Summary
Previously we identified that the Escherichia coli protein MqsR (YgiU) functions as a toxin and that it is involved in the regulation of motility by quorum sensing signal autoinducer-2 (AI-2). Furthermore, MqsR is directly associated with biofilm development and is linked to the development of persister cells. Here we show that MqsR and MqsA (YgiT) are a toxin/antitoxin (TA) pair, which, in significant difference to other TA pairs, regulates additional loci besides its own. We have recently identified that MqsR functions as an RNase. However, using three sets of whole-transcriptome studies and two nickel-enrichment DNA binding microarrays coupled with cell survival studies in which MqsR was overproduced in isogenic mutants, we identified eight genes (cspD, clpX, clpP, lon, yfjZ, relB, relE and hokA) that are involved in a mode of MqsR toxicity in addition to its RNase activity. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) showed that (i) the MqsR/MqsA complex (and MqsA alone) represses the toxin gene cspD, (ii) MqsR overproduction induces cspD, (iii) stress induces cspD, and (iv) stress fails to induce cspD when MqsR/MqsA are overproduced or when mqsRA is deleted. Electrophoretic mobility shift assays show that the MqsA/MqsR complex binds the promoter of cspD. In addition, proteases Lon and ClpXP are necessary for MqsR toxicity. Together, these results indicate the MqsR/MqsA complex represses cspD which may be derepressed by titrating MqsA with MqsR or by degrading MqsA via stress conditions through proteases Lon and ClpXP. Hence, we demonstrate that the MqsR/MqsA TA system controls cell physiology via its own toxicity as well as through its regulation of another toxin, CspD.
Introduction
Toxin/antitoxin (TA) systems are ubiquitous (Gerdes et al., 2005) in bacterial chromosomes and in low-copy-number plasmids where they appear to stabilize these systems (Engelberg-Kulka et al., 2006). TA systems typically consist of pairs of genes located in one operon, which encode two components, a stable toxin that can cause cell death by disrupting an essential cellular process and a labile antitoxin that can bind and form a tight complex with the toxin and neutralize its activity (Brown and Shaw, 2003). Influencing these TA systems may provide new antimicrobial agents; for example, agents that bind the antitoxin may free the toxin to cause bacteriostatic or bacteriocidal effects (Nieto et al., 2007). Many chromosomal TA systems have been characterized in Escherichia coli including (listed as toxin/antitoxin) MazF/MazE (Aizenman et al., 1996), RelE/RelB (Gotfredsen and Gerdes, 1998), ChpB/ChpS (Masuda et al., 1993), YoeB/YefM (Cherny and Gazit, 2004), YafQ/DinJ (Motiejūnaitė et al., 2007) and YhaV/PrlF (Schmidt et al., 2007). Although the mechanism of toxicity at the molecular level is slightly different, MazF (Gerdes et al., 2005), RelE (Gerdes et al., 2005), ChpB (Gerdes et al., 2005), YoeB (Christensen et al., 2004) and YhaV (Schmidt et al., 2007) prevent translation by cleaving RNAs; the mode of translation inhibition by YafQ is currently still unclear (Tsilibaris et al., 2007). CcdB of the F plasmid inhibits gyrase while antitoxin CcdA actively dissociates the CcdB:gyrase complex in a process termed rejuvenation (De Jonge et al., 2009).
The role of TA systems in cell physiology is not well understood, and indeed nine hypothetical biological functions of TA systems have been proposed (Magnuson, 2007) (the last suggested by us): addictive genomic debris, stabilization of genomic parasites, selfish alleles, gene regulation, growth control, persister formation, programmed cell arrest, programmed cell death and anti-phage measures (Pecota and Wood, 1996). In regard to programmed cell death, there have been few clear examples of toxin-mediated cell death in a physiologically relevant situation (Pandey and Gerdes, 2005; Magnuson, 2007). Our hypothesis is that TA system toxicity in bacteria is linked to biofilm formation, dispersal and quorum sensing (QS). For example, we found recently that the deletion of five E. coli TA systems influences biofilm development through fimbriae and dispersal (Kim et al., 2009). Also, we identified that the toxin Hha controls cell death and biofilm dispersal via its activation of prophage lytic genes (e.g. rzpD, yfjZ, appY and alpA) and several proteases (e.g. Lon and ClpXP) which may, in turn, activate toxins by degrading antitoxins (García-Contreras et al., 2008). Similarly, the putative holin Cid and anti-holin Lrg system of Staphylococcus aureus links cell death and lysis to biofilm development (Bayles, 2007). Hence, TA systems are directly related to biofilm formation.
The benefits of cell death for biofilm dispersal have been shown clearly using prophage. Autolysis via prophage Pf4 allows Pseudomonas aeruginosa cells to disperse from the biofilm matrix (Webb et al., 2003) and undergo phenotypic variation (Webb et al., 2004). Also, in the case of Pseudoalteromonas tunicata, it uses the autolytic protein AlpP for autolysis for biofilm dispersal (Mai-Prochnow et al., 2006). Furthermore, prophage control biofilm architecture, dispersal and virulence (Rice et al., 2009). In E. coli, we have shown that cryptic prophage CP4-57 affects motility, metabolism and biofilm formation (Wang et al., 2009). Hence, cell death via prophage has been linked to biofilm formation and may be necessary for dispersal. Thus, we propose a similar role for TA systems in that they may slow metabolism to influence biofilm formation.
Gerdes (2000) proposed that a realistic suicide hypothesis for a TA system must include a regulatory network system for sensing cell density such as QS signals. However, until now, a QS system has not been related previously to a TApair. We discovered that MqsR (formerly YgiU) is induced in biofilms (Ren et al., 2004a) and that MqsR with antitoxin MqsA (YgiT) regulates directly or indirectly motility related to QS (González Barrios et al., 2006). This QS is directly associated with biofilm development as it mediates the ability of autoinducer 2 (AI-2) to increase E. coli biofilm formation (González Barrios et al., 2006). MqsR sequences are conserved in many genera including Yersinia pseudotuberculosis, Y. pestis, Bordetella bronchiseptica and Pseudomonas fluorescens. MqsR/MqsA influence biofilm formation via qseBC, which controls motility through flhDC (González Barrios et al., 2006). In addition, mqsR is the most highly induced gene in persister cells as compared with non-persisters (Shah et al., 2006). Furthermore, MqsR was hypothesized to be a toxin in conjunction with antitoxin MqsA, since deletion of mqsA is lethal (Baba et al., 2006; Shah et al., 2006). Previously, we showed that MqsR is a toxin (Zhang et al., 2008), and its three-dimensional structure revealed it is an RNase similar to RelE and YoeB (Brown et al., 2009). Furthermore, MqsA binds DNA via its helix–turn–helix (HTH) C-terminal domain (Brown et al., 2009). Unlike most TApairs including RelE/RelB, YoeB/YefM and MazF/MazE, the MqsR toxin gene precedes the antitoxin gene in the bicistronic operon (Shah et al., 2006). Furthermore, the MqsR/MqsATApair is unique (Brown et al., 2009) in that (i) the antitoxin is larger than the toxin, (ii) both toxin and antitoxin are basic (typically the toxin is basic while the antitoxin is acidic), (iii) the mqsRA sequences are not homologous to any member of a recognized TA system, (iv) the antitoxin binds the toxin at its N-terminus and requires a metal, zinc, for structural stability, (v) the antitoxin is structured throughout its entire sequence, and (vi) the antitoxin binds more than its own promoter (e.g. mqsRA, mcbR and spy) via its C-terminal domain. Also, MqsR cleaves mRNA at GCU sites (Yamaguchi et al., 2009). Interestingly, six of the 14 MqsR-resistant mRNAs that lack MqsR-specific GCU sequences (pheL, tnaC, trpL, yciG, ygaQ and ralR) (Yamaguchi et al., 2009) have been shown by us to be regulated in biofilms (Domka et al., 2007).
Here, using whole-transcriptome analyses, regulator/DNA binding assays and isogenic mutants, we show eight proteins are involved in MqsR toxicity including toxin CspD; i.e. the MqsR/MqsA complex regulates cspD transcription. We also present evidence that MqsR toxicity is dependent on Lon and ClpXP protease activity.
Results
MqsR is toxic, MqsA reduces toxicity and MqsR/MqsA influence biofilm formation
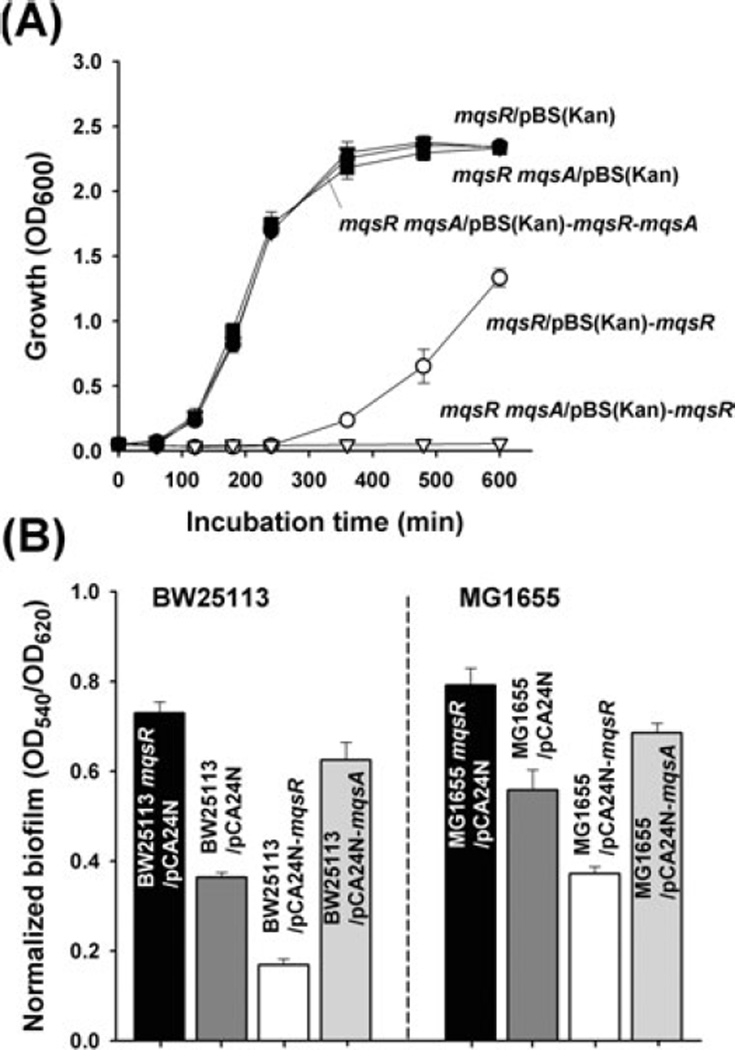
We have shown previously that overproduction of MqsR is toxic (Zhang et al., 2008) and that MqsA diminishes the toxicity of MqsR in both BW25113 and MG1655 strains (Brown et al., 2009). More clearly, to explore the impact of MqsR toxicity on cell physiology, we constructed a mqsR mqsA double mutant and tested cell survival with or without MqsR production. Note that it is not possible to delete solely the gene that encodes the antitoxin, mqsA (Baba et al., 2006; Shah et al., 2006); similar results have been found with other antitoxins such as HigA of Vibrio cholerae (Budde et al., 2007). As expected, production of MqsR in the mqsR mqsA double mutant that lacks the antitoxin was much more toxic compared with production of MqsR in the single mqsR mutant (Fig. 1A). Also, overproduction of MqsR is not toxic when MqsA is also overproduced (Fig. 1A).
Fig. 1.
Growth curves for BW25113 mqsR and BW25113 mqsR mqsA containing pBS(Kan) (empty plasmid), pBS(Kan)-mqsR and pBS(Kan)-mqsR-mqsA at 37°C in LB with 1 mM IPTG induction (A). Normalized biofilm formation (total biofilm/growth) with 96-well polystyrene plates at 37°C in LB with 1 mM IPTG induction for 24 h for E. coli BW25113 mqsR, BW25113, MG1655 mqsR and MG1655 containing pCA24N, pCA24N-mqsA, pCA24N-mqsR (B). Growth data are the average of two independent cultures, biofilm data are the average of 10 replicate wells from two independent cultures, and one standard deviation is shown.
Since mqsR was discovered as an induced gene during biofilm formation (eightfold) (Ren et al., 2004a), we also tested whether MqsR/MqsA are linked to biofilm development using two K-12 strains, BW25113 and MG1655. Consistently, producing the antitoxin MqsA increased normalized biofilm formation (biofilm/growth) whereas overproducing MqsR reduced biofilm formation (Fig. 1B). Corroborating these effects, deleting mqsR increased biofilm formation in both strains (Fig. 1B). Taken together, these results confirm that MqsR is a toxin, that MqsA is the essential antitoxin for reducing MqsR toxicity, and that altering the ratio of MqsR/MqsA influences biofilm formation.
Identification of genes related to MqsR toxicity via whole-transcriptome analysis
We investigated the genes controlled by MqsR using whole-transcriptome analyses to investigate further its mode of toxicity beyond that as an RNase. We reasoned that the MqsR/MqsA complex may alter cell physiology in two ways when a stress is encountered: (i) unbound MqsR (after degradation of labile MqsA) decreases protein production via its degradation of mRNAs, and (ii) specific loci may be repressed/derepressed if MqsR/MqsA or MqsA alone serves to stimulate/repress transcription via the DNA-binding function of MqsA. In addition, we showed previously that MqsA regulates more than its own operon (Brown et al., 2009) as it also binds the promoters of mcbR and spy. To take advantage of the isogenic Keio library of single-gene knockouts (Baba et al., 2006), the remainder of the experiments were performed with BW25113.
Using planktonic cells, an initial whole-transcriptome study showed that upon mqsR deletion (BW25113 mqsR versus BW25113 at a turbidity of 0.5 at 600 nm), 239 genes are repressed by more than twofold while 76 genes are induced (Table 1). Among these, motility-related genes, including 16 flagella genes, six curli genes and six chemotaxis genes, were repressed by deleting mqsR (Table 1). These results were consistent with our initial report that AI-2 signalling and motility/curli are strongly influenced by MqsR/MqsA(González Barrios et al., 2006) and suggest (i) that MqsA, in the absence of MqsR, represses motility genes directly, (ii) that MqsA represses a positive regulator of motility, or (iii) that MqsR degrades RNA of a positive regulator for motility. Significantly, mqsR deletion repressed cspD, which encodes a stress-induced DNA replication inhibitor (Yamanaka et al., 2001), and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) corroborated that cspD is repressed upon inactivating mqsR (3 ± 1-fold). This suggests MqsA represses cspD in the absence of MqsR. To confirm the hypothesis that MqsA represses cspD in the absence of MqsR, we quantified the transcript levels of cspD in BW25113 mqsR mqsA overproducing MqsA using qRT-PCR. As expected, cspD was repressed (2 ± 1-fold) by the overproduction of MqsA in the absence of MqsR. Therefore, MqsA appears to regulate cspD directly.
Table 1.
Partial list of differentially expressed genes for (i) planktonic BW25113 mqsR versus BW25113 at a turbidity of 0.5 at 600 nm, (ii) planktonic BW25113/pCA24N-mqsR versus BW25113/pCA24N with 2 mM IPTG added at a turbidity of 0.5 (grown for 3 h), and (iii) planktonic BW25113 mqsR/pCA24N-mqsR versus BW25113 mqsR/pCA24N grown to a turbidity of 0.5 and then 2 mM IPTG was added for 15 min.
| Fold changea | |||||
|---|---|---|---|---|---|
| Group and gene |
b number |
mqsR vs. WT |
pCA24N-mqsR vs. pCA24N (3 h) |
pCA24N-mqsR vs. pCA24N (15 min) |
Description of encoded protein |
| Stress response related | |||||
| dnaK | b0014 | 2.1 | 1.5 | 2.3 | Chaperone Hsp70, DNA biosynthesis, autoregulated heat shock proteins |
| dnaJ | b0015 | 2.6 | 1.6 | 2.6 | Chaperone with DnaK, heat shock protein |
| clpP | b0437 | 1.5 | 2.3 | 1.9 | ATP-dependent proteolytic subunit of ClpA-ClpP serine protease |
| lon | b0439 | 1.7 | 2.0 | 2.8 | DNA-binding, ATP-dependent protease LA, lon deletion mutants form long cells |
| htpG | b0473 | 2.5 | 2.0 | 3.7 | Heat shock chaperone, HSP90 family, has ATPase activity |
| cstA | b0598 | −2.0 | 4.3 | 1.2 | Carbon starvation protein |
| dps | b0812 | 1.6 | 3.5 | 1.0 | Global regulator, starvation conditions |
| cspD | b0880 | −2.1 | 6.5 | −1.3 | Stress-induced DNA replication inhibitor |
| umuC | b1184 | −2.0 | −1.8 | 1.0 | SOS mutagenesis and repair |
| cspC | b1823 | 1.1 | 5.7 | 1.1 | Cold shock protein |
| htpX | b1829 | 1.5 | 5.3 | 1.9 | Heat shock protein, integral membrane protein |
| clpB | b2592 | 2.8 | 1.8 | 4.6 | ATP-dependent protease and chaperone, protein disaggregation chaperone |
| rpoS | b2741 | 1.1 | 3.7 | 1.2 | RNA polymerase, sigma S (sigma38) factor |
| sspA | b3229 | 1.1 | 3.0 | 1.2 | Regulator of transcription, stringent starvation protein A |
| yrfH | b3400 | 2.6 | 1.4 | 3.7 | Hsp15, DNA/RNA-binding heat shock protein |
| yrfI | b3401 | 2.6 | 1.0 | 3.5 | Hsp33, redox-regulated chaperone |
| ibpB | b3686 | 5.7 | 1.6 | 7.5 | Heat shock protein |
| ibpA | b3687 | 4.9 | 2.1 | 8.6 | Heat shock protein |
| hslU | b3931 | 2.1 | 1.6 | 3.2 | Heat shock protein HslVU, ATPase subunit, homologous to chaperones |
| hslV | b3932 | 2.5 | 1.6 | 3.5 | Heat shock protein HslVU, proteasome-related peptidase subunit |
| osmY | b4376 | 4.6 | 1.7 | −1.4 | Hyperosmotically inducible periplasmic protein |
| Transcription, translation and regulation related | |||||
| fur | b0683 | 1.1 | 3.2 | 1.3 | Ferric iron uptake global transcriptional repressor |
| bssR | b0836 | −3.3 | −1.3 | 1.2 | Repressor of biofilm formation by indole transport regulation |
| bssS | b1060 | 1.2 | 3.0 | 2.5 | Repressor of biofilm formation by indole transport regulation; global regulator, e.g. of AI-2 transport and motility genes |
| mqsA | b3021 | 3.5 | 3.7 | 2.5 | Antitoxin part in MqsR-MqsA TA system |
| relE | b1563 | −1.2 | 2.8 | 1.2 | Toxin part in RelE-RelB TA system |
| relB | b1564 | −1.2 | 2.8 | 1.2 | Antitoxin part in RelE-RelB TA system |
| yhdM | b3292 | 1.5 | 1.7 | 2.0 | Zn-responsive activator of zntA transcription |
| rpmF | b1089 | −1.1 | 4.0 | 1.2 | 50S ribosomal subunit protein L32 |
| rpsV | b1480 | 1.1 | 5.3 | 1.1 | 30S ribosomal subunit protein S22 |
| rpsI | b3230 | 1.1 | 3.2 | 1.2 | 30S ribosomal subunit protein S9 |
| rplN | b3310 | 1.1 | 1.9 | 2.1 | 50S ribosomal subunit protein L14 |
| rplB | b3317 | −1.3 | 1.9 | 2.1 | 50S ribosomal subunit protein L2, binds Zn (II) |
| rplW | b3318 | 1.1 | 1.7 | 2.3 | 50S ribosomal subunit protein L23 |
| rplD | b3319 | 1.0 | 1.6 | 2.0 | 50S ribosomal subunit protein L4, erythromycin sensitivity |
| rplC | b3320 | 1.2 | 1.6 | 2.1 | 50S ribosomal subunit protein L3 |
| rpsG | b3341 | 1.1 | 1.9 | 2.0 | 30S ribosomal subunit protein S7 |
| Cell wall/membrane and motility/chemotaxis | |||||
| csgG | b1037 | −2.1 | −2.1 | 1.1 | Curli production assembly/transport component, 2nd curli operons |
| csgF | b1038 | −2.5 | −2.1 | 1.1 | Curli production assembly/transport component, 2nd curli operons |
| csgE | b1039 | −2.6 | −2.1 | 1.0 | Curli production assembly/transport component, 2nd curli operons |
| csgD | b1040 | −2.0 | −1.6 | 1.2 | Putative 2-component transcriptional regulator for 2nd curli operons |
| csgB | b1041 | −2.5 | −2.0 | 1.0 | Minor curlin subunit precursor, similar to CsgA |
| csgA | b1042 | −2.0 | −1.6 | 1.0 | Curlin major subunit, coiled surface structures, cryptic |
| csgC | b1043 | −2.1 | −1.6 | 1.0 | Putative curli production protein |
| flgN | b1070 | −2.5 | −1.5 | 1.1 | Protein of flagellar biosynthesis |
| flgM | b1071 | −2.3 | −1.3 | 1.0 | Anti-FliA (anti-sigma) factor, also known as RflB protein |
| flgA | b1072 | −2.5 | −1.5 | −1.2 | Flagellar biosynthesis, assembly of basal-body periplasmic P ring |
| flgB | b1073 | −2.1 | −2.0 | −1.5 | Flagellar biosynthesis, cell-proximal portion of basal-body rod |
| flgC | b1074 | −2.0 | −1.8 | −1.5 | Flagellar biosynthesis, cell-proximal portion of basal-body rod |
| flgD | b1075 | −2.1 | −2.1 | −1.3 | Flagellar biosynthesis, initiation of hook assembly |
| flgF | b1077 | −2.1 | −2.8 | −1.4 | Flagellar biosynthesis, cell-proximal portion of basal-body rod |
| flgI | b1080 | −2.5 | −2.4 | 1.0 | Homologue of Salmonella P-ring of flagella basal body |
| flgK | b1082 | −2.3 | −1.8 | −1.1 | Flagellar biosynthesis, hook–filament junction protein 1 |
| flgL | b1083 | −2.5 | −2.0 | 1.0 | Flagellar biosynthesis; hook–filament junction protein |
| flhE | b1878 | −2.0 | −2.2 | −1.2 | Flagellar protein |
| flhB | b1880 | −2.0 | −2.6 | 1.0 | Putative part of export apparatus for flagellar proteins |
| cheZ | b1881 | −2.3 | −1.8 | 1.0 | Chemotactic response, CheY protein phophatase |
| cheY | b1882 | −2.6 | −2.2 | −1.1 | Chemotaxis regulator transmits chemoreceptor signals to flagellar motor components |
| cheB | b1883 | −2.6 | −2.2 | −1.1 | Response regulator for chemotaxis (CheA sensor), protein methylesterase |
| cheR | b1884 | −2.3 | −1.5 | −1.1 | Response regulator for chemotaxis, protein glutamate methyltransferase |
| cheW | b1887 | −3.3 | −2.4 | −1.1 | Positive regulator of CheA protein activity |
| cheA | b1888 | −3.3 | −2.0 | 1.0 | Sensory transducer kinase between CheB and CheY |
| fliC | b1923 | −4.9 | 1.1 | −1.1 | Flagellar biosynthesis, flagellin, filament structural protein |
| fliD | b1924 | −2.8 | −1.7 | 1.0 | Flagellar biosynthesis, filament capping protein; enables filament assembly |
| fliS | b1925 | −2.8 | −2.1 | −1.2 | Flagellar biosynthesis, repressor of class 3a and 3b operons (RflA activity) |
| fliT | b1926 | −2.5 | −1.7 | 1.0 | Flagellar biosynthesis, repressor of class 3a and 3b operons (RflA activity) |
| hokE | b4415 | −2.0 | −3.7 | 1.2 | Small toxic membrane polypeptide |
| hokA | b4455 | −2.1 | −4.9 | −1.1 | Small toxic membrane polypeptide |
| hokD | b1562 | −1.5 | 9.8 | 1.0 | Polypeptide destructive to membrane potential |
| Metabolism related | |||||
| gltL | b0652 | −4.0 | −1.1 | 1.1 | ATP-binding protein of glutamate/aspartate transport system |
| gltK | b0653 | −5.3 | 1.1 | −1.1 | Glutamate/aspartate transport system permease |
| gltJ | b0654 | −6.5 | 1.1 | 1.0 | Glutamate/aspartate transport system permease |
| gltI | b0655 | −4.9 | 2.0 | 1.2 | Putative periplasmic binding transport protein |
| putA | b1014 | −2.0 | 6.1 | 1.2 | Proline dehydrogenase, P5C dehydrogenase |
| putP | b1015 | −4.3 | 6.1 | 1.4 | Major sodiumproline symporter |
| add | b1623 | 3.5 | 2.4 | −1.3 | Adenosine deaminase |
| katE | b1732 | 2.0 | −1.3 | −1.3 | Catalase, hydroperoxidase HPII(III) |
| yeiN | b2165 | −6.5 | −1.2 | −1.2 | Hypothetical protein |
| yeiC | b2166 | −6.5 | −1.1 | −1.2 | Putative kinase |
| glpT | b2240 | −1.7 | −4.0 | −1.4 | sn-glycerol-3-phosphate permease |
| glpA | b2241 | −1.4 | −4.3 | −1.5 | Anaerobic glycerol-3-phosphate dehydrogenase subunit A |
| glpB | b2242 | −1.3 | −4.9 | −1.7 | sn-glycerol-3-phosphate dehydrogenase (anaerobic) |
| glpC | b2243 | −1.4 | −7.5 | −1.8 | sn-glycerol-3-phosphate dehydrogenase (anaerobic), K-small subunit |
| glpD | b3426 | 1.1 | −10.6 | 1.2 | sn-glycerol-3-phosphate dehydrogenase (aerobic) |
| glpK | b3926 | 1.1 | −4.0 | −1.3 | Glycerol kinase |
| glpF | b3927 | 1.0 | −5.3 | 1.1 | Facilitated diffusion of glycerol |
| dsdA | b2366 | 4.0 | −1.1 | −2.0 | d-serine dehydratase (deaminase) |
| cysK | b2414 | 1.3 | 7.0 | −1.1 | Cysteine synthase A, o-acetylserine sulfhydrolase A |
| cysA | b2422 | −1.5 | 5.7 | −1.1 | ATP-binding component of sulfate permease A protein |
| eutG | b2453 | −1.6 | −4.6 | −1.1 | Ethanolamine, similar to iron-containing alcohol dehydrogenase |
| pheL | b2598 | −1.9 | 5.7 | −1.2 | Leader peptide of chorismate mutase-P-prephenate dehydratase |
| relA | b2784 | 1.0 | 1.5 | −1.1 | (p)ppGpp synthetase I (GTP pyrophosphokinase); regulation of RNA synthesis, stringent factor |
| agaY | b3137 | −1.3 | −5.3 | 1.0 | Tagatose-bisphosphate aldolase 2 |
| glvB | b3682 | −1.8 | −4.0 | −1.1 | PTS system, arbutin-like IIB component |
| tnaC | b3707 | −1.1 | 13.9 | −3.0 | Tryptophanase leader peptide |
| tnaA | b3708 | 1.3 | 5.3 | −1.3 | Tryptophanase |
| rbsA | b3749 | −3.7 | −1.3 | 1.3 | ATP-binding component of d-ribose high-affinity transport system |
| rbsC | b3750 | −3.0 | 1.0 | 1.3 | d-ribose high-affinity transport system |
| ptsA | b3947 | −1.6 | −4.0 | 1.2 | PEP-protein phosphotransferase system enzyme I |
| phnM | b4095 | −1.5 | −4.6 | −1.6 | Phosphonate metabolism |
| phnL | b4096 | −1.8 | −4.0 | −1.6 | ATP-binding component of phosphonate transport |
| phnH | b4100 | −1.6 | −4.6 | −1.4 | Phosphonate metabolism |
| phnE | b4104 | −1.4 | −4.3 | −1.4 | Membrane channel protein component of Pn transporter |
| Transport related | |||||
| ydgF | b1600 | −1.2 | −1.4 | −2.1 | Multidrug resistance efflux protein, with MdtI |
| tdcC | b3116 | −1.8 | −1.5 | −2.1 | Serine/threonine:H+ symport permease |
| malP | b3417 | 1.7 | −1.1 | −2.6 | Maltodextrin phosphorylase |
| malG | b4032 | 1.6 | −1.5 | −2.0 | Maltose transport complex, inner membrane-spanning subunit |
| malF | b4033 | 1.3 | 1.0 | −2.0 | Maltose transport complex, inner membrane-spanning subunit |
| malE | b4034 | 1.5 | 1.1 | −3.5 | Maltose-binding protein, periplasmic, substrate recognition for active transport of and chemotaxis toward maltose and maltodextrin |
| malK | b4035 | 1.1 | 1.1 | −3.2 | Maltose transport complex, ATP-binding subunit |
| malL | b4036 | 1.9 | 1.2 | −4.3 | Maltoporin, maltose high-affinity uptake system |
| malM | b4037 | 1.6 | 1.3 | −3.0 | Periplasmic protein in maltose transport system, function unknown |
| cadB | b4132 | −1.6 | −3.0 | −3.0 | Lysine-cadaverine antiporter |
| frdC | b4152 | −1.4 | −1.4 | −3.0 | Fumarate reductase membrane anchor polypeptide |
| frdA | b4154 | −1.3 | 1.8 | −2.3 | Fumarate reductase flavoprotein subunit |
| yjfF | b4231 | −1.8 | −3.0 | −2.1 | Putative ABC transporter permease protein, function unknown |
| fecE | b4287 | 2.5 | −2.1 | −2.5 | ATP-binding component of citrate-dependent iron(III) transport protein |
| fecD | b4288 | 2.1 | −3.2 | −2.3 | Citrate-dependent iron transport, membrane-bound protein |
| fecC | b4289 | 2.5 | −2.5 | −2.5 | Citrate-dependent iron(III) transport protein, cytosolic |
| fecB | b4290 | 2.3 | −2.6 | −3.0 | Citrate-dependent iron transport, periplasmic protein |
| fecA | b4291 | 1.9 | −2.1 | −2.0 | Outer membrane ferric citrate receptor, ferric citrate uptake |
| Bacteriophage related | |||||
| rzpD | b0556 | −2.1 | −4.6 | −1.2 | Putative bacteriophage lambda murein endopeptidase |
| Small RNA related | |||||
| ffs | b0455 | −1.1 | 5.3 | 1.7 | 4.5S RNA, component of ribonucleoprotein particle |
| rtT | b4425 | 1.1 | 5.7 | 1.6 | rtT RNA, may modulate the stringent response |
| rye | b4438 | −1.1 | 4.0 | −1.1 | Novel sRNA, function unknown |
Genes considered differentially expressed based on the standard deviation for expression ratio for all genes are shown in boldface.
All experiments were performed with LB medium at 37°C.
In addition, hokA and hokE, encoding small toxic polypeptides that damage the membrane (Pedersen and Gerdes, 1999), were repressed by deleting mqsR; hence, it is possible that HokA and HokE are also repressed by MqsA. Also repressed upon mqsR deletion were gltL, gltK, gltJ and gltI (encoding the glutamate/aspartate transport system), cstA (encoding the carbon starvation protein) (Schultz and Matin, 1991), bssR (encoding a repressor of biofilm formation involved in indole transport) (Domka et al., 2006), and umuC (encoding a protein involved in SOS mutagenesis and repair) (Shinagawa et al., 1983). Lastly, a number of genes encoding chaperone network proteins including ibpA (fivefold), ibpB (sixfold), hslU (twofold), hslV (threefold), dnaK (twofold) and dnaJ (threefold) were induced upon the deletion of mqsR.
In a second whole-transcriptome study in which MqsR was produced for 3 h rather than deleted [BW25113/pCA24N-mqsR versus BW25113/pCA24N at a turbidity of 0.5 at 600 nm with 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) induction], MqsR induced 132 genes and repressed 299 genes by more than threefold (Table 1). tnaA (encodes a tryptophanase) and tnaC (encodes a tryptophanase leader peptide), both linked to indole synthesis, were induced by 5- and 14-fold respectively. Consistent with the mqsR deletion microarray results, cspD was induced significantly (sevenfold) upon overexpressing mqsR. Hence, these results corroborate that CspD plays an important role in MqsR-mediated toxicity. Also, the RelE (threefold) and RelB (threefold) TA systems (Gotfredsen and Gerdes, 1998) along with HokD (10-fold) (Gerdes et al., 1986) were induced (Table 1). In addition, genes encoding nutrient starvation-related proteins including cstA (fourfold) (Schultz and Matin, 1991), rpoS (encoding the stress and stationary-phase sigma S factor, fourfold) (Lange and Hengge-Aronis, 1991), and dps (stress response DNA-binding protein; fourfold) (Almirón et al., 1992) were strongly induced (Table 1) upon overexpressing mqsR as a result of its toxicity. mqsA was also induced by fourfold. Induction of these genes during MqsR production was confirmed using qRT-PCR. Consistent with the microarray data, transcription of relB, relE, hokD, cstA, rpoS, dps and mqsA were induced by 3 ± 1-, 4 ± 1-, 5 ± 1-, 4 ± 1-, 4 ± 1-, 3 ± 1- and 8 ± 1-fold respectively. Hence, over-expressing mqsR for 3 h dramatically and consistently induces the transcription of starvation- and stress-related genes as well as mqsA, the gene that encodes its own antitoxin. Also, the transcription of bssS (threefold), the regulator of biofilm formation through signal secretion (Domka et al., 2006), was induced by MqsR production for 3 h.
Overexpressing mqsR repressed significantly glpA (fourfold), glpB (fivefold), glpC (eightfold), glpD (11-fold), glpF (fivefold), glpK (fourfold) and glpT (fourfold) (Table 1); these genes are related to glycerol-3-phosphate (G3P) metabolism via G3P dehydrogenase. This result further supports that MqsR regulates AI-2 uptake systems in E. coli (González Barrios et al., 2006) by affecting glycerol and G3P metabolism. Among these genes, glpD encodes a component of the multi-drug tolerance mechanism for persister formation (Spoering et al., 2006), which is one of the hypothetical functions of TA systems (Magnuson, 2007). GlpD overproduction induced tolerance to ampicillin and ofloxacin, whereas the deletion of glpD repressed the production of persister cells (Spoering et al., 2006); hence, MqsR may be linked to persister formation through GlpD.
To identify additional genes that are influenced by MqsR, we performed a third whole-transcriptome analysis for MqsR in which mqsR was induced for only for 15 min (BW25113 mqsR/pCA24N-mqsR versus BW25113 mqsR/pCA24N grown to a turbidity of 0.5 at 600 nm and 2 mM IPTG added for 15 min). This type of array, a pulsed expression of the regulatory gene which may destabilize mRNA, was used recently to find changes due to overproduction of regulatory protein CsrA, which was found to influence mRNA stability and led to its association to cyclic-di-GMP (Jonas et al., 2008). By producing MqsR for a short period, 46 genes were induced and 48 genes were repressed by twofold (Table 1). As expected, mqsR was induced (955-fold). Consistent with the 3 h mqsR overexpression microarray data, mqsA was also induced threefold. Therefore, it appears that MqsR, in association with MqsA, induces mqsRA and acts as a positive regulator of its own transcription.
Genes influenced by production of MqsR in the pulsed expression microarray study included genes that are related to stress-associated chaperones and heat shock proteins including ibpA, ibpB, clpB, yrfH, yrfI, htpG, dnaJ, dnaK, hslV and hslU (Table 1). In contrast to the mqsR deletion microarray data, these stress-related, protein-encoding genes were induced by MqsR after the short induction period (15 min). Thus, these genes are most likely affected due to MqsR RNase activity. lon (threefold) and clpP (twofold), which encode antitoxin-degrading proteases (Christensen et al., 2004), were also induced by the overexpression of mqsR for 15 min. In contrast, transport-related gene clusters including those for the maltose transport system (malE, malF, malG, malK, malL and malM) and ferric citrate transport system genes (fecA, fecB, fecC, fecD and fecE) were repressed significantly by overexpressing mqsR for 15 min. Hence, this short-term experiment linked MqsR to Lon and ClpP proteases.
MqsR and MqsA autoregulate their transcription as positive regulators
To corroborate that MqsR stimulates its own transcription as predicted by the whole-transcriptome analysis (Table 1) and to investigate whether MqsA stimulates transcription of the mqsRA operon, we confirmed the enhanced transcription of mqsR and mqsA via qRT-PCR respectively. As expected, a dramatic induction was observed not only for transcripts of mqsA by producing MqsR (37 ± 2-fold), but also for transcripts of mqsR by producing MqsA (57 ± 1-fold). Also, the deletion of mqsR resulted in a sevenfold decrease in the transcription of mqsA. A similar induction in transcription of a TA operon also occurs upon producing the toxin RelE (toxin part of the RelE/RelB TA pair) (Overgaard et al., 2008). Also, we showed previously that the MqsR/MqsA complex and MqsA alone bind directly the mqsRA promoter (Brown et al., 2009). Together taken, the availability of MqsR or MqsA alters the balance of free and bound MqsR and MqsA respectively. This disproportional ratio between MqsR and MqsA may affect to the binding affinity of MqsR/MqsA complex and positively stimulates its own transcription. Typically, most toxin/antitoxin complexes bind their own promoters and act as autorepressors (Jensen and Gerdes, 1995).
Putative MqsR/MqsA DNA-binding regions
Previously, we found that deletion of mqsR affects the expression of many genes as well as affects biofilm formation through motility and QS (González Barrios et al., 2006); hence, based on the interaction between MqsR and MqsA (Brown et al., 2009), it is evident that direct DNA binding is achieved via MqsA. To identify MqsR/MqsA-binding sequences regions under biofilm conditions, we performed a nickel-enrichment DNA microarray in which we cross-linked the MqsR/MqsA complex to DNA using formaldehyde and purified MqsR/MqsA with the bound DNA via a His6-affinity tag on MqsR using biofilms cells (E. coli AG1/pCA24N-mqsR versus E. coli AG1/pCA24N with 2 mM IPTG induction for 24 h). This analysis identified 37 regions of DNA (10 intergenic regions and 27 genes) bound to MqsR/MqsA including cspD (which agrees with two of the whole-transcriptome analyses and qRT-PCR), mcbR, spy, bssR, and other genes encoding transport proteins such as ykfD and feoA (Table S1).
Among the identified sequences, MqsR/MqsA appears to bind bssR in vivo (Table S1). BssR is repressor of biofilm formation and regulates uptake and export of the stationary-phase signal indole (Domka et al., 2006). This gene was also repressed by deleting mqsR (deletion microarray, Table 1) which should increase MqsA; hence, it is possible that MqsR/MqsA represses bssR transcription. This observation may be associated with the reduction of biofilm formation via MqsR overproduction (Fig. 1B); i.e. overproducing toxin MqsR should result in the degradation of MqsA which would derepress bssR and thereby reduce biofilm formation.
Previously, we showed that McbR (formerly YncC) stimulates biofilm formation by reducing mucoidy via colanic acid production and that mcbR is dramatically repressed by the deletion of mqsR and inactivation of mqsA (González Barrios et al., 2006; Zhang et al., 2008). In addition, we showed that the MqsR/MqsA complex or MqsA alone binds tightly the promoter region of mcbR (Brown et al., 2009). Therefore, it appears MqsA may induce transcription of mcbR which likely explains the increased biofilm formation detected upon overproducing MqsA.
To verify that MqsA alone (not bound to MqsR) regulates these genes, we performed a second nickel-enrichment DNA microarray in which we purified MqsA together with bound DNA via a His6-affinity tag on MqsA (BW25113/pBAD-mqsA versus BW25113/pBAD-Myc-His C with 0.5% l-arabinose to induce mqsA added at a turbidity of 0.8 at 600 nm and cells grown for 24 h). Consistent with the DNA sequences bound by the MqsR/MqsA complex in the initial nickel-enrichment DNA microarray, we identified that seven sequences, including cspD, mcbR and spy, are also enriched using MqsA alone (Table S1). Corroborating these results, using qRT-PCR, we found that MqsA overproduction dramatically induced the transcriptions of mcbR and spy by 20 ± 1- and 15 ± 1-fold respectively. Therefore, MqsA alone or in a complex with MqsR induces transcription of mcbR and spy and represses transcription of cspD by binding their promoter regions directly.
MqsR/MqsA binds the cspD promoter directly via MqsA
Since the DNA microarrays, nickel-enrichment DNA microarray and qRT-PCR results indicated the MqsR/MqsA complex is related to the expression of cspD, we investigated if the MqsR/MqsA complex (via the C-terminus of MqsA) binds directly to the promoter of cspD using electrophoretic mobility shift assays (EMSA). Previously, we confirmed that the MqsR/MqsA complex or MqsA alone tightly bind to the promoter regions of mcbR and spy as well as its own promoter (Brown et al., 2009). Consistently, in this study, we found direct binding of the MqsR/MqsA complex to the promoter regions for cspD (Fig. 2A). However, MqsR/MqsA does not bind to the bssR promoter region (Fig. 2A). As a negative control, we show that the N-terminal domain of MqsA does not bind to cspD (Fig. 2B); hence, the C-terminal domain of MqsA is necessary and sufficient to bind the promoter of cspD. This result agrees with earlier results showing that MqsR alone (Yamaguchi et al., 2009) and MqsR associated with the N-terminus of MqsA (Brown et al., 2009) do not bind DNA, and that the C-terminal domain of MqsA in the MqsR/MqsA complex or MqsA alone is responsible for promoter binding (Brown et al., 2009). Taken together, transcription of cspD is be controlled by the direct binding of the MqsR/MqsA complex or MqsA via the MqsA C-terminal domain.
Fig. 2.
Electrophoretic mobility shift assay (EMSA) to confirm the specific DNA binding of the MqsR/MqsA complex. MqsR/MqsA (full length of MqsA) (A) and MqsR/MqsA-N (N-terminal of MqsA) (B) were tested for binding to the promoter regions of cspD (PcspD) and bssR (PbssR). Lane 1: labelled promoter regions (6 ng), lane 2: protein complex (200 ng) and labelled promoter regions (6 ng), and lane 3: protein complex (200 ng) with labelled (6 ng) and unlabelled (specific competitor) promoter regions (600 ng). The unspecific competitor poly(dI-dC) was used for all samples.
MqsR/MqsA regulates PcspD under stress conditions
Since TA pairs are induced under stress (Engelberg-Kulka et al., 2006) and since MqsA binds the promoter region of cspD to repress its transcription and this repression is derepressed by overexpressing mqsR, we investigated further the impact of the MqsR/MqsA complex and stress regulation on cspD. Using qRT-PCR and a BW25113 mqsR mqsA mutant overproducing MqsR/MqsA, we found, similar to overproduction of MqsA alone, that cspD was repressed by overproducing MqsR/MqsA (3 ± 1-fold). Also, under conditions of oxidative stress (30 mM H2O2) (Lee et al., 2007), cspD was strongly induced (5 ± 1-fold) in wild-type BW25113. Under these conditions, clpX (6 ± 1-fold), clpP (6 ± 1-fold), lon (8 ± 1-fold) and mqsR (4 ± 1-fold) were also induced. These results show that environmental stress not only induces proteases that degrade antitoxins (ClpXP and Lon), but it also induces production of both the MqsR and CspD toxins. Critically, cspD was not induced by oxidative stress in the BW25113 mqsR mqsA mutant.
We also reasoned that since MqsA represses cspD, if there is excess MqsA, then cspD would remain repressed even under stress conditions where cspD is induced strongly. We found cspD was repressed by the overproduction of MqsR/MqsA under stress conditions [4 ± 1-fold by qRT-PCR, using BW25113 mqsR mqsA/pBS(Kan)-mqsR-mqsA versus BW25113 mqsR mqsA/pBS(Kan) in LB with 1 mM IPTG at a turbidity of 1 at 600 nm and 30 mM H2O2 for 15 min]. Hence, the significant induction of cspD by H2O2 stress is eliminated when the MqsR/MqsA complex or MqsA alone is overproduced. Therefore, cspD transcription is directly and strongly linked to the MqsR/MqsA TA system.
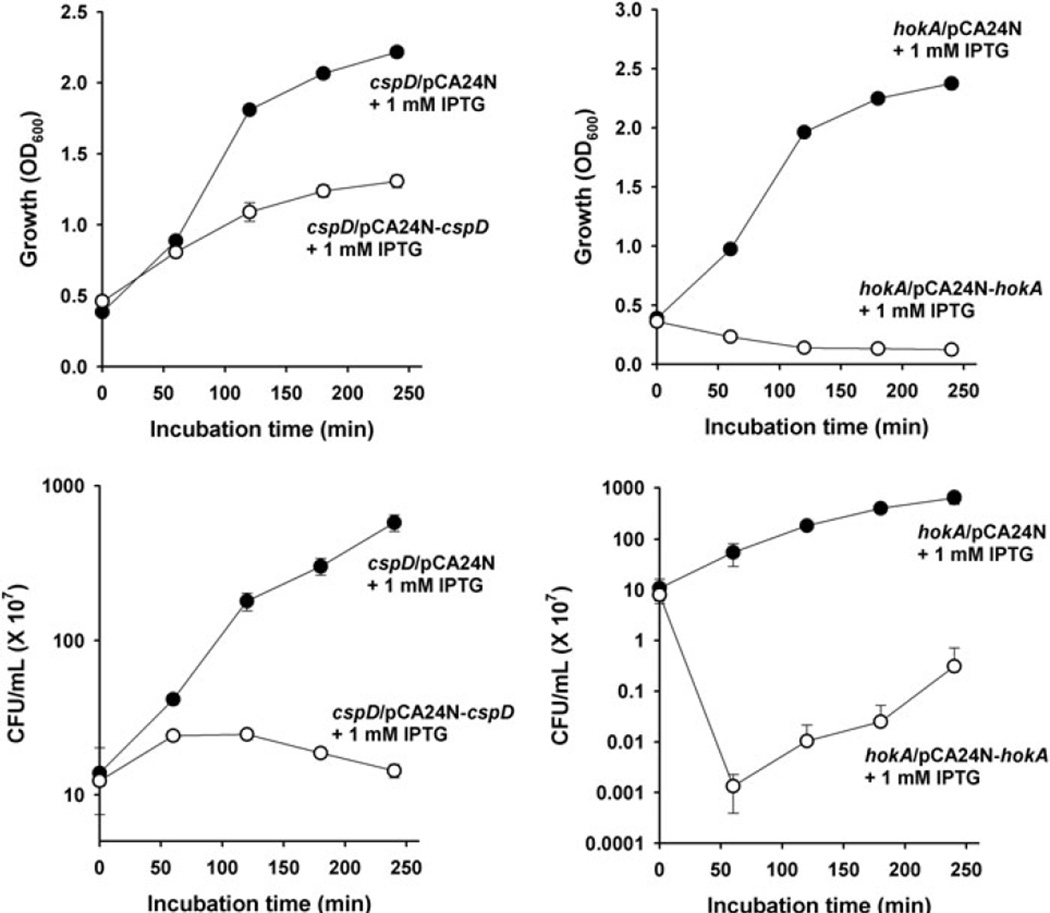
CspD and HokA are toxic
To investigate whether MqsR/MqsA induction of CspD and HokA leads to toxicity, growth and cell viability were assayed by producing these proteins in strains that lacked the genes that encode these two toxins. As expected, cell growth and viability were dramatically arrested for CspD and HokA production (Fig. 3 and Fig. S1). Therefore, stimulation of the production of CspD and HokA via MqsR/MqsA is toxic.
Fig. 3.
Growth curves and cell viability (cfu ml−1) for BW25113 cspD/pCA24N and BW25113 cspD/pCA24N-cspD (A) and for BW25113 hokA/pCA24N and BW25113 hokA/pCA24N-hokA (B). Cells were grown to a turbidity of 0.5 at 600 nm at 37°C in LB medium, then 1 mM IPTG was added to induce the pCA24N-based genes. Data are the average of two independent cultures, and one standard deviation is shown.
Taken together, our results indicate that under normal growth, cspD is repressed by the MqsR/MqsA TA complex. Under stress, MqsA is degraded by proteases (ClpXP and Lon), and cspD transcription is derepressed by the breakdown of the MqsA. The combination of both toxins CspD and MqsR, the latter functioning as an RNase, subsequently leads to a reduction in metabolism.
Confirmation of proteins linked to MqsR toxicity
In an effort to identify proteins other than CspD that may be related to MqsR toxicity as well as to demonstrate MqsR toxicity is associated with CspD, we examined four other toxin-related proteins (RelB, RelE, HokA and HokD) and six stress response-related proteins (CstA, HslU, IbpA, IbpB, UmuC and UmuD) based on the three sets of whole-transcriptome data and the two nickel-enrichment DNA microarray data. In addition, based on the toxin-associated genes we identified to be associated with toxin Hha and its antitoxin TomB (García-Contreras et al., 2008), we also examined ClpX, ClpP, Lon, RzpD and YfjZ. clpX encodes the AAA+ ATPase that binds the serine protease ClpP, clpP encodes a serine protease and lon encodes an ATP-dependent protease. rzpD encodes a putative murein endopeptidase, yfjZ encodes the antitoxin component of the putative TA pair YpjF/YfjZ, cspD encodes a DNA replication inhibitor, relB encodes the antitoxin for the RelE/RelB TA systems, relE encodes the toxin for RelE/RelB, and hokA and hokD encode small toxic membrane polypeptides. cstA encodes a carbon starvation protein, umuC and umuD encode a SOS mutagenesis protein, and ibpA, ibpB and hslU encode heat shock proteins.
Among these 16 genes, deletion of eight genes (cspD, clpX, clpP, lon, yfjZ, relB, relE and hokA) prevented MqsR from inhibiting cell growth (clpX, clpP, lon, yfjZ and cspD) or reduced its ability to inhibit growth (relB, relE and hokA) (Fig. 4A), as well as abolished the ability of MqsR to reduce biofilm formation (Fig. 4B); hence, the eight proteins encoded by these genes may be directly involved in MqsR toxicity as well as biofilm formation. If this hypothesis is correct, then overproduction of the toxin MqsR should induce these genes. We tested this by overexpressing mqsR and measuring transcription via qRT-PCR (using RNA samples from BW25113/pCA24N-mqsR grown to a turbidity of 0.5 with 2 mM IPTG for induction in LB for 3 h); we found that all eight genes were induced significantly: clpX (2 ± 1-fold), clpP (3 ± 1-fold), lon (4 ± 1-fold), yfjZ (3 ± 1-fold), hokA (3 ± 1-fold), cspD (5 ± 1-fold), relB (3 ± 1-fold) and relE (4 ± 1-fold). Therefore, MqsR, via MqsA, must influence the transcription of these genes.
Fig. 4.
Growth curves (A) and normalized biofilm formation (total biofilm/growth; B) of isogenic deletion strains (clpX, clpP, lon, yfjZ, cspD, relB, relE and hokA) with production of MqsR via pCA24N-mqsR and IPTG at 37°C in LB medium. Growth data are the average of two independent cultures, biofilm data are the average of 10 replicate wells from two independent cultures, and one standard deviation is shown.
Discussion
We have shown previously that TA systems are important for biofilm formation using MqsR (Ren et al., 2004a; González Barrios et al., 2006; Zhang et al., 2008) and the E. coli Δ5 strain, which lacks five TA systems (listed as toxin/antitoxin: MazF/MazE, RelE/RelB, ChpB, YoeB/YefM and YafQ/DinJ) (Kim et al., 2009). In the current study, we confirm that MqsR is a toxin paired with its antitoxin MqsA and show that the toxicity of this system is related to its direct regulation of CspD as well as related to the activities ClpX, ClpP, Lon, YfjZ, RelB, RelE and HokA. In addition, our results begin to explain how the MqsR/MqsA complex acts as a global regulatory protein (Table 1 and Table S1) that controls more than its own transcription (González Barrios et al., 2006). We confirm that MqsR/MqsA binds specific promoter regions including cspD (Fig. 2), in addition to its own promoter (Brown et al., 2009). This direct interaction is mediated via the C-terminal HTH DNA-binding domain of MqsA, as we have shown via multiple gel shift assays. These results are consistent with our previous report that the MqsR/MqsA complex or MqsA alone binds to the promoter regions of mcbR and spy as well as to its own promoter (Brown et al., 2009). In addition, Yamaguchi and colleagues (2009) found that there are two palindrome sequences in the promoter region of the mqsRA locus and that the MqsR/MqsA complex or MqsA alone can bind to these two palindromic sequences. Moreover, we found here that the promoter regions cspD, mcbR and spy, that are controlled by the MqsR/MqsA TA pair as confirmed by the gel shift assay (Fig. 2 and Brown et al., 2009) and two nickel enrichment DNA microarray studies (Table S1), also have similar regions with T-rich palindromic spacers (TCAAATTTTTGA for cspD; CATCATTGTTCTGCTG for mcbR; and AGTGTTTTTTACACT for spy). Also, the 58 bp and 74 bp promoter fragments of mcbR and spy, respectively, containing these palindromes, are bound by MqsA in the gel shift assays (data not shown). Hence, these palindrome sequences are important for recognition of specific promoter regions by the C-terminal part of MqsA. Yamaguchi and colleagues (2009) also found that the binding activity of MqsA is enhanced in the presence of MqsR compared with MqsA alone. This result is supported by our qRT-PCR result that the overexpression of MqsR/MqsA repressed more cspD transcription compared with that of MqsA alone.
Here, the lines of evidence that indicate the MqsR/MqsA TA pair directly regulate toxin CspD are (i) growth and biofilm results that show CspD is involved in MqsR-related toxicity (Fig. 4), (ii) MqsR/MqsA and MqsA regulate cspD transcription as shown by two sets of whole-transcriptome studies (Table 1) and qRT-PCR, (iii) the cspD promoter is bound directly by the MqsR/MqsA complex as well as by MqsA alone as shown by two sets of nickel-enrichment DNA microarray studies (Table S1) and by EMSA (Fig. 2), and (iv) qRT-PCR shows that induction of cspD by H2O2 stress is abolished by high levels of the MqsR/MqsA complex and by MqsA alone as well as by the absence of the mqsRA locus. Hence, the MqsR/MqsA TA system is a global regulator through its RNase activity (Brown et al., 2009) and through its regulation of the CspD toxin.
CspD inhibits chromosomal replication in nutrient-depleted cells (Yamanaka et al., 2001). As previously shown (Yamanaka et al., 2001) and reconfirmed in this work (Fig. 3A), CspD production is toxic. Furthermore, we linked MqsR to the nutrient starvation genes cstA, rpoS and dps (Table 1); hence, it is likely that MqsR induces starvation-like responses. TA systems are induced by starvation including depletion of amino acids and glucose, and the TA systems coordinately reduce DNA replication and translation (Gerdes, 2000). Hence, MqsR toxicity should be linked, either directly or indirectly, to induction of other TA systems including RelE/RelB. Corroborating this hypothesis, we established that toxin Hha induces genes of other TA pairs, such as RelE (threefold)/RelB (threefold), YoeB (sevenfold)/YefM (sevenfold) and YafQ (twofold)/DinJ (fourfold) (García-Contreras et al., 2008). Lastly, MqsR toxicity also involves the membrane toxin peptide HokA (Fig. 4A). Hence, the mechanism of MqsR toxicity is complex.
ClpXP is an important protease system for stress-induced environments and degrades RpoS (Zhou et al., 2001) and Dps (Stephani et al., 2003); hence, ClpXP induction by MqsR may lead to induction of stress-induced protection proteins regulated by RpoS and Dps which leads to toxicity. Moreover, other groups reported that labile antitoxins including MazE (Aizenman et al., 1996) and RelB (Christensen et al., 2001) are degraded by the ATP-dependent ClpP and Lon proteases respectively. Therefore, we propose that ClpXP might be involved in MqsR toxicity via MqsA-specific degradation. This is based on the observations that clpP is induced by mqsR overexpression for 15 min (the whole-transcriptome study; Table 1) and that the deletion of clpP abolishes MqsR toxicity (Fig. 4A). In addition, many genes encoding chaperones such as ibpA, ibpB, hslU, hslV, dnaK and dnaJ were directly influenced by MqsR; therefore, MqsR may prevent access of cells to key chaperone proteins. RpoS expression in E. coli is stimulated by a secreted extracellular signal (Holland and Rather, 2008); hence, the specific relationship among RpoS, ClpXP and AI-2 may be considered as a key factor for AI-2-related MqsR toxicity.
We showed previously that the balance between toxins and antitoxins has an important role in biofilm development (Kim et al., 2009). When bacteria encounter nutrient deficient or stressful conditions, toxins are activated by specific degradation of their antitoxins through cellular proteases (Gerdes, 2000), and it is this altered ratio between toxins and antitoxins that can influence biofilm formation (Kim et al., 2009). Consistent with this finding, production of MqsA increased biofilm formation whereas that of MqsR reduced biofilm formation (Fig. 1B); hence, we found additional evidence that the balance between the toxin and antitoxin influences biofilm formation. Similarly, for the RelE/RelB TA pair, excess of free RelE displaces or breaks the interaction between a C-terminal part of RelB and the binding site of RelE under environmental change, and thereby derepresses its own relBE transcription (Overgaard et al., 2008). Analogously, cspD is regulated by the ratio of MqsR and MqsA and involves MqsA decay through proteases Lon and ClpXP as a result of stress conditions. Hence, CspD production may be related to toxicity and biofilm formation via MqsR.
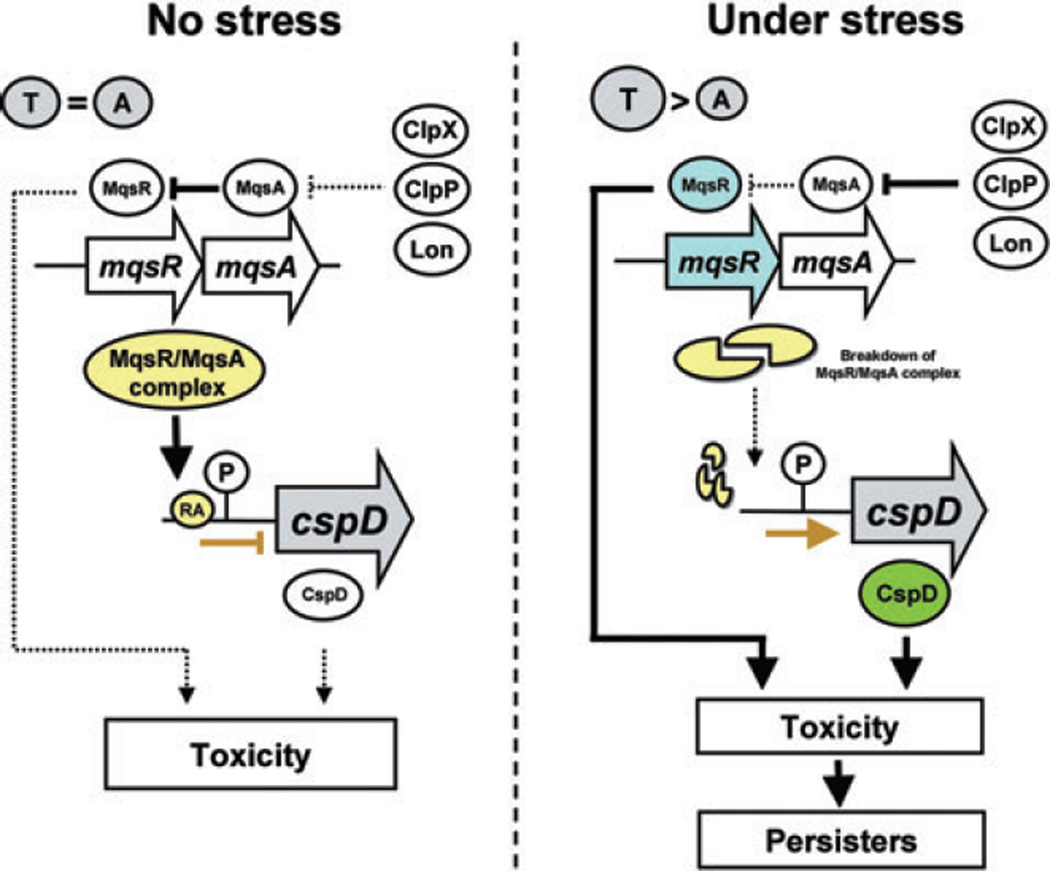
The induction of Hok-like proteins including HokA is very toxic to E. coli (Fig. 3B), resulting in loss of membrane potential, cell respiration arrest and efflux of small molecules (Gerdes et al., 1997). After inducing cell death via HokA, cell viability (cfu ml−1) and growth may be restored (Fig. 3B); a similar phenomenon occurs after induction of MqsR (Brown et al., 2009). Hence, our data suggest that MqsR potentially increases persister cell formation (Shah et al., 2006) via HokA. In addition, since mqsR transcription is strongly induced by several stresses including hydrogen peroxide (H2O2) (our qRT-PCR result and Zheng et al., 2001) and heat (50°C) (Richmond et al., 1999) and since mqsR is the most highly induced gene under persistence conditions (Shah et al., 2006), it appears that mqsRA transcription may be activated under stress conditions to increase persistence. Recently, we found that the deletion of the mqsRA locus repressed persister formation, that overproduction of MqsR/MqsA increases persistence, and that CspD also influences MqsR-related persister production (Kim and Wood, 2010). Taken together, these observations suggest that MqsR toxicity is due to both its RNase activity (Brown et al., 2009) and its interaction with MqsA to form a complex that acts as a DNA regulator which regulates at least other toxins like CspD (Fig. 5). Moreover, the CspD toxin is associated with persister production through MqsR (Kim and Wood, 2010).
Fig. 5.
Schematic of MqsR toxicity via CspD. cspD is repressed by the MqsR/MqsA complex. Under stress, MqsA is degraded by proteases (ClpXP and Lon), and cspD transcription is derepressed by the breakdown of the MqsR/MqsA complex. Stress also elevates the level of free-MqsR toxin. → indicates induction, ⊥ indicates repression, and dotted lines indicate regulatory pathways that are not active whereas solid lines indicate active regulatory pathways.
To our knowledge, this is first report that a TA system regulates the transcription of other toxins as well as its own, via direct binding to the promoter region. Given that MqsR is associated with MqsA as a specific regulator of QS that is directly associated with biofilm formation (González Barrios et al., 2006), our results support the idea that TA systems may form an important part of a regulatory network that senses cell density (Gerdes, 2000) and controls cell physiology.
Experimental procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. For isogenic mutants and overexpression of specific genes, we used the Keio collection (Baba et al., 2006) and ASKA library (Kitagawa et al., 2005), respectively, from the Genome Analysis Project in Japan. Strains BW25113 mqsR and MG1655 mqsR were verified via PCR, and the rest of the Keio knockouts were verified by Baba and colleagues (2006).
Table 2.
Bacterial strains and plasmids used in this study.
| Strains and plasmids | Genotype/relevant characteristics | Source |
|---|---|---|
| E. coli K-12 strains | ||
| TG1 | supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rk− mk−) [F’ traD36 proAB lacIqZΔM15] | Gibson (1984) |
| AG1 | recA1 endA1 gyrