Abstract
The loss of skeletal muscle mass (atrophy) that accompanies disuse and systemic diseases is highly debilitating. Although the pathogenesis of this condition has been primarily studied in mammals, Drosophila is emerging as an attractive system to investigate some of the mechanisms involved in muscle growth and atrophy. In this review, we highlight the outstanding unsolved questions that may benefit from a combination of studies in both flies and mammals. In particular, we discuss how different environmental stimuli and signaling pathways influence muscle mass and strength and how a variety of disease states can cause muscle wasting.
Keywords: skeletal muscle growth, muscle atrophy, animal models of muscle wasting, proteostasis
INTRODUCTION
Skeletal muscle accounts for approximately 40%–50% of the body mass in humans. Maintenance of muscle mass and strength through proper nutrition and exercise is critical to maintain full activity, prevent obesity and decrease the risk of heart disease, diabetes, and cancer (Pate et al., 1995). Lack of contractile activity and a variety of systemic diseases, as well as inadequate nutrition, all lead to fiber atrophy (i.e. loss of cell proteins), and consequently, a decrease in functional capacity. In addition, there is a progressive loss of muscle mass and strength in the aged, often termed “sarcopenia” (Nair, 2005), which is a major contributor to the frailty in the elderly and increases the risk of age-related diseases, injury and death. General wasting of all muscles occurs with fasting and disuse and is an integral feature of a number of systemic diseases, including many cancers, cardiac failure, renal failure, sepsis, AIDS, as well as burns and traumatic injury, and muscle loss correlates with a poor prognosis. In these conditions, the skeletal muscles are inherently normal, and the loss of mass is a response to physiological or pathological stimuli (e.g. fasting or cancer), rather than to any intrinsic genetic defect. By contrast, there are a variety of primary muscle diseases (termed myopathies) that result from inherent structural or enzymatic defects (e.g. muscular dystrophies) or from inflammatory diseases (e.g. myosites; (Askanas and Engel, 2002). Many mutations that perturb muscle function by affecting the contractile apparatus, energy metabolism or membrane integrity can lead to skeletal and cardiac myopathies.
Extensive studies in vertebrates have shown that skeletal muscle is a highly adaptive tissue that responds to exercise, nutrient supply, and endocrine factors with alterations in fiber composition and size (Figure 1). Similar observations have been made in Drosophila where muscles account for 40 to 50% of the body mass, and muscle growth depends on nutrient supply sensed via the Insulin/Akt/TOR pathway (as in mammals). For example, under normal nutrient conditions, Drosophila muscles during larval development undergo a striking ~50-fold increase in fiber size in only 5 days (Demontis and Perrimon, 2009; Figure 2A).
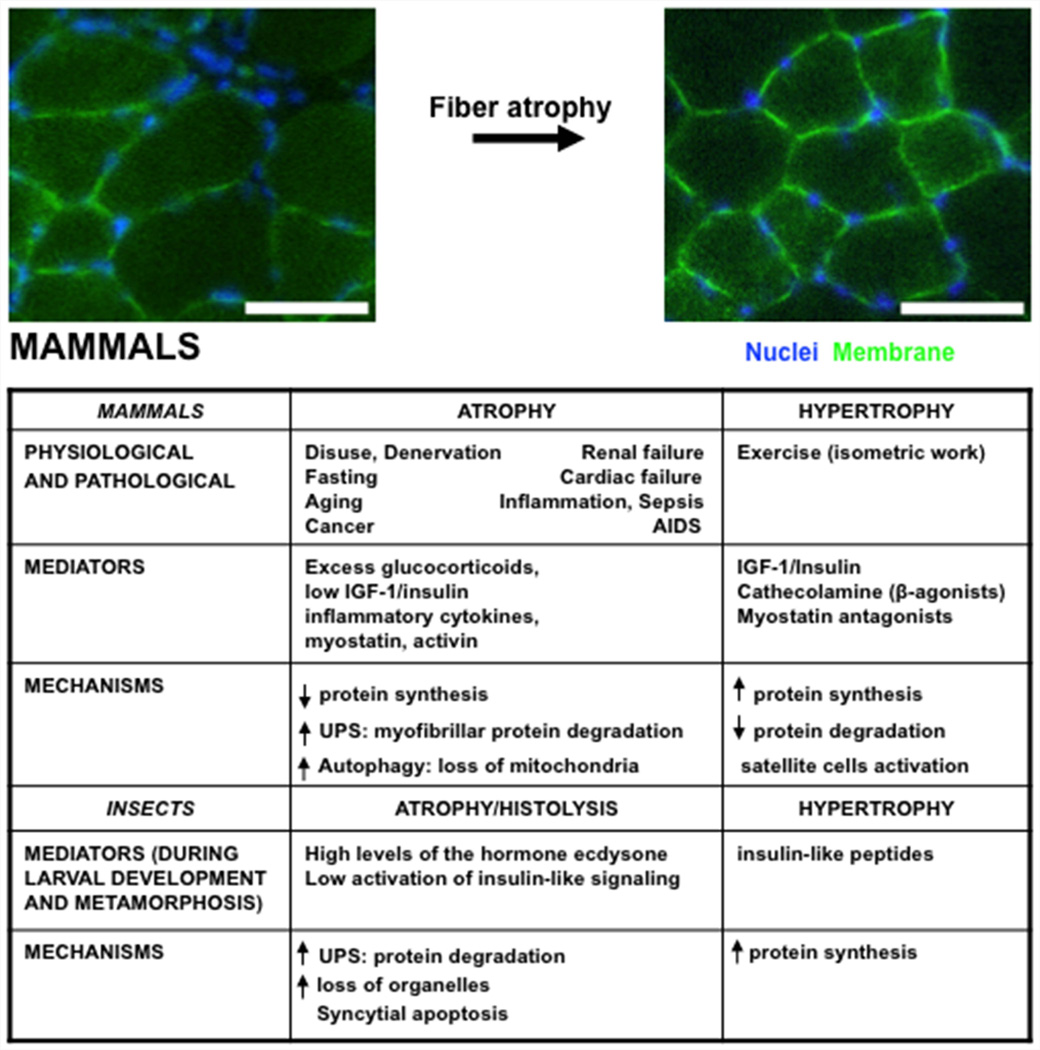
Figure 1.
Environmental stimuli and signaling pathways increasing proteolysis and depressing protein synthesis and leading to muscle atrophy. Representative fields of a transverse section of muscle fibers from fed mice or weight-matched mice deprived of food for 48h are shown. Nuclei and membranes are indicated in blue (Hoechst) and green, respectively. Scale bar is 50 µm.
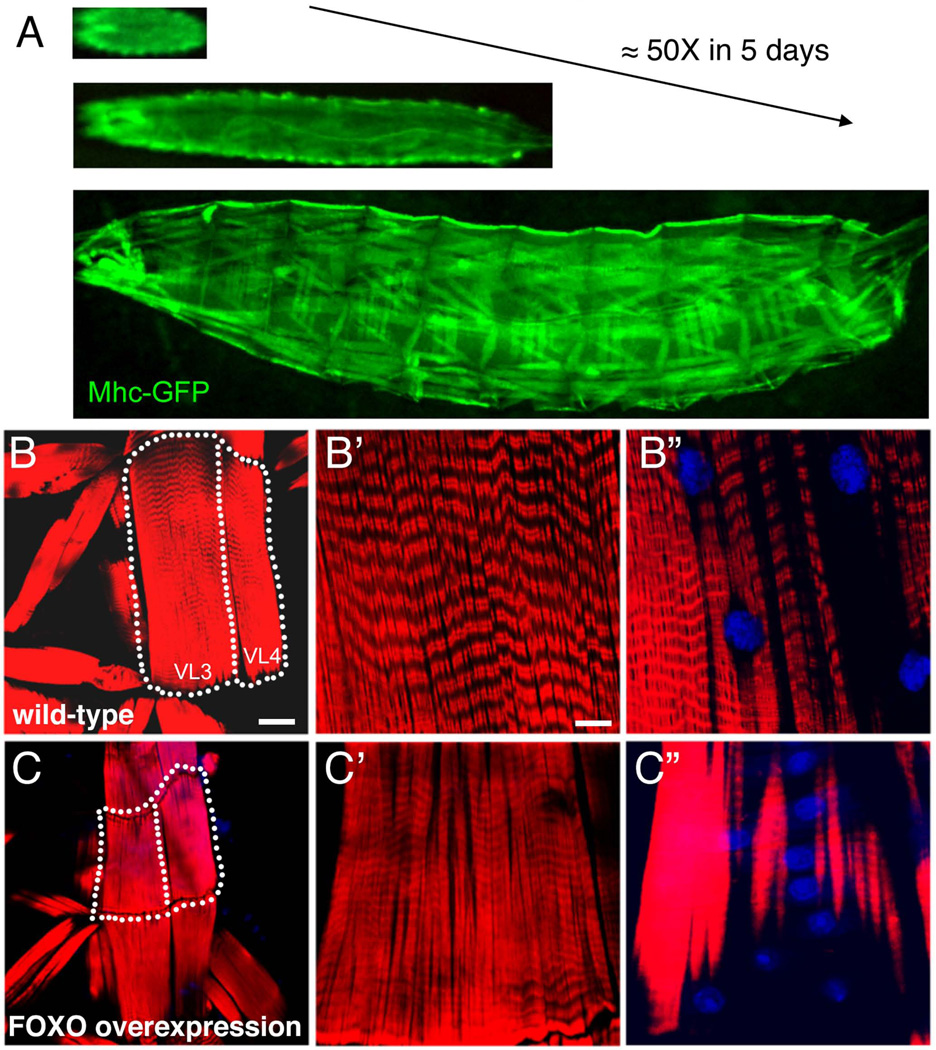
Figure 2.
