecific labeling in these cells. The pattern of LMP1 is known to vary with regard to stage of the gHV life cycle, and the positive labeling of some epithelial cells with LMP1 and not EBNA2 is of unknown significance at this time.
Figure 3.
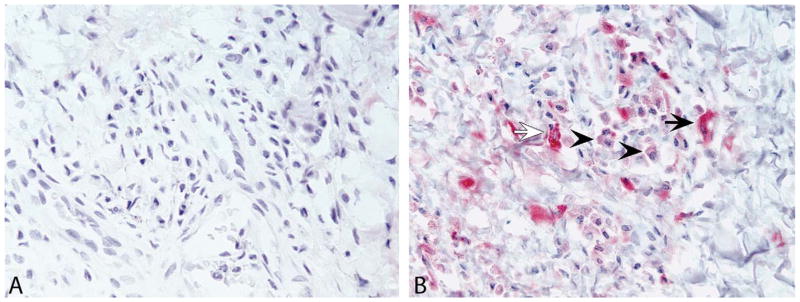
Immunohistochemistry for EBV antigens EBNA2 and LMP1 in sections of genital lesions from a Mauritian-origin cynomolgus macaque (Case No. 14). Representative images show negative immunolabeling for the Epstein-Barr virus (EBV) nuclear antigen EBNA2 (A) and moderate to strong cytoplasmic labeling for latent membrane protein 1 (LMP1; B) in low numbers of intralesional spindle-shaped stromal cells (arrows) and plasma cells (arrowheads). Images were taken at 40× objective magnification.
Discussion
In this study, we provide an initial characterization of a distinctive genital lesion type in Mauritian-origin cynomolgus macaques. Lesions included hyperplastic and inflammatory elements but lacked clear neoplastic morphology, including atypia characteristic of genital PV infection. We identified gHV DNA from the LCV genus within these lesions but not PV or PXV DNA. Further work is needed to establish epidemiologic and molecular features of these lesions and investigate the potential etiologic role of LCV infection. This information should contribute to improved care and management of this unique population of animals.
Based on location and gross appearance, the observed lesions were originally suspected to be of PV origin. This idea was supported by evidence showing that rhesus and cynomolgus macaques are naturally infected with a range of PVs associated with preneoplastic and neoplastic changes, including exophytic genital and cutaneous papillomas (Ostrow et al. 1990; Wood et al. 2004, 2007; Chen et al. 2009; Wood et al. 2011). A viral etiology was also supported by one prior case report in a male cynomolgus macaque of Mauritian origin (Gordon, Reim, and McClain 2000). In this previous study, a condylomatous penile lesion grossly similar to those described here showed unidentified intranuclear viral particles on electron microscopy but was negative for PV on PCR and in situ hybridization. Viral particles visualized in this previous report were round with an estimated diameter of 80 nm to 120 nm, consistent with a HV. This latter report suggests that these lesions may represent a longstanding pathologic entity among Mauritian-origin macaques.
Herpesviridae is a diverse family of large, double-stranded DNA viruses comprised of alpha, beta, and gamma subfamilies. Classification of HVs is based on viral genome sequence, biology, and cellular tropism, and associated disease outcomes in the host (Damania, Choi, and Jung 2000). Taxonomy of HVs reflects the host from which the virus was initially isolated (Davison et al. 2009), and phylogenetic congruence among host species and viral isolates supports the idea of coevolution/cospeciation (Ehlers et al. 2010; McGeoch, Rixon, and Davison 2006). The Gammaherpesvirinae subfamily is divided into 4 genera within 2 genogroups. The gamma-1 HV genogroup is comprised of LCVs, while the gamma-2 genogroup includes the gHV genera Rhadinovirus, Macavirus, and Perca-virus (Davison et al. 2009). Within the gamma-1 group is the human gHV Epstein-Barr virus (EBV, HHV4) along with other LCVs isolated from Old and New World NHPs (Carville and Mansfield 2008). The latter group includes MfasLCV-1, which was previously isolated from a single cynomolgus macaque (Ehlers et al. 2003). The gamma-1 HVs are characterized by their ability to establish latent infections of B-lymphocytes, which are typically controlled by the host immune system of healthy individuals. Lytic infection can also occur in epithelial cells but is less common (Damania, Choi, and Jung 2000).
gHV infections have been observed in association with various proliferative and neoplastic lesions in both NHPs and humans, particularly in immunosuppressed hosts (Carville and Mansfield 2008; Katoh et al. 2002; Klein, Klein, and Kashuba 2010). The only human LCV, EBV, is classified as an oncogenic virus and implicated in lymphoid and epithelial malignancies, including Burkitt’s lymphoma, nasopharyngeal and gastric carcinomas, and Hodgkin’s disease–related lymphomas (Katoh et al. 2002). Infection of rhesus macaques with gHV has also been associated with malignancy, especially among immuno-compromised animals in SIV challenge-vaccine models, which exhibit increased susceptibility for developing B-cell associated malignancies (Feichtinger et al. 1990). Similarly, HIV-positive human patients exhibit increased incidence of EBV coinfection and increased risk of B-cell derived malignancies when infected with multiple EBV isolates (Yao et al. 1996). Immunosuppressed hosts infected with EBV are also prone to develop lymphoproliferative disorders, which have been modeled in EBV-positive cynomolgus and rhesus macaques (Schmidtko et al. 2002; Rivailler et al. 2002).
Despite low MHC diversity, Mauritian-origin cynomolgus macaques are not immunocompromised, in contrast to SIV-infected rhesus macaques (Kutok et al. 2004; Marr-Belvin et al. 2008). Nevertheless, the characteristic genital lesions described in this report to date have been observed only in macaques of Mauritian-origin, suggesting a potential host genetic and/or immunologic component to the development of lesions. Further work is needed to characterize potential associations with specific MHC haplotypes among Mauritian macaques and investigate potential etiologies, including LCV. Additional questions include the background rate of LCV exposure/infection in unaffected Mauritian macaques, the potential role of chronic inflammation in gene activation of latent virus, cellular localization of virus within lesions and adjacent normal tissue, and colony management strategies to reduce lesion incidence.
Supplementary Material
Supplemental Figure 1. Immunolabeling for latent membrane protein 1 (LMP1) within intralesional spindle-shaped stromal cells (arrows) and plasma cells (arrowhead) from additional Mauritian-origin cynomolgus macaques (Case Nos. 7 and 10). Nonspecific staining was also present in blood (asterisk). Images were taken at 40× objective magnification.
Supplemental Table S1. Viral isolate National Centre for Biotechnology Information (NCBI) accession numbers.
Supplemental Table S2. Nucleotide alignment of isolates in (S2A) DNA Polymerase (DPOL) region, (S2B) pB region, and (2SC) major DNA binding protein (MDBP) region.
Supplemental Table S3. Distribution of viral isolates in cynomolgus macaques from Mauritius.
Acknowledgments
The authors thank Jean Gardin, Matt Dwyer, Hermina Borgerink, Benjamin Smith, Anne Dunn, and Danielle Bottalico for technical support.
Abbreviations
- AAALAC
Assessment and Accreditation of Laboratory Animal Care
- DPOL
DNA Polymerase
- EBNA
EBV nuclear antigen
- EBV
Epstein-Barr virus
- gB
glycoprotein B
- gHV
Gammaherpesvirinae
- GTR
general time reversible
- HV
herpesvirus
- S/HIV
simian/human immunodeficiency virus
- IHC
immunohistochemistry
- LCV
lymphocryptovirus
- LMP1
latent membrane protein 1
- MCV
molluscum contagiosum virus
- MDBP
Major DNA Binding Protein
- ML
maximum likelihood
- MHC
major histocompatibility complex
- NHP
nonhuman primate
- ORF
open reading frame
- PCR
polymerase chain reaction
- PV
papillomavirus
- PXV
poxvirus
- RRV
rhesus rhadinovirus
- WFU
Wake Forest University
Footnotes
The authors disclosed receipt of the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article. Marie Claire Domaingue and Patricia Koenig are employed by Biodia Co Ltd. David Elmore has served as a consultant to Biodia Co Ltd.
Supplemental tables and figures can be found at http://tpx.sagepub.com/supplemental
The contents are solely the responsibility of the authors and do not necessarily represent the views of the NIAID, NCRR, NCI, or NIH.
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by Public Health Service awards K01 RR21322 and P40 RR021380 from the National Center for Research Resources (NCRR) and R21 AI083962 from the National Institute of Allergy and Infectious Diseases (NIAID), the Einstein-Montefiore Center for AIDS funded by the National Institutes of Health (NIH; AI51519), the Einstein Cancer Research Center (P30CA013330) from the National Cancer Institute (NCI), and the WFU School of Medicine Translational Science Institute.
References
- Banks M, King DP, Daniells C, Stagg DA, Gavier-Widen D. Partial characterization of a novel gammaherpesvirus isolated from a European badger (Meles meles) J Gen Virol. 2002;83:1325–30. doi: 10.1099/0022-1317-83-6-1325. [DOI] [PubMed] [Google Scholar]
- Blancher A, Bonhomme M, Crouau-Roy B, Terao K, Kitano T, Saitou N. Mitochondrial DNA sequence phylogeny of 4 populations of the widely distributed cynomolgus macaque (Macaca fascicularis fascicularis) J Hered. 2008;99:254–64. doi: 10.1093/jhered/esn003. [DOI] [PubMed] [Google Scholar]
- Blaschke S, Hannig H, Buske C, Kaup FJ, Hunsmann G, Bodemer W. Expression of the simian Epstein-Barr virus-encoded latent membrane protein-1 in malignant lymphomas of SIV-infected rhesus macaques. J Med Virol. 2001;65:114–20. [PubMed] [Google Scholar]
- Brown ST, Nalley JF, Kraus SJ. Molluscum contagiosum. Sex Transm Dis. 1981;8:227–34. doi: 10.1097/00007435-198107000-00012. [DOI] [PubMed] [Google Scholar]
- Burk RD, Ho GY, Beardsley L, Lempa M, Peters M, Bierman R. Sexual behavior and partner characteristics are the predominant risk factors for genital human papillomavirus infection in young women. J Infect Dis. 1996;174:679–89. doi: 10.1093/infdis/174.4.679. [DOI] [PubMed] [Google Scholar]
- Carville A, Mansfield KG. Comparative pathobiology of macaque lymphocryptoviruses. Comp Med. 2008;58:57–67. [PMC free article] [PubMed] [Google Scholar]
- Castle PE, Schiffman M, Gravitt PE, Kendall H, Fishman S, Dong H, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Lorincz A, Schussler JE, Burk RD. Comparisons of HPV DNA detection by MY09/11 PCR methods. J Med Virol. 2002;68:417–23. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- Chen Z, van Doorslaer K, DeSalle R, Wood CE, Kaplan JR, Wagner JD, Burk RD. Genomic diversity and interspecies host infection of alpha12 Macaca fascicularis papillomaviruses (MfPVs) Virology. 2009;393:304–10. doi: 10.1016/j.virol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B, Choi JK, Jung JU. Signaling activities of gammaherpesvirus membrane proteins. J Virol. 2000;74:1593–601. doi: 10.1128/jvi.74.4.1593-1601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–77. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Borchers K, Grund C, Frolich K, Ludwig H, Buhk HJ. Detection of new DNA polymerase genes of known and potentially novel herpesviruses by PCR with degenerate and deoxyinosine-substituted primers. Virus Genes. 1999;18:211–20. doi: 10.1023/a:1008064118057. [DOI] [PubMed] [Google Scholar]
- Ehlers B, Ochs A, Leendertz F, Goltz M, Boesch C, Matz-Rensing K. Novel simian homologues of Epstein-Barr virus. J Virol. 2003;77:10695–99. doi: 10.1128/JVI.77.19.10695-10699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Spiess K, Leendertz F, Peeters M, Boesch C, Gatherer D, McGeoch DJ. Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus. J Gen Virol. 2010;91:630–42. doi: 10.1099/vir.0.017251-0. [DOI] [PubMed] [Google Scholar]
- Feichtinger H, Putkonen P, Parravicini C, Li SL, Kaaya EE, Bottiger D, Biberfeld G, Biberfeld P. Malignant lymphomas in cynomolgus monkeys infected with simian immunodeficiency virus. Am J Pathol. 1990;137:1311–15. [PMC free article] [PubMed] [Google Scholar]
- Florese RH, Wiseman RW, Venzon D, Karl JA, Demberg T, Larsen K, Flanary L, Kalyanaraman VS, Pal R, Titti F, Patterson LJ, Heath MJ, O’Connor DH, Cafaro A, Ensoli B, Robert-Guroff M. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: Influence of Mauritian MHC haplotypes on susceptibility/ resistance to SHIV(89.6P) infection. Vaccine. 2008;26:3312–21. doi: 10.1016/j.vaccine.2008.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein T, Lowenstine LJ, Lipscomb TP, Mazet JA, Novak J, Stott JL, Gulland FM. Infection with a novel gammaherpesvirus in northern elephant seals (Mirounga angustirostris) J Wildl Dis. 2006;42:830–35. doi: 10.7589/0090-3558-42.4.830. [DOI] [PubMed] [Google Scholar]
- Gordon HP, Reim DA, McClain SA. Condyloma acuminatum in a cynomolgus monkey (Macaca fascicularis) Contemp Top Lab Anim Sci. 2000;39:30–33. [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multi-platform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–24. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Gulley ML, Glaser SL, Craig FE, Borowitz M, Mann RB, Shema SJ, Ambinder RF. Guidelines for interpreting EBER in situ hybridization and LMP1 immunohistochemical tests for detecting Epstein-Barr virus in Hodgkin lymphoma. Am J Clin Pathol. 2002;117:259–67. doi: 10.1309/MMAU-0QYH-7BHA-W8C2. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Hure MC, Goldstein T, Aldridge BM, Gulland FM, Saliki JT, Buckles EL, Lowenstine LJ, Stott JL. Otarine herpesvirus-1: A novel gammaherpesvirus associated with urogenital carcinoma in California sea lions (Zalophus californianus) Vet Microbiol. 2002;86:131–37. doi: 10.1016/s0378-1135(01)00497-7. [DOI] [PubMed] [Google Scholar]
- Klein G, Klein E, Kashuba E. Interaction of Epstein-Barrvirus (EBV) with human B-lymphocytes. Biochem Biophys Res Commun. 2010;396:67–73. doi: 10.1016/j.bbrc.2010.02.146. [DOI] [PubMed] [Google Scholar]
- Kutok JL, Klumpp S, Simon M, MacKey JJ, Nguyen V, Middeldorp JM, Aster JC, Wang F. Molecular evidence for rhesus lymphocryptovirus infection of epithelial cells in immunosuppressed rhesus macaques. J Virol. 2004;78:3455–61. doi: 10.1128/JVI.78.7.3455-3461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackovich JK, Brown DR, Homer BL, Garber RL, Mader DR, Moretti RH, Patterson AD, Herbst LH, Oros J, Jacobson ER, Curry SS, Klein PA. Association of herpesvirus with fibro-papillomatosis of the green turtle Chelonia mydas and the loggerhead turtle Caretta caretta in Florida. Dis Aquat Organ. 1999;37:89–97. doi: 10.3354/dao037089. [DOI] [PubMed] [Google Scholar]
- Marr-Belvin AK, Carville AK, Fahey MA, Boisvert K, Klumpp SA, Ohashi M, Wang F, O’Neil SP, Westmoreland SV. Rhesus lymphocryptovirus type 1-associated B-cell nasal lymphoma in SIV-infected rhesus macaques. Vet Pathol. 2008;45:914–21. doi: 10.1354/vp.45-6-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Mee ET, Berry N, Ham C, Sauermann U, Maggiorella MT, Martinon F, Verschoor EJ, Heeney JL, Le Grand R, Titti F, Almond N, Rose NJ. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics. 2009;61:327–29. doi: 10.1007/s00251-009-0369-8. [DOI] [PubMed] [Google Scholar]
- Ostrow RS, McGlennen RC, Shaver MK, Kloster BE, Houser D, Faras AJ. A rhesus monkey model for sexual transmission of a papillomavirus isolated from a squamous cell carcinoma. Proc Natl Acad Sci U S A. 1990;87:8170–74. doi: 10.1073/pnas.87.20.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–18. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rivailler P, Jiang H, Cho YG, Quink C, Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: Genetic validation for an Epstein-Barr virus animal model. J Virol. 2002;76:421–26. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakulwira K, Theamboonlers A, Oraveerakul K, Chaiyabutr N, Bhattarakosol P, Poovorawan Y. Orangutan herpesvirus. J Med Primatol. 2004;33:25–29. doi: 10.1111/j.1600-0684.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- Schmidtko J, Wang R, Wu CL, Mauiyyedi S, Harris NL, Della Pelle P, Brousaides N, Zagachin L, Ferry JA, Wang F, Kawai T, Sachs DH, Cosimi BA, Colvin RB. Posttransplant lymphoproliferative disorder associated with an Epstein-Barr-related virus in cynomolgus monkeys. Transplantation. 2002;73:1431–39. doi: 10.1097/00007890-200205150-00012. [DOI] [PubMed] [Google Scholar]
- Sussman R, Tattersall I. Distribution, abundance, and putative ecological strategy of Macaca fascicularis on the island of Mauritius, southwestern Indian Ocean. Folia Primatol. 1986;46:28–43. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–39. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trama JP, Adelson ME, Mordechai E. Identification and genotyping of molluscum contagiosum virus from genital swab samples by real-time PCR and Pyrosequencing. J Clin Virol. 2007;40:325–29. doi: 10.1016/j.jcv.2007.09.007. [DOI] [PubMed] [Google Scholar]
- VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–71. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CE, Borgerink H, Register TC, Scott L, Cline JM. Cervical and vaginal epithelial neoplasms in cynomolgus monkeys. Vet Pathol. 2004;41:108–15. doi: 10.1354/vp.41-2-108. [DOI] [PubMed] [Google Scholar]
- Wood CE, Chen Z, Cline JM, Miller BE, Burk RD. Characterization and experimental transmission of an oncogenic papillomavirus in female macaques. J Virol. 2007;81:6339–45. doi: 10.1128/JVI.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CE, Tannehill-Gregg SH, Chen Z, Doorslaer K, Nelson DR, Cline JM, Burk RD. Novel betapapillomavirus associated with hand and foot papillomas in a cynomolgus macaque. Vet Pathol. 2011;48:731–36. doi: 10.1177/0300985810383875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao QY, Tierney RJ, Croom-Carter D, Dukers D, Cooper GM, Ellis CJ, Rowe M, Rickinson AB. Frequency of multiple Epstein-Barr virus infections in T-cell-immunocompromised individuals. J Virol. 1996;70:4884–94. doi: 10.1128/jvi.70.8.4884-4894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Immunolabeling for latent membrane protein 1 (LMP1) within intralesional spindle-shaped stromal cells (arrows) and plasma cells (arrowhead) from additional Mauritian-origin cynomolgus macaques (Case Nos. 7 and 10). Nonspecific staining was also present in blood (asterisk). Images were taken at 40× objective magnification.
Supplemental Table S1. Viral isolate National Centre for Biotechnology Information (NCBI) accession numbers.
Supplemental Table S2. Nucleotide alignment of isolates in (S2A) DNA Polymerase (DPOL) region, (S2B) pB region, and (2SC) major DNA binding protein (MDBP) region.
Supplemental Table S3. Distribution of viral isolates in cynomolgus macaques from Mauritius.