Developmental growth of skeletal muscles in Drosophila larvae is inhibited by FOXO overexpression. A) Skeletal muscle size dramatically increases by 50-fold in the larval stage of development, which lasts 5 days. Muscle growth results from enhanced protein synthesis without any addition of muscle nuclei. Larvae expressing a GFP-tagged Myosin Heavy Chain (Mhc) protein in body wall muscles are shown. B) The size increase of body wall muscles is inhibited by overexpressing the transcription factor FOXO in muscles (C, Dmef2-Gal4 UAS-foxo versus Dmef2-Gal4 in B). Ventral Longitudinal muscles 3 and 4 (VL3 and VL4) from animals at the end of larval development are outlined for comparison in B and C. The dramatic increase in muscle mass observed during Drosophila larval development provides a sensitive setup for the identification of evolutionarily conserved genes regulating muscle mass. Micrographs in B’-B” and C’-C” outline sarcomeres and nuclei within muscle fibers (F-actin, red; Nuclei, blue). Scale bar is 40 µm in B-C and 10 µm in B’-B” and C’-C”. See Demontis and Perrimon, 2009, for more information.
Similarities between vertebrate and Drosophila muscles are both structural and functional. Both are composed of tandem arrays of sarcomeres containing the thin and thick filaments, which, in a typical muscle twitch, slide past each other in response to calcium release from the sarcoplasmic reticulum (SR, the specialized endoplasmic reticulum (ER) of muscles), resulting in force generation (Taylor, 2006). In addition, in both insects and mammals, muscle fibers can be either glycolytic or oxidative. For example, Drosophila direct and indirect flight muscles, which promote wing motion indirectly by compressing the thorax, can function for extended periods during flight and are primarily oxidative. By contrast, body wall muscles of the larva and leg muscles of adult flies, which are used only intermittently, rely mainly on glycolysis (Taylor, 2006). These distinct patterns of energy metabolism resemble the differences between type I and type IIb fibers in mammalian muscles. Type I slow fibers are non-fatiguing, primarily burn fatty acids and glucose oxidatively, and are dark in color because they are rich in mitochondria, myoglobin, and blood supply. By contrast, the easily fatigued, fast type IIb fibers are primarily glycolytic, and have a low mitochondrial content and capillarity density (Taylor, 2006). Most mammalian muscles, especially in humans, are composed of mixtures of fiber types that are recruited in an ordered fashion, but overall fiber composition is adapted to the specific functions of the muscle. For example, in rodents the antigravity soleus muscle, which is continually used in standing, is composed primarily of oxidative fibers and is quite resistant to fatigue. In typical mixed muscles, the slower oxidative fibers are used in all contractions, but the easily fatigued larger glycolytic fibers are recruited only with maximal efforts (Brooke and Kaiser, 1970)
Here in this review, we discuss how Drosophila with its extensive genetic toolkit and short life cycle provides a powerful experimental system to address some of the outstanding unsolved questions about muscle atrophy. Specifically, we review the mechanisms of skeletal muscle atrophy and hypertrophy that may be similar in Drosophila and mammals, and discuss emerging insights and outstanding questions that may benefit from studies in both species (see Tables I and II).
Table I.
A list of prominent yet unsolved questions for future research on muscle atrophy in Drosophila and mammals.
| OUTSTANDING QUESTIONS FOR FUTURE RESEARCH |
|---|
IN DROSOPHILA
|
IN MAMMALS
|
IN BOTH
|
Table II.
Similarities and differences in muscle atrophy and hypertrophy in insects and mammals. Future research will benefit from studies in both species because of their complementary advantages.
| COMPARING ADAPTIVE PROPERTIES OF MUSCLES IN MAMMALS AND DROSOPHILA | |
| SIMILARITIES | MAJOR DIFFERENCES |
|
|
| ADVANTAGES FOR STUDYING MUSCLE ATROPHY AND HYPERTROPHY | |
| IN DROSOPHILA | IN MAMMALS |
|
|
MODULATION OF MUSCLE MASS BY ENVIRONMENTAL STIMULI AND TRANSCRIPTION FACTORS
In mammals, muscles are the major protein reservoir in the body, and during fasting, amino acids generated by net protein degradation are released into the venous blood to provide substrates for hepatic gluconeogenesis. Thus, under starvation conditions, muscle proteolysis is critical for maintaining the supply of glucose, in particular to the brain, and of amino acids essential for continued protein synthesis. Myofibrillar proteins comprise about two thirds of muscle dry weight, and changes in muscle size are due primarily to changes in the content of the contractile apparatus (Cohen et al., 2009; Solomon and Goldberg, 1996). In most types of muscle atrophy, the loss of mass is driven by an increase in protein degradation and to a lesser extent by a decrease in protein synthesis (Figure 1). Protein degradation occurs via both the ubiquitin proteasome system (UPS) and the autophagy/lysosome pathway, while protein synthesis is regulated primarily by Insulin/IGF-1 (Insulin-like Growth Factor 1) acting through the Akt/TOR/FoxO pathway (Glass, 2010; Figure 3). During atrophy, both UPS and autophagy/lysosome degradative systems are activated, and key components are induced by transcription factors of the FoxO family (Sandri et al., 2004; Zhao et al., 2007) and NF-κB (Cai et al., 2004; Hunter et al., 2002).
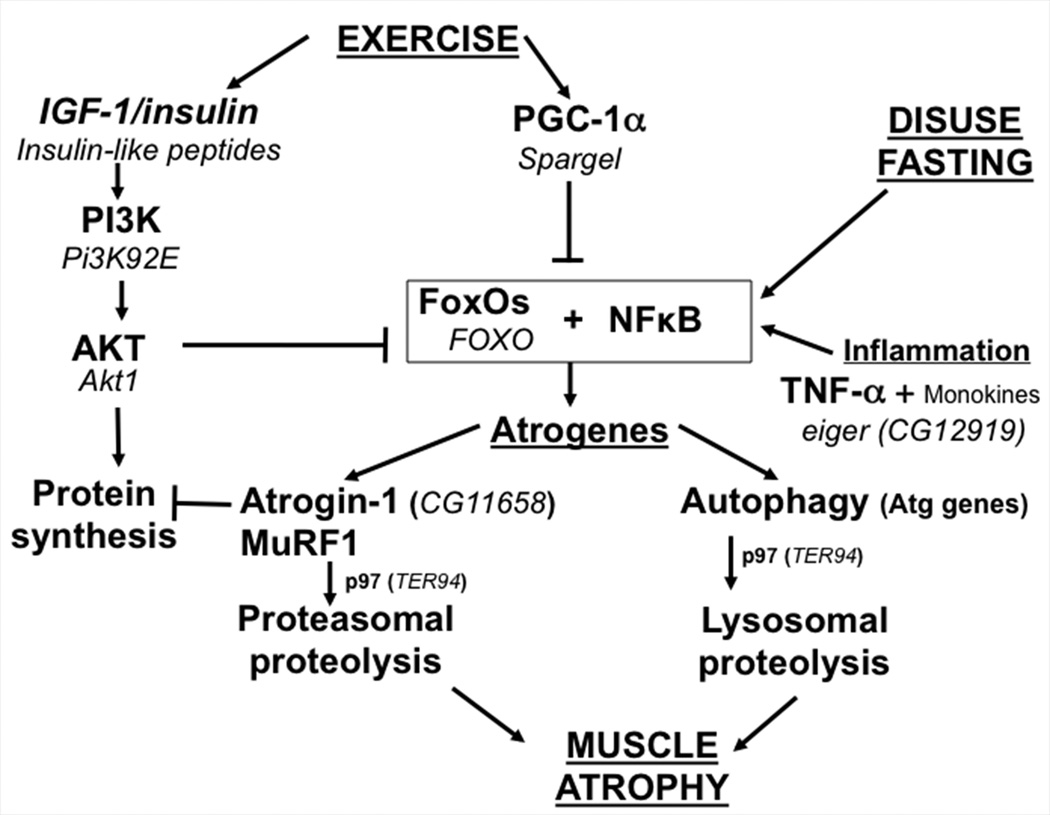
Figure 3.
Signaling pathways increasing proteolysis, depressing protein synthesis and leading to muscle atrophy. A comparison of the molecular players in Drosophila (indicated in Italics) and mammals is shown.
In mammals, systemic muscle wasting is induced in response to fasting or diseases (i.e. untreated diabetes, AIDS, cancer and heart failure) and by various circulating hormones, cytokines, and excess of glucocorticoids (Figure 1). In addition, atrophy of specific muscles occurs upon decreased usage (e.g. with nerve injury or immobilization in a cast). These very different physiological stimuli may involve some common signaling mechanisms. Contractile activity causes release of IGF-1 (Insulin-like Growth Factor 1), which functions as an important autocrine growth factor. IGF-1 is also a circulating hormone released from the liver in response to pituitary growth hormone. The autocrine production of IGF-1 by muscle falls with disuse and following loss of innervation (Zeman et al., 2009), in animals lacking pituitary growth hormone or during chronic heart failure (Schulze and Spate, 2005). So the loss of IGF-1, through both endocrine and autocrine mechanisms, contributes to the atrophy process mainly by attenuating signaling by the IGF-1/Akt pathway in both systemic catabolic states and locally with disuse. Reduced Akt signaling decreases protein synthesis in muscle as in other cells and leads to enhanced muscle proteolysis via activation of FoxO transcription factors (Glass, 2010; Figure 3). Similarly, upon fasting when circulating insulin and IGF-1 levels fall, autocrine production of IGF-1 by muscle also decreases. As a result, the IGF-1/Akt pathway is markedly depressed leading to rapid protein loss. Furthermore, in fasting and other catabolic states, the release of glucocorticoids by the adrenal glands increases and further promotes proteolysis and inhibits muscle protein synthesis.
In addition to FoxOs, other transcription factors (i.e. Smad 2 and 3, NF-κB, JunB and Myogenin) have been shown to influence muscle mass. For example, the Smad transcription factors mediate the catabolic effects of myostatin, a circulating TGF-family member that inhibits normal muscle growth (Lee, 2004). Myostatin binds to the Activin RIIB (ActRIIB) receptors and activates transcription by Smad 2 and 3. Conversely, inhibition of ActRIIB receptors or of Smad 2 and 3 causes muscle hypertrophy (Sartori et al., 2009; Figure 4). NF-κB has also been shown to be an important factor in muscle atrophy (Bonetto et al., 2011; Cai et al., 2004), although its exact mechanism appears unclear (Mourkioti et al., 2006). Endurance exercise induces the production of the transcriptional co-activator PGC-1α, which promotes mitochondrial gene expression leading to oxidative phosphorylation and subsequently differentiation of oxidative muscle fibers (Handschin and Spiegelman, 2008). In addition, JunB, which has long been known as a rapid-response gene that triggers cell proliferation, is also important in determining the size and growth rate of muscle fibers in adult mammals, which are postmitotic cells. JunB overproduction can both promote hypertrophy and inhibit muscle atrophy (Raffaello et al., 2010; Sandri et al., 2006; Wang et al., 2005) by inhibiting FoxO3, and perhaps other catabolic processes.
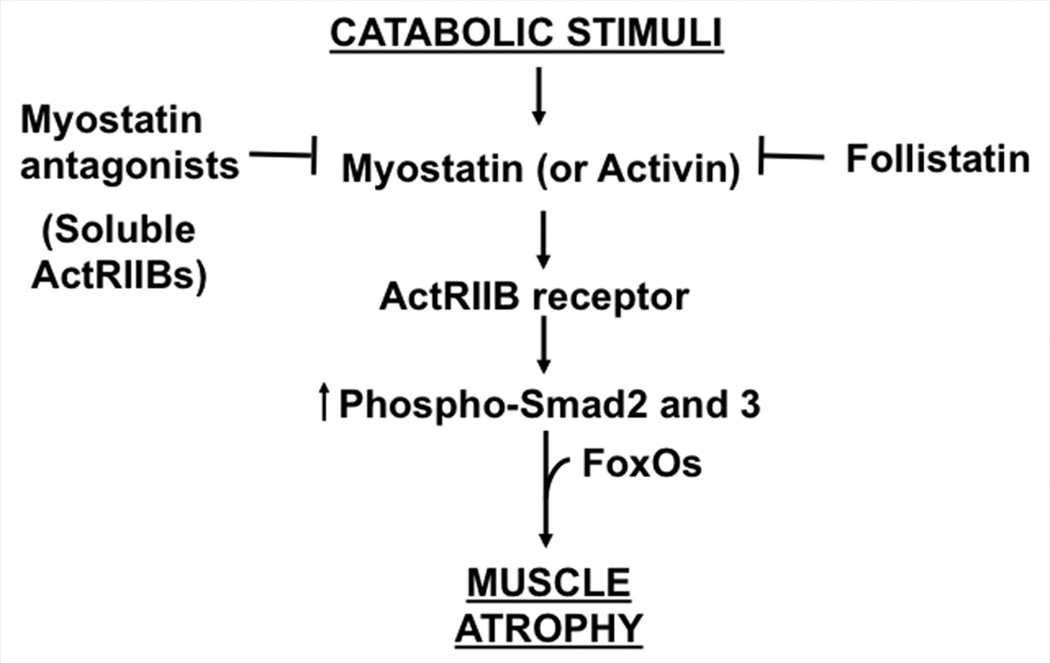
Figure 4.
Myostatin signaling pathway as a basis to counteract muscle wasting. Molecules to inhibit this pathway are currently in human clinical trials to cure the muscle wasting associated with Duchenne Muscular Dystrophy.
A number of other transcription factors (e.g. Myogenin, MyoD and MEF2) are important in embryonic differentiation of muscle, but are not critical in post-natal muscle growth. However, in the adult Myogenin participates in the induction of atrophy following denervation (Moresi et al., 2010). While multiple transcription factors can promote or inhibit muscle atrophy, their specific roles, modes of activation, and functional interactions occurring during different types of muscle atrophy remain unclear. This area represents an important gap in our knowledge and possibly could lead to the development of rational interventions to combat muscle wasting.
The powerful genetic toolkit available in Drosophila may be of particular advantage to analyze the genetic interactions among transcription factors inducing muscle atrophy or hypertrophy and to identify novel regulators of muscle mass. In Drosophila, muscles undergo dramatic size increase during larval development, where fiber growth also relies heavily on nutrient sensing via the Insulin/Akt/TOR pathway (Demontis and Perrimon, 2009). In the larval muscles, the activities of the transcription factors FOXO, Myc and Mnt are integrated to achieve normal muscle growth. In particular, FOXO inhibits muscle growth at least in part by decreasing Myc (diminutive) gene expression and its transcriptional activity (Demontis and Perrimon, 2009). Conversely, overexpression of the Insulin receptor results in increased Myc gene expression, which promotes nucleolar biogenesis and primes muscle growth. In addition, RNAi-mediated knock-down of Myc inhibits muscle growth, as does overexpression of Mnt (Demontis and Perrimon, 2009), which antagonizes Myc activity by binding to the common interaction partner Max (Bellosta and Gallant, 2010). Finally, genome-wide RNAi analyses, as recently applied to the study of muscle morphogenesis (Schnorrer et al., 2010), may uncover new regulators of fiber size in Drosophila. Orthologs of these genes may also be important in human biology and thus represent new therapeutic targets.
DENERVATION
Like nutrient deprivation, loss of muscle stimulation by nerves, as occurs during inactivity, immobilization, paralysis, nerve injury and neuromuscular diseases, such as amyotrophic lateral sclerosis (ALS), cause profound muscle atrophy in mammals (Figure 1). Different muscle fiber types have distinct propensities to atrophy upon denervation, with type I oxidative fibers being more sensitive to denervation-induced atrophy than type II (Herbison et al., 1979) even though type II fibers are lost preferentially in various systemic wasting states, e.g. glucocorticoids treatment (Goldberg and Goodman, 1969), fasting (Li and Goldberg, 1976), sepsis (Tiao et al., 1997) or cancer (Acharyya et al., 2004; Baracos et al., 1995). Importantly, activation of the Insulin/Akt pathway is sufficient to counteract the loss of muscle mass associated with denervation (Bodine et al., 2001b). Acutely, the weight loss and transcriptional response to denervation and pure disuse (e.g. with spinal isolation) are very similar, but with time, denervated muscles show a more profound atrophy (Sacheck et al., 2007). So although the post-natal maintenance of muscle mass is clearly dependent on continual neuronal activity, which causes contractile activity, it remains possible that trophic factors released by innervating motor neurons may also be important in determining muscle properties. Interestingly, some muscles, such as the extraocular muscles of primates, are extremely resistant to denervation atrophy when compared to limb muscles (Porter et al., 1989). Differences in fiber-type composition and innervation pattern may explain the resistance of extraocular muscles to wasting following denervation.
Whether denervation influences muscle mass in the adult fly, and whether it can be counteracted by Insulin/Akt signaling has not yet been studied, probably because of the difficulties in altering physiological load and measuring the sizes of specific muscles in this organism. However, muscle-nerve interactions have been thoroughly examined during the developmental phase of metamorphosis in Drosophila, when these interactions are important to define proper muscle size and patterning. In particular, surgical severing of the mesothoracic nerve with microbeam lasers leads to denervation that can retard the formation of some muscle fibers, like the dorso-longitudinal flight muscles, by regulating the rate of proliferation of the pool of myoblasts, the muscle stem cells that are precursors of the adult musculature (Fernandes and Keshishian, 1998). Similarly, nerve-muscle interactions are necessary for achieving proper muscle size in the moth Manduca sexta, where denervation during development also decreases the number of proliferating myoblasts (Bayline et al., 2001). In addition, denervation results in muscle patterning defects in Drosophila, with a failure to form dorsal ventral muscles, indicating that the differentiation of specific muscle fibers is perturbed (Fernandes and Keshishian, 1998). The interconnection between neuronal activity and muscle patterning has also been extensively studied in adult mammals, where motoneurons influence fiber type differentiation (Grinnell, 1995) even in the adult. Moreover, prolonged electrical stimulation of type II fibers causes the muscle to acquire many characteristics of type I fibers (Goldspink, 1985). Despite these observations, the transcriptional mechanisms underlying muscle-nerve interactions remain to be clarified. Knowledge gained from studies in Drosophila should help define the nature of muscle-nerve interactions, which are presumably related to the mechanisms functioning in mammalian adult differentiated muscles.
GLUCOCORTICOIDS, INFLAMMATION, AND OXIDATIVE STRESS
Among the major stimuli that can trigger muscle atrophy in mammals are the glucocorticoids (e.g. cortisol) or its synthetic analogs (e.g. dexamethasone), which at high pharmacological doses cause atrophy preferentially of type IIb glycolytic fibers (Dahlmann et al., 1986; Goldberg and Goodman, 1969). These steroids are released from the adrenal gland in stressful states and are essential for muscle wasting during fasting, renal failure, diabetes and sepsis (Menconi et al., 2007; Schakman et al., 2008). Glucocorticoids act to reduce muscle size by inhibiting amino acid transport into muscle, protein synthesis, and enhancing proteolysis. Consequently, they promote the release from muscles of amino acids that can be burned directly or serve as substrates in the liver for glucose production. Although not normally catabolic, normal or slightly increased levels of glucocorticoids can induce muscle wasting when there is a fall in Insulin/IGF-1 signaling, as occurs during fasting, diabetes and insulin-resistant states (most diseases; Hu et al., 2009). Because of their ability to also reduce inflammation, proliferation of white cells and immune responses, glucocorticoids are widely used to treat rheumatoid arthritis, allergic reactions, asthma, transplant rejection, lupus, and some forms of cancer. The induction of muscle (and bone) wasting is a serious adverse effect of high doses of these drugs and often limits their use in the clinic. Interestingly, lack of contractile activity enhances the tendency of muscles to atrophy in response to glucocorticoids (Goldberg and Goodman, 1969). For example, although the dark oxidative soleus is relatively resistant to these agents, the denervated soleus is highly susceptible to cortisone-induced atrophy, which helps explain the marked loss of body mass in the bed-ridden patients. Thus, an outstanding question is how the sensitivity of different muscles to glucocorticoids or other catabolic stimuli is modulated at the molecular level by contractile work or other anabolic stimuli.
Although no hormones are known that function similarly to glucocorticoids upon stress in insects, a class of steroid hormones called ecdysteroids (including ecdysone) has been implicated in the complete breakdown and death of muscle cells (also termed histolysis) during pupal metamorphosis. Throughout this process, most larval muscles degenerate completely in response to ecdysone and presumably provide amino acids for the development of the adult tissues. Although glucocorticoids do not cause death of muscles in mammals, the rapid loss of muscle mass and breakdown of myofibrils in response to structurally related molecules (glucocorticoids and ecdysteroids) suggests some similar mechanisms of action. Accordingly, larval muscles undergo marked atrophy before apoptosis is evident in response to ecdysone (Bayline et al., 1998; Hegstrom and Truman, 1996). However, not all insect muscles undergo histolysis in response to ecdysteroids. An example of such diverse responses to ecdysteroids comes from pioneering studies in the moth Manduca sexta. During pupal metamorphosis, the larval muscles are either maintained, modified, or degenerate (Bayline et al., 1998; Hegstrom and Truman, 1996). In particular, the large intersegmental muscles (ISMs) from Manduca initially atrophy and loose approximately 40% of their mass starting three days before adult eclosion, while at the same time, the flight muscles of the adult musculature are actively growing in mass. Subsequently, ISMs undergo cell death and are completely lost within 30 hours of adult life, while newly-formed flight muscles do not (Schwartz, 1992).
Studies in Manduca have benefited from the large sizes of these muscles, but the genetic analysis of this process is easier in Drosophila, where different muscles also respond in distinct fashion to ecdysone. By 8 hours after puparium formation, most muscles in the head and thoracic segments undergo histolysis while the abdominal muscles are preserved (Fernandes et al., 1991). Among the thoracic muscles, the larval oblique muscles escape histolysis and serve as template for the formation of the dorsal longitudinal muscles (DLMs), a group of indirect flight muscles (IFMs) of the adult (Farrell et al., 1996; Roy and VijayRaghavan, 1998; Dutta et al., 2004). Subsequently, most larval muscles in the abdomen undergo histolysis. For example, the dorsal external oblique muscles (DEOMs) degenerate in the late prepupal stage (Wasser et al., 2007). However, some other abdominal muscles initially persist, undergo atrophy (day 1 and 2 of pupal stage), and subsequently increase in mass (hypertrophy; late pupal stage) to form the temporary dorsal internal oblique muscles (DIOMs) of the adult. DIOMs are needed for eclosion and degenerate only after the adult has emerged (Kimura and Truman, 1990). Interestingly, the nuclear proteins EAST and Chromator have antagonistic functions and respectively accelerate and delay the atrophy of DIOMs during pupal metamorphosis. In addition, Chromator partially blocks the breakdown of DEOMs muscles (Wasser et al., 2007).
The mechanisms by which distinct insect muscles respond differently to ecdysone are still not understood, and thus this may represent a valuable model to better understand the specialized responses of distinct muscles to catabolic stimuli. Algorithms recently became available to facilitate the analysis of images from high-throughput RNAi screens for muscle histolysis (Chinta et al., 2012). In mammals, there are several sexually dimorphic muscles (e.g. the levator anus that is also involved in the control of the penis) that show much greater sensitivity to the anabolic actions of testosterone than typical muscles (Herbst and Bhasin, 2004). The precise basis for their greater sensitivity to these steroids is largely unknown.
In addition to glucocorticoids, pro-inflammatory cytokines including TNF-α (Tumor Necrosis Factor α), IL-1 (Interleukin 1), IL-6, activin, and myostatin have all been reported to contribute to muscle wasting (cachexia) in mammals during sepsis and in cancer-bearing animals at least in part by activating the NF-κB, STAT3 and/or the Smad transcription factors (Bonetto et al., 2011). Some of these cytokines and the pathways they activate are conserved in Drosophila. For example, Drosophila eiger (CG12919) is a TNF superfamily ligand that activates a canonical TNF signaling cascade (Moreno et al., 2002). In addition, the JAK/STAT pathway, which is activated in mammals by various members of the Interleukin family, is conserved in Drosophila where it is activated by the Unpaired family of cytokine-like ligands (outstretched, unpaired 2, and unpaired 3). Studies on the effects of these signaling pathways on Drosophila muscle growth and remodeling may provide useful models of inflammation-induced atrophy.
Another type of circulating factor that can increase muscle size or counteract atrophy are cathecolamines and their synthetic analogs, clenbuterol or β2 agonists, which activate β2 adrenergic receptors (whose Drosophila homolog is octopamine receptor 2). These agents cause production of cyclic-AMP and activation of PKA that can ultimately cause cardiac and skeletal muscle hypertrophy (Navegantes et al., 2001). In fact, clenbuterol has often been used illegally to induce growth of cattle and enhance food production but, because residues of cathecolamines are retained in the meat, this practice is dangerous and is banned. Also, because of the side effects of cathecolamines (e.g. blood pressure and cardiac function), their ability to inhibit atrophy has not been exploited as a therapy for muscle wasting.
Increased production of reactive oxygen species (ROS) has been proposed to play a role in the muscle degeneration of the queen fire ant, Solenopsis spp., where histolysis follows the mating flight and insemination (Davis et al., 1993). Similarly, in mammals there are multiple observations suggesting a possible role of oxidative stress in triggering muscle atrophy. First, the transcription factor ATF4, which promotes the expression of oxidative stress responsive genes, is upregulated in most types of atrophy (Lecker et al., 2004) as are the metallothioneins, heavy metal-binding components that can serve antioxidant roles (Sacheck et al., 2007). In addition, during disuse, ROS have been proposed to activate several transcription factors involved in muscle atrophy in mammals, including NF-κB and FoxO (Dodd et al., 2010), but other modes of activation have also been demonstrated. Genetic manipulations that enhance muscle ROS levels, such as knock-out of the cytoplasmic anti-oxidant enzyme Sod1 (superoxide dismutase 1) in mice leads to oxidative damage to proteins and muscle wasting (Jang and Van Remmen, 2011; Muller et al., 2006). On the other hand, mice devoid of the mitochondrial superoxide dismutase 2 in muscle have elevated oxidative stress, but do not exhibit muscle wasting (Kuwahara et al., 2010; Lustgarten et al., 2009). Thus, oxidative stress does not appear to be sufficient to induce atrophy, and it remains unclear if these signs of oxidative stress are a key factor in the atrophy process or an incidental factor.
PHYSICAL ACTIVITY AND INACTIVITY
Muscle wasting in adult mammals is generally viewed as a reversible process, but this reversibility has not been rigorously studied, e.g. at different ages. The loss of muscle on fasting is rapidly reversed by refeeding and can even lead to “overshoot” growth. Similarly, the loss of body mass with acute illness is rapidly reversed upon recovery, especially in children where it has been termed “catch-up” growth. In the elderly, this capacity to reverse wasting seems to be more limited but this impression and its possible cellular basis have not been systematically studied. The extent of muscle wasting induced by food deprivation in mammals can be counteracted by physical activity, especially isometric exercise which stimulates the IGF-1/Akt pathway and builds muscle mass and strength. In fact, in starving rats where there is rapid muscle wasting, generally increased activity of the soleus (e.g. induced by loss of a synergist) can induce hypertrophy (Goldberg, 1968). Both strength and endurance exercise increase the activity of the transcriptional co-activator PGC-1α (Gibala et al., 2009; Terada et al., 2002), which stimulates in turn mitochondrial biogenesis and oxidative metabolism characteristic of type I fibers but not hypertrophy (Handschin and Spiegelman, 2008). Type I aerobic fibers are relatively resistant to atrophy induced by various circulating catabolic factors, including fasting, glucocorticoids, cancer and sepsis, apparently due to the presence of PGC-1α. Its expression is decreased in many, perhaps all, types of atrophy (Sacheck et al., 2007; Sandri et al., 2006), and PGC-1α and its homolog PGC-1β, directly antagonize the induction of the atrophy gene program by both FoxO3 (Sandri et al., 2006) and NF-κB, and thus suppress the acceleration of protein degradation (Brault et al., 2010; Figure 3). Presumably, these effects help account for the ability of exercise to retard atrophy. Following disuse and upon restoration of activity, the increased PGC-1α levels and IGF-1/PI3K-Akt signaling inactivate FoxO leading to decreased expression of atrogenes. These genes are induced or suppressed coordinately during various types of atrophy due to uremia, diabetes, cancer, denervation, disuse and fasting and include components of both the UPS and autophagy/lysosome pathways (Zhao et al., 2007). Among these atrogenes, two muscle-specific ubiquitin ligases, MuRF1 and atrogin-1, are dramatically induced by FoxO3 (Sandri et al., 2004) and are essential for muscle atrophy (Bodine et al., 2001a; Gomes et al., 2001). In addition to a role for PGC-1α in repressing FoxO activity, PGC-1α4 (a PGC-1α splice variant preferentially produced after resistance exercise) protects muscles from atrophy by inducing IGF-1 while repressing myostatin expression (Ruas et al., 2012).
The mechanisms linking contractile activity to the action of these transcription factors in atrophy is still not clear even though this area is critical and determines whether a cell grows or atrophies. One well-studied exception is NF-AT, a key transcription factor that is activated by increases in Ca2+ levels. In skeletal muscle, it is involved in fiber type specification and influences PGC-1α expression (McCullagh et al., 2004) and in cardiac muscle it can trigger hypertrophy (van Rooij et al., 2002). An additional potential link between physical activity and protein turnover is the dihydropyridine receptor (DHPR) α1S subunit, which can serve as a molecular sensor of muscle activity (Pietri-Rouxel et al., 2010). DHPR α1S knock-down induces muscle atrophy via FoxO3A activation, which leads to upregulation of autophagy-related genes and the induction of autophagy. However, the loss of DHPR presumably represents a form of disuse since it should reduce contractile activity (i.e. disrupt excitation-contraction coupling).
Thus far, studies on the effects of muscular contractions have been lacking in insects. Although genetic studies in Drosophila in principle might help elucidate the interconnections between exercise and muscle mass, experimental approaches for exercising this organism and methods for measuring changes in muscle mass or metabolic adaptations have not been described, in contrast to the extensive literature on mammals. Recently, a mechanized platform for the physical training of flies has been described (Piazza et al., 2009), in which a repeated tapping of the container where the flies are housed activates the innate instinct of the flies to climb (negative geotaxis). This promising system may be useful to dissect the genetic basis underlying the effect of physical exercise on muscles. For example, an interaction between physical exercise and spargel, the PGC-1α homolog that promotes mitochondrial production in Drosophila (Tiefenbock et al., 2010), has been found with this system, highlighting that spargel is required for full exercise capacity in Drosophila and the induction of physiological effects deriving from exercise (Tinkerhess et al., 2012).
While endurance exercise clearly decreases the loss of muscle mass in mice and in humans, no morphological signs of muscle deterioration have been observed in flightless Drosophila mutants or in response to transient local paralysis of adult flies induced with the temperature-sensitive dynamin mutant shibire (which are depleted of synaptic vesicles; Deak, 1976). Such studies did not identify decreases in muscle mass of the kinds seen with disuse in small mammals where fiber diameters decrease by 10–40% within 1–2 weeks without any clear structural deterioration. This apparent lack of clear responses to disuse is important to define more rigorously since Drosophila muscles may differ radically from mammals and lack the capability to alter muscle mass in response to changes in contractile activity. While such an apparent difference may be due to insufficient studies, it is also possible that because of their brief lifespan and very different lifestyle, Drosophila may not have evolved similar regulatory mechanisms as mammals for mobilizing amino acids from muscle proteins. So disuse may not result in obvious muscle atrophy in flies.
ROLE OF PROTEIN SYNTHESIS AND DEGRADATION PATHWAYS IN MUSCLE ATROPHY
Despite the greater diversity of possible regulatory factors in mammals (ranging from hormones to cytokines), the final cellular mechanisms regulating muscle size appear similar in Drosophila and mammals (Figures 1, 2 and 3). In Drosophila, the Insulin/Akt-responsive transcription factor FOXO mediates most of the gene expression changes induced by nutrient starvation in larval muscles (Teleman et al., 2008) and its activation is sufficient to stunt developmental muscle growth (Demontis and Perrimon, 2009; Figure 2B and C). Similarly, in mammals, various types of muscle atrophy share a common transcriptional program mainly driven by FoxO3 (Lecker et al., 2004; Sandri et al., 2004) and possibly other FoxO family members, like FoxO1. MuRF1 and atrogin-1 are key atrogenes induced by FoxO3 (Sandri et al., 2004) that are necessary for rapid muscle atrophy (Bodine et al., 2001a; Gomes et al., 2001). While proteins comprising the thick filaments of the myofibrils have been clearly identified as MuRF1 substrates (Cohen et al., 2009), how atrogin-1 causes muscle loss is less clear, but probably involves the degradation of growth-related proteins including the transcription factor MyoD, and the translation initiation factor eIF3-f, which in turn reduces protein synthesis (Lagirand-Cantaloube et al., 2008; Figure 3). Additional E3s are also important in muscle size control. These include TRAF6, responsible for activating many atrophy-related signaling cascades (Paul et al., 2010), TRIM32, involved in the degradation of thin filaments and the desmin cytoskeleton (Cohen et al., 2012; Kudryashova et al., 2005) and Cbl-b, which inhibits IGF-1 signaling by promoting the degradation of the Insulin Receptor Substrate IRS-1 (Nakao et al., 2009). Homologs of atrogin-1 (Drosophila CG11658), TRIM32 (Drosophila abba), TRAF6 (Drosophila CG10961) and Cbl-b (Drosophila Cbl) are encoded in the Drosophila genome, suggesting possible similarities in the pathways governing proteolysis and muscle atrophy in fruit flies and mammals. For example, CG11658 expression levels increase in muscle wasted (mute) mutants, lacking a component of the histone locus body and characterized by a severe loss of muscle mass and integrity during development (Bulchand et al., 2010). Moreover, Drosophila TRIM32 (abba) is required for integrity of the costamere, a structure that connects the sarcomeres to the overlying sarcolemma providing stability during muscle contraction, and its mutation results in unbundling of myofibrils and progressive muscle wasting (LaBeau-DiMenna et al., 2012).
However, no homologs of MuRF1, a E3 ligase that ubiquitinates myofibrillar proteins, are found in flies. Very recently, the p97/VCP ATPase complex, which forms distinct complexes involving different adaptors (e.g. p47 and Ufd-1) and distinct components of the thick (i.e. Myosin Light chains) and thin (i.e. actin) filaments, has been shown to have a major role in multiple types of atrophy, where it seems to catalyze the extraction of ubiquitinated proteins from the myofibrils prior to proteasomal degradation (Piccirillo and Goldberg, 2012). Thus, Drosophila homologs of p97 (Drosophila TER94) and Ufd-1 (Drosophila CG6233) could participate to a conserved mechanism for muscle protein degradation, although p97 also functions in other disassociation and degradative processes (e.g. destruction of misfolded proteins in the ER). Interestingly, human mutations in p97 cause an inclusion body myopathy and all amino acid residues altered in the disease are perfectly conserved in Drosophila TER94 (Ritson et al., 2010). It will clearly be of interest to define the biochemical mechanisms for disassembly and degradation of the sarcomeric apparatus during pupal metamorphosis and if they involve the p97 complex.
Studies in Manduca have highlighted the involvement of the UPS in ecdysone-induced muscle histolysis. In particular, levels of ubiquitin conjugates dramatically increase at eclosion, during which loss of muscle proteins is maximal. Histolysis is also characterized by the coordinated induction of ubiquitin activating enzymes (E1), several ubiquitin conjugating enzymes (E2s), ubiquitin ligases (E3s), and ubiquitin itself (Haas et al., 1995; Schwartz et al., 1990) together with heightened expression in the abdominal intersegmental muscles (ISMs) of MS73/Rpt3 and S10b/Rpt4, which encode ATPase subunits of the 26S proteasome (Dawson et al., 1995; Hastings et al., 1999; Jones et al., 1995; Low et al., 1997). Thus, activation of the UPS seems to play a fundamental role in the loss of skeletal muscle mass during insect metamorphosis similar to that in atrophy in mammals. These insect muscles shrink in size markedly (atrophy) before the onset of apoptosis, presumably due to increased proteolysis. It should be noted that the UPS is also required to maintain muscle mass and integrity during larval development (Haas et al., 2007) and for viability in mammals, due to its many critical roles in cellular quality control and regulating metabolism. In other words, different degrees of protein breakdown have beneficial or detrimental effects on muscle mass, depending on what proteins are digested.
In addition to the UPS, autophagy contributes to the histolysis of several tissues during metamorphosis in Drosophila, suggesting that it may be part of the catabolic response activated in muscles in response to ecdysone signaling (Baehrecke, 2003).
Many atrogenes induced by FoxO3 encode for proteins of the autophagy/lysosome system (e.g. Cathepsin L, LC3, GabarapL1; Lecker et al., 2004; Zhao et al., 2007), which contributes to muscle atrophy in mammals especially through the disposal of mitochondria (Romanello et al., 2010), as well as soluble cell proteins. Importantly, Mul1 is an E3 ubiquitin ligase recently implicated in FoxO induced-mitophagy (Lokireddy et al., 2012), the autophagic destruction of mitochondria, while the ubiquitin ligase Parkin is needed for mitophagy of damaged mitochondria (Yang and Yang, 2011). Both Mul1 and Parkin are conserved in Drosophila (CG1134 and parkin, respectively) and may play a role in muscle wasting in this organism.
The loss of mitochondria must contribute importantly to the decreased endurance during atrophy. Nevertheless, the changes in mitochondrial numbers and functional capacity during different types of atrophy have surprisingly not been studied extensively. Because ubiquitination can also target larger structures to autophagic vacuoles, several ubiquitin-binding proteins (including p97, p62, NBR1, NDP52 and parkin) exist in cells that bind insoluble or organellar ubiquitinated proteins and facilitate their docking to the autophagic vacuole via LC3. Their functions in muscle merit further study to better understand the coordination between different protein degradation pathways during atrophy and their roles in clearing different cellular constituents (reviewed in Korolchuk et al., 2010), especially since certain components (p97 cofactors) are essential in proteolysis by both degradative systems.
In addition to the activation of protein degradation pathways, muscle wasting upon fasting, glucocorticoid treatment and other disease states result from an inhibition of protein synthesis that is triggered by decreased IGF-1/Akt/TOR signaling and inhibited by glucocorticoids (Hu et al., 2009). Interestingly, ecdysteroids inhibit Insulin signaling in Drosophila (Colombani et al., 2005), suggesting that muscle histolysis during metamorphosis may also rely on inhibition of Akt signaling and the activation of FOXO. However, mutant flies lacking the only Drosophila foxo gene are viable and have only minor developmental defects (Junger et al., 2003), suggesting that FOXO is not necessary for muscle histolysis during metamorphosis. By contrast, mammalian muscle contains three closely related FoxO genes and their respective roles are still uncertain. FoxO1, 3 and 4 are coordinately activated upon nutrient deprivation, but may have distinct roles in specific catabolic states and may regulate distinct genes (Moylan et al., 2008), although FoxO1 and 3 both induce atrogin-1, MuRF1 and Mul1. FoxO3a and FoxO4 knock-out mice (Hosaka et al., 2004) are viable and analysis of FoxOs-deficient mice (Paik et al., 2007) may clarify whether specific FoxO transcription factors are necessary for the induction of atrophy in response to different stimuli. Thus far, it is only known that overexpression of FoxO-dominant negative mutants, which block all FoxOs in adult muscles, can prevent multiple types of atrophy (Sandri et al., 2004). It would be interesting to test whether FOXO is able to alter the expression of the Drosophila homolog of atrogin-1 as in mammals.
SYNCYTIAL APOPTOSIS
Additional mechanisms responsible for modifying muscle mass during development and adulthood include the poorly defined pathway of syncytial apoptosis. In this process, loss of individual nuclei within a fiber is more commonly observed than the death of the entire fiber, possibly via caspase-independent mechanisms acting via endonuclease-G (Primeau et al., 2002; Sandri and Carraro, 1999) or nucleophagy, the selective autophagic degradation of nuclear components (Park et al., 2009). In addition, segmental necrosis, the death of a portion of a fiber without the overall loss of the fiber, is a characteristic feature of many myopathies, especially Duchenne muscular dystrophy (Wallace and McNally, 2009).
The genetic regulation of syncytial apoptosis, and whether it differs from segmental necrosis is unclear and may benefit from genetic studies to elucidate the underlying regulatory pathways. In insects, syncytial apoptosis occurs during metamorphosis, when some nuclei within the same fiber degenerate and are lost, while those in proximity to the site of innervation are spared (Bayline et al., 1998; Hegstrom and Truman, 1996). Therefore, in insects the local induction of syncytial apoptosis may vary even within a single fiber depending on the proximity to the site of innervation.
While apoptosis occurs upon muscle histolysis during insect metamorphosis, fiber cell death has not been observed upon inhibition of larval muscle growth in Drosophila (Demontis and Perrimon, 2009) and during rapid atrophy in mammals (Bruusgaard and Gundersen, 2008). In animal models of uremia, fasting, diabetes and cancer, the pro-apoptotic gene Bnip3 is induced but it also functions in mitophagy (Mammucari et al., 2007; Lecker et al., 2004). However, segmental apoptosis of fibers has been reported during aging in mammals and Drosophila (Marzetti et al., 2012; Zheng et al. 2005). Future studies in Drosophila and mammals should elucidate the pathways governing muscle syncytial apoptosis, and how this poorly understood process impacts on the loss of muscle mass and strength.
Importantly, other pathways of programmed cell death including necroptosis, which depends on the activity of the serine/threonine kinase RIP1, have not been examined in muscles. Notably, TNF-α, which helps trigger muscle wasting in inflammatory states (Glass, 2010), although perhaps indirectly, is able to induce necroptosis in other cell types (Galluzzi and Kroemer, 2008). The possible relevance of necroptosis during muscle wasting awaits further investigation.
ROLE OF MUSCLE STEM CELLS
In vertebrates, satellite cells are the small mononucleated muscle stem cells localized between the sarcolemma and the basal lamina of muscle fibers. In the adult, most stem cells are in a quiescent state, but can become dramatically activated upon injury or specific stimuli like IGF-1, which promotes their proliferation and differentiation during hypertrophy (Biressi and Rando, 2010). For example, after damage, satellite cells fuse to one another or to an undamaged fiber, promoting muscle regeneration, through a process that partially recapitulates myoblast fusion during development. Interestingly, apoptosis of satellite cells but not pre-existing fibers has been observed during rapid atrophy (Bruusgaard and Gundersen, 2008). Unfortunately, since satellite cells are heterogeneous, few in number (less than 2% of the nuclear content of muscle), and much less susceptible to in vivo electroporation than muscle fibers, their specific role in atrophy is still largely unknown. Although satellite cells are not needed for muscle re-growth following disuse-induced atrophy (Jackson et al., 2012), further studies are necessary to test whether satellite cells can possibly reverse or prevent muscle atrophy in other contexts, as has been recently suggested (Thornell, 2011).
Important roles in hypertrophy have often been postulated, especially in providing additional nuclei to prevent a fall in concentration of nuclei when fibers enlarge. However, it remains controversial whether stem cell proliferation is essential during hypertrophy as it clearly is upon regeneration (Zammit et al., 2002). New techniques have recently become available to dissect their possible contributions to muscle adaptations. These include the ex vivo transfection of muscle stem cells followed by intramuscular injection and the in vivo administration of adenoviral vectors where transgene expression is driven under muscle stem cell specific promoters of the Pax3 and Pax7 genes (Biressi and Rando, 2010).
Currently, there is no evidence of satellite cells in any Drosophila adult muscles, and it is not clear whether they do not exist or simply have not yet been identified. If present they may be related to the twist-expressing adult muscle precursors (AMPs), muscle progenitor cells that escape histolysis and form all adult muscles in Drosophila (Figeac et al., 2007). Interestingly, even if the Drosophila homologs of the mammalian satellite cells markers Pax3 and Pax7, paired and gooseberry, respectively, are not expressed in AMPs, these muscle-committed stem-like cells, like quiescent satellite cells, have high levels of Notch activation (Figeac et al., 2011). This finding suggests that studies on twist-expressing muscle progenitors in Drosophila may provide useful information on the regulation of satellite cell function in mammals.
EFFECTS OF MUSCLE MASS ON BODY METABOLISM IN DROSOPHILA AND MAMMALS
It is now clear that the skeletal muscle has major effects on the metabolism of the organism, its growth, aging, and resistance to disease (Demontis and Perrimon, 2010; Demontis et al., 2013). These organismal effects involve muscle-derived signaling factors but also arise from indirect effects of muscle mass and its high metabolic demand. Skeletal muscle consumes a major fraction of nutrients, and with intense exercise, its energetic demand increases dramatically. Consequently, multiple physiological mechanisms exist to provide glucose or fatty acids to muscle. For example, when muscle growth was induced in adult mice by increasing the expression of the Akt1 kinase in type IIb fibers, the transgenic mice displayed resistance to both diet-induced obesity and hepatic steatosis, at least in part via a stimulation of fatty acid oxidation in the liver (Izumiya et al., 2008). In addition, postnatal loss of the transcription factor myogenin in muscles results in smaller body size of mice, suggesting that Myogenin acts to influence the post-natal growth of muscles and also other tissues via un unknown mechanism (Knapp et al., 2006). As in mammals, in developing Drosophila larvae the extent of muscle growth influences whole-organism growth and metabolism. In particular, muscle-specific inhibition of the Insulin receptor/Akt/TOR pathway and of Myc activity decreases not only muscle growth, but also feeding behavior and the growth of non-muscle tissues, while muscle-specific activation of the Insulin receptor has the opposite effects (Demontis and Perrimon, 2009).
A remarkable and very promising intervention to increase muscle mass and reduce atrophy in mammals is through inhibition of the TGF-β family members, like Myostatin and Activin, all of which signal via the ActRIIB complex (Figure 4). Activin is produced normally in multiple cell types and also in cancerous cells and plays an important role in reproduction (Xia and Schneyer, 2009), while Myostatin is expressed predominantly in skeletal muscles (Zhou et al., 2010; Zimmers et al., 2002). Myostatin normally acts to limit prenatal and postnatal muscle growth and mutations in the Myostatin gene or its receptor result in increased muscle mass in mice, cattle, dogs and humans (Lee, 2004). Developmental inhibition of Myostatin results in both muscle hyperplasia and hypertrophy, while postnatal inhibition increases muscle mass via hypertrophy, with no accompanying hyperplasia in several species (McPherron 2010). Suppression of the Myostatin-based pathway (as in Figure 4) has been also proven effective in counteracting muscle wasting in mice with chronic kidney disease (Zhou et al., 2010), following treatment with glucocorticoids (Gilson et al., 2007), as well as in cancer cachexia (Benny Klimek et al., 2010; Zhou et al., 2010). This effect on muscle size resulted at least in part from the inhibition of the FOXO-induced activation of atrogenes and ubiquitin-dependent proteolysis and perhaps also through the subsequent stimulation of muscle stem cell proliferation. Remarkably, inhibition of Activin and Myostatin signaling via blockade of the ActRIIB receptors not only dramatically reverses cancer-induced weight loss in certain cancer models, but also improves the survival of mice without slowing tumor growth (Zhou et al., 2010). Thus, improving muscle mass aside from enhancing the quality of life can strikingly influence cancer-related mortality, at least in mice.
PERSPECTIVES
In this review, we have highlighted a number of important similarities and differences between developmental and post-natal mechanisms regulating muscle size in Drosophila and mammals. The emerging evidence that multiple signaling pathways influence muscle size and are intricately interconnected should have important implications in understanding human diseases and applications in combating muscle wasting. Future studies in both Drosophila and mammals should further dissect the pathways governing muscle growth and atrophy in health and disease to discover novel drugs and therapeutic interventions.
Acknowledgments
Supported by funding to ALG from the Muscular Dystrophy Association and the NIH (AR055255), to RP from Italian Association for Cancer Research AIRC-Start Up (11423), to FD from St. Jude Children’s Hospital/ALSAC, and to NP from the NIH (R01AR057352) and HHMI.
REFERENCES
- Acharyya S, Ladner KJ, Nelsen LL, Damrauer J, Reiser PJ, Swoap S, Guttridge DC. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest. 2004;114:370–378. doi: 10.1172/JCI20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askanas V, Engel WK. Inclusion-body myositis and myopathies: different etiologies, possibly similar pathogenic mechanisms. Curr Opin Neurol. 2002;15:525–531. doi: 10.1097/00019052-200210000-00002. [DOI] [PubMed] [Google Scholar]
- Baehrecke EH. Autophagic programmed cell death in Drosophila. Cell Death Differ. 2003;10:940–945. doi: 10.1038/sj.cdd.4401280. [DOI] [PubMed] [Google Scholar]
- Baracos VE, DeVivo C, Hoyle DH, Goldberg AL. Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am J Physiol. 1995;268:E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- Bayline RJ, Duch C, Levine RB. Nerve-muscle interactions regulate motor terminal growth and myoblast distribution during muscle development. Dev Biol. 2001;231:348–363. doi: 10.1006/dbio.2001.0158. [DOI] [PubMed] [Google Scholar]
- Bayline RJ, Khoo AB, Booker R. Innervation regulates the metamorphic fates of larval abdominal muscles in the moth, Manduca sexta. Dev Genes Evol. 1998;208:369–381. doi: 10.1007/s004270050193. [DOI] [PubMed] [Google Scholar]
- Bellosta P, Gallant P. Myc Function in Drosophila. Genes Cancer. 2010;1:542–546. doi: 10.1177/1947601910377490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benny Klimek ME, Aydogdu T, Link MJ, Pons M, Koniaris LG, Zimmers TA. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem Biophys Res Commun. 2010;391:1548–1554. doi: 10.1016/j.bbrc.2009.12.123. [DOI] [PubMed] [Google Scholar]
- Biressi S, Rando TA. Heterogeneity in the muscle satellite cell population. Semin Cell Dev Biol. 2010;21:845–854. doi: 10.1016/j.semcdb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001a;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001b;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Bonetto A, Aydogdu T, Kunzevitzky N, Guttridge DC, Khuri S, Koniaris LG, Zimmers TA. STAT3 Activation in Skeletal Muscle Links Muscle Wasting and the Acute Phase Response in Cancer Cachexia. PLoS One. 2011;6:e22538. doi: 10.1371/journal.pone.0022538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke MH, Kaiser KK. Muscle fiber types: how many and what kind? Arch Neurol. 1970;23:369–379. doi: 10.1001/archneur.1970.00480280083010. [DOI] [PubMed] [Google Scholar]
- Bruusgaard JC, Gundersen K. In vivo time-lapse microscopy reveals no loss of murine myonuclei during weeks of muscle atrophy. J Clin Invest. 2008;118:1450–1457. doi: 10.1172/JCI34022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S, Menon SD, George SE, Chia W. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J Cell Sci. 2010;123:2697–2707. doi: 10.1242/jcs.063172. [DOI] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE, Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Chinta R, Tan JH, Wasser M. The study of muscle remodeling in Drosophila metamorphosis using in vivo microscopy and bioimage informatics. BMC Bioinformatics. 2012;13:S14. doi: 10.1186/1471-2105-13-S17-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol. 2012;198:575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- Dahlmann B, Rutschmann M, Reinauer H. Effect of starvation or treatment with corticosterone on the amount of easily releasable myofilaments in rat skeletal muscles. Biochem J. 1986;234:659–664. doi: 10.1042/bj2340659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis WL, Jacoby BH, Jones RG, Goodman DB. Superoxide formation preceding flight muscle histolysis in Solenopsis: fine structural cytochemistry and biochemistry. Histochem J. 1993;25:478–490. doi: 10.1007/BF00159283. [DOI] [PubMed] [Google Scholar]
- Dawson SP, Arnold JE, Mayer NJ, Reynolds SE, Billett MA, Gordon C, Colleaux L, Kloetzel PM, Tanaka K, Mayer RJ. Developmental changes of the 26 S proteasome in abdominal intersegmental muscles of Manduca sexta during programmed cell death. J Biol Chem. 1995;270:1850–1858. doi: 10.1074/jbc.270.4.1850. [DOI] [PubMed] [Google Scholar]
- Deak II. Use of Drosophila mutants to investigate the effect of disuse on the maintenance of muscle. J Insect Physiol. 1976;22:1159–1165. doi: 10.1016/0022-1910(76)90127-x. [DOI] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development. 2009;136:983–993. doi: 10.1242/dev.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Piccirillo R, Goldberg AL, Perrimon N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell. 2013 doi: 10.1111/acel.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd SL, Gagnon BJ, Senf SM, Hain BA, Judge AR. Ros-mediated activation of NF-kappaB and Foxo during muscle disuse. Muscle Nerve. 2010;41:110–113. doi: 10.1002/mus.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Anant S, Ruiz-Gomez M, Bate M, VijayRaghavan K. Founder myoblasts and fibre number during adult myogenesis in Drosophila. Development. 2004;131:3761–3772. doi: 10.1242/dev.01249. [DOI] [PubMed] [Google Scholar]
- Farrell ER, Fernandes J, Keshishian H. Muscle organizers in Drosophila: the role of persistent larval fibers in adult flight muscle development. Dev Biol. 1996;176:220–229. doi: 10.1006/dbio.1996.0129. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Bate M, Vijayraghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Fernandes JJ, Keshishian H. Nerve-muscle interactions during flight muscle development in Drosophila. Development. 1998;125:1769–1779. doi: 10.1242/dev.125.9.1769. [DOI] [PubMed] [Google Scholar]
- Figeac N, Daczewska M, Marcelle C, Jagla K. Muscle stem cells and model systems for their investigation. Dev Dyn. 2007;236:3332–3342. doi: 10.1002/dvdy.21345. [DOI] [PubMed] [Google Scholar]
- Figeac N, Jagla T, Aradhya R, Da Ponte JP, Jagla K. Specification and behavior of AMPs, muscle-committed transient Drosophila stem cells. Fly (Austin) 2011;5:7–9. doi: 10.4161/fly.5.1.13710. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Gibala MJ, McGee SL, Garnham AP, Howlett KF, Snow RJ, Hargreaves M. Brief intense interval exercise activates AMPK and p38 MAPK signaling and increases the expression of PGC-1alpha in human skeletal muscle. J Appl Physiol. 2009;106:929–934. doi: 10.1152/japplphysiol.90880.2008. [DOI] [PubMed] [Google Scholar]
- Gilson H, Schakman O, Combaret L, Lause P, Grobet L, Attaix D, Ketelslegers JM, Thissen JP. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology. 2007;148:452–460. doi: 10.1210/en.2006-0539. [DOI] [PubMed] [Google Scholar]
- Glass DJ. Signaling pathways perturbing muscle mass. Curr Opin Clin Nutr Metab Care. 2010;13:225–229. doi: 10.1097/mco.0b013e32833862df. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Role of insulin in work-induced growth of skeletal muscle. Endocrinology. 1968;83:1071–1073. doi: 10.1210/endo-83-5-1071. [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Goodman HM. Relationship between cortisone and muscle work in determining muscle size. J Physiol. 1969;200:667–675. doi: 10.1113/jphysiol.1969.sp008715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink G. Malleability of the motor system: a comparative approach. J Exp Biol. 1985;115:375–391. doi: 10.1242/jeb.115.1.375. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell AD. Dynamics of nerve-muscle interaction in developing and mature neuromuscular junctions. Physiol Rev. 1995;75:789–834. doi: 10.1152/physrev.1995.75.4.789. [DOI] [PubMed] [Google Scholar]
- Haas AL, Baboshina O, Williams B, Schwartz LM. Coordinated induction of the ubiquitin conjugation pathway accompanies the developmentally programmed death of insect skeletal muscle. J Biol Chem. 1995;270:9407–9412. doi: 10.1074/jbc.270.16.9407. [DOI] [PubMed] [Google Scholar]
- Haas KF, Woodruff E, 3rd, Broadie K. Proteasome function is required to maintain muscle cellular architecture. Biol Cell. 2007;99:615–626. doi: 10.1042/BC20070019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings RA, Eyheralde I, Dawson SP, Walker G, Reynolds SE, Billett MA, Mayer RJ. A 220-kDa activator complex of the 26 S proteasome in insects and humans. A role in type II programmed insect muscle cell death and cross-activation of proteasomes from different species. J Biol Chem. 1999;274:25691–25700. doi: 10.1074/jbc.274.36.25691. [DOI] [PubMed] [Google Scholar]
- Hegstrom CD, Truman JW. Steroid control of muscle remodeling during metamorphosis in Manduca sexta. J Neurobiol. 1996;29:535–550. doi: 10.1002/(SICI)1097-4695(199604)29:4<535::AID-NEU9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Herbison GJ, Jaweed MM, Ditunno JF. Muscle atrophy in rats following denervation, casting, inflammation, and tenotomy. Arch Phys Med Rehabil. 1979;60:401–404. [PubMed] [Google Scholar]
- Herbst KL, Bhasin S. Testosterone action on skeletal muscle. Curr Opin Clin Nutr Metab Care. 2004;7:271–277. doi: 10.1097/00075197-200405000-00006. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Wang H, Lee IH, Du J, Mitch WE. Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice. J Clin Invest. 2009;119:3059–3069. doi: 10.1172/JCI38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RB, Stevenson E, Koncarevic A, Mitchell-Felton H, Essig DA, Kandarian SC. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. Faseb J. 2002;16:529–538. doi: 10.1096/fj.01-0866com. [DOI] [PubMed] [Google Scholar]
- Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JR, Mula J, Kirby TJ, Fry CS, Lee JD, Ubele MF, Campbell KS, McCarthy JJ, Peterson CA, Dupont-Versteegden EE. Satellite cell depletion does not inhibit adult skeletal muscle regrowth following unloading-induced atrophy. Am J Physiol Cell Physiol. 2012;303:C854–C861. doi: 10.1152/ajpcell.00207.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YC, Van Remmen H. Age-associated alterations of the neuromuscular junction. Exp Gerontol. 2011;46:193–198. doi: 10.1016/j.exger.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Haire MF, Kloetzel PM, Mykles DL, Schwartz LM. Changes in the structure and function of the multicatalytic proteinase (proteasome) during programmed cell death in the intersegmental muscles of the hawkmoth, Manduca sexta. Dev Biol. 1995;169:436–447. doi: 10.1006/dbio.1995.1159. [DOI] [PubMed] [Google Scholar]
- Jones SW, Hill RJ, Krasney PA, O'Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. Faseb J. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura KI, Truman JW. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci. 1990;10:403–401. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp JR, Davie JK, Myer A, Meadows E, Olson EN, Klein WH. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006;133:601–610. doi: 10.1242/dev.02249. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Kuwahara H, Horie T, Ishikawa S, Tsuda C, Kawakami S, Noda Y, Kaneko T, Tahara S, Tachibana T, Okabe M, Melki J, Takano R, Toda T, Morikawa D, Nojiri H, Kurosawa H, Shirasawa T, Shimizu T. Oxidative stress in skeletal muscle causes severe disturbance of exercise activity without muscle atrophy. Free Radic Biol Med. 2010;48:1252–1262. doi: 10.1016/j.freeradbiomed.2010.02.011. [DOI] [PubMed] [Google Scholar]
- LaBeau-DiMenna EM, Clark KA, Bauman KD, Parker DS, Cripps RM, Geisbrecht ER. Thin, a Trim32 ortholog, is essential for myofibril stability and is required for the integrity of the costamere in Drosophila. Proc Natl Acad Sci U S A. 2012;109:17983–17988. doi: 10.1073/pnas.1208408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. Embo J. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. Faseb J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- Li JB, Goldberg AL. Effects of food deprivation on protein synthesis and degradation in rat skeletal muscles. Am J Physiol. 1976;231:441–448. doi: 10.1152/ajplegacy.1976.231.2.441. [DOI] [PubMed] [Google Scholar]
- Lokireddy S, Wijesoma IW, Teng S, Bonala S, Gluckman PD, McFarlane C, Sharma M, Kambadur R. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Low P, Bussell K, Dawson SP, Billett MA, Mayer RJ, Reynolds SE. Expression of a 26S proteasome ATPase subunit, MS73, in muscles that undergo developmentally programmed cell death, and its control by ecdysteroid hormones in the insect Manduca sexta. FEBS Lett. 1997;400:345–349. doi: 10.1016/s0014-5793(96)01413-5. [DOI] [PubMed] [Google Scholar]
- Lustgarten MS, Jang YC, Liu Y, Muller FL, Qi W, Steinhelper M, Brooks SV, Larkin L, Shimizu T, Shirasawa T, McManus LM, Bhattacharya A, Richardson A, Van Remmen H. Conditional knockout of Mn-SOD targeted to type IIB skeletal muscle fibers increases oxidative stress and is sufficient to alter aerobic exercise capacity. Am J Physiol Cell Physiol. 2009;297:C1520–C1532. doi: 10.1152/ajpcell.00372.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Calvani R, Bernabei R, Leeuwenburgh C. Apoptosis in skeletal myocytes: a potential target for interventions against sarcopenia and physical frailty - a mini-review. Gerontology. 2012;58:99–106. doi: 10.1159/000330064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lomo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc Natl Acad Sci U S A. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC. Metabolic functions of Myostatin and GDF11. Immunol Endocr Metab Agents Med Chem. 2010;10:217–231. doi: 10.2174/187152210793663810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi M, Fareed M, O'Neal P, Poylin V, Wei W, Hasselgren PO. Role of glucocorticoids in the molecular regulation of muscle wasting. Crit Care Med. 2007;35:S602–S608. doi: 10.1097/01.CCM.0000279194.11328.77. [DOI] [PubMed] [Google Scholar]
- Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkioti F, Kratsios P, Luedde T, Song YH, Delafontaine P, Adami R, Parente V, Bottinelli R, Pasparakis M, Rosenthal N. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116:2945–2954. doi: 10.1172/JCI28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moylan JS, Smith JD, Chambers MA, McLoughlin TJ, Reid MB. TNF induction of atrogin-1/MAFbx mRNA depends on Foxo4 expression but not AKT-Foxo1/3 signaling. Am J Physiol Cell Physiol. 2008;295:C986–C993. doi: 10.1152/ajpcell.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FL, Song W, Liu Y, Chaudhuri A, Pieke-Dahl S, Strong R, Huang TT, Epstein CJ, Roberts LJ, 2nd, Csete M, Faulkner JA, Van Remmen H. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Nair KS. Aging muscle. Am J Clin Nutr. 2005;81:953–963. doi: 10.1093/ajcn/81.5.953. [DOI] [PubMed] [Google Scholar]
- Nakao R, Hirasaka K, Goto J, Ishidoh K, Yamada C, Ohno A, Okumura Y, Nonaka I, Yasutomo K, Baldwin KM, Kominami E, Higashibata A, Nagano K, Tanaka K, Yasui N, Mills EM, Takeda S, Nikawa T. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol Cell Biol. 2009;29:4798–4811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navegantes LC, Resano NM, Migliorini RH, Kettelhut IC. Catecholamines inhibit Ca(2+)-dependent proteolysis in rat skeletal muscle through beta(2)-adrenoceptors and cAMP. Am J Physiol Endocrinol Metab. 2001;281:E449–E454. doi: 10.1152/ajpendo.2001.281.3.E449. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- Pate RR, Pratt M, Blair SN, Haskell WL, Macera CA, Bouchard C, Buchner D, Ettinger W, Heath GW, King AC, et al. Physical activity and public health. A recommendation from the Centers for Disease Control and Prevention and the American College of Sports Medicine. Jama. 1995;273:402–407. doi: 10.1001/jama.273.5.402. [DOI] [PubMed] [Google Scholar]
- Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. 2010;191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza N, Gosangi B, Devilla S, Arking R, Wessells R. Exercise-training in young Drosophila melanogaster reduces age-related decline in mobility and cardiac performance. PLoS One. 2009;4:e5886. doi: 10.1371/journal.pone.0005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo R, Goldberg AL. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. Embo J. 2012;31:3334–3350. doi: 10.1038/emboj.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piétri-Rouxel F, Gentil C, Vassilopoulos S, Baas D, Mouisel E, Ferry A, Vignaud A, Hourdé C, Marty I, Schaeffer L, Voit T, Garcia L. DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. Embo J. 2010;29:643–654. doi: 10.1038/emboj.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JD, Burns LA, McMahon EJ. Denervation of primate extraocular muscle. A unique pattern of structural alterations. Invest Ophthalmol Vis Sci. 1989;30:1894–908. [PubMed] [Google Scholar]
- Primeau AJ, Adhihetty PJ, Hood DA. Apoptosis in heart and skeletal muscle. Can J Appl Physiol. 2002;27:349–395. doi: 10.1139/h02-020. [DOI] [PubMed] [Google Scholar]
- Raffaello A, Milan G, Masiero E, Carnio S, Lee D, Lanfranchi G, Goldberg AL, Sandri M. JunB transcription factor maintains skeletal muscle mass and promotes hypertrophy. J Cell Biol. 2010;191:101–113. doi: 10.1083/jcb.201001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, Lee VM, Forman MS, Taylor JP. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M. Mitochondrial fission and remodelling contributes to muscle atrophy. Embo J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, VijayRaghavan K. Patterning muscles using organizers: larval muscle templates and adult myoblasts actively interact to pattern the dorsal longitudinal flight muscles of Drosophila. J Cell Biol. 1998;141:1135–1145. doi: 10.1083/jcb.141.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, Lanza IR, Rasbach KA, Okutsu M, Nair KS, Yan Z, Leinwand LA, Spiegelman BM. A PGC-1alpha Isoform Induced by Resistance Training Regulates Skeletal Muscle Hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. Faseb J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- Sandri M, Carraro U. Apoptosis of skeletal muscles during development and disease. Int J Biochem Cell Biol. 1999;31:1373–1390. doi: 10.1016/s1357-2725(99)00063-1. [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:C1248–C1257. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- Schakman O, Gilson H, Thissen JP. Mechanisms of glucocorticoid-induced myopathy. J Endocrinol. 2008;197:1–10. doi: 10.1677/JOE-07-0606. [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Schönbauer C, Langer CC, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A, Keleman K, Dickson BJ. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- Schubiger M, Wade AA, Carney GE, Truman JW, Bender M. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development. 1998;125:2053–2062. doi: 10.1242/dev.125.11.2053. [DOI] [PubMed] [Google Scholar]
- Schulze PC, Spate U. Insulin-like growth factor-1 and muscle wasting in chronic heart failure. Int J Biochem Cell Biol. 2005;37:2023–2035. doi: 10.1016/j.biocel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Schwartz LM. Insect muscle as a model for programmed cell death. J Neurobiol. 1992;23:1312–1326. doi: 10.1002/neu.480230918. [DOI] [PubMed] [Google Scholar]
- Schwartz LM, Myer A, Kosz L, Engelstein M, Maier C. Activation of polyubiquitin gene expression during developmentally programmed cell death. Neuron. 1990;5:411–419. doi: 10.1016/0896-6273(90)90080-y. [DOI] [PubMed] [Google Scholar]
- Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- Taylor MV. Comparison of muscle development in Drosophila and vertebrates. In: Sink H, editor. Muscle development in Drosophila. Georgetown, TX / New York, NY: Landes Bioscience / Springer; 2006. pp. 169–203. [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional Control of Protein Biosynthetic Capacity by Insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun. 2002;296:350–354. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- Thornell LE. Sarcopenic obesity: satellite cells in the aging muscle. Curr Opin Clin Nutr Metab Care. 2011;14:22–27. doi: 10.1097/MCO.0b013e3283412260. [DOI] [PubMed] [Google Scholar]
- Tiao G, Lieberman M, Fischer JE, Hasselgren PO. Intracellular regulation of protein degradation during sepsis is different in fast- and slow-twitch muscle. Am J Physiol. 1997;272:R849–R856. doi: 10.1152/ajpregu.1997.272.3.R849. [DOI] [PubMed] [Google Scholar]
- Tiefenbock SK, Baltzer C, Egli NA, Frei C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. Embo J. 2010;29:171–183. doi: 10.1038/emboj.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkerhess MJ, Healy L, Morgan M, Sujkowski A, Matthys E, Zheng L, Wessells RJ. The Drosophila PGC-1α homolog spargel modulates the physiological effects of endurance exercise. PLoS One. 2012;7:e31633. doi: 10.1371/journal.pone.0031633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Doevendans PA, de Theije CC, Babiker FA, Molkentin JD, de Windt LJ. Requirement of nuclear factor of activated T-cells in calcineurin-mediated cardiomyocyte hypertrophy. J Biol Chem. 2002;277:48617–48626. doi: 10.1074/jbc.M206532200. [DOI] [PubMed] [Google Scholar]
- Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol. 2009;71:37–57. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005;19:1715–1722. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Schneyer AL. The biology of activin: recent advances in structure, regulation and function. J Endocrinol. 2009;202:1–12. doi: 10.1677/JOE-08-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yang WY. Spatiotemporally controlled initiation of Parkin-mediated mitophagy within single cells. Autophagy. 2011;7:1230–1238. doi: 10.4161/auto.7.10.16626. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Heslop L, Hudon V, Rosenblatt JD, Tajbakhsh S, Buckingham ME, Beauchamp JR, Partridge TA. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp Cell Res. 2002;281:39–49. doi: 10.1006/excr.2002.5653. [DOI] [PubMed] [Google Scholar]
- Zeman RJ, Zhao J, Zhang Y, Zhao W, Wen X, Wu Y, Pan J, Bauman WA, Cardozo C. Differential skeletal muscle gene expression after upper or lower motor neuron transection. Pflugers Arch. 2009;458:525–535. doi: 10.1007/s00424-009-0643-5. [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proc Natl Acad Sci USA. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, Rosenfeld R, Chen Q, Boone T, Simonet WS, Lacey DL, Goldberg AL, Han HQ. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 2010;142:531–543. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee SJ. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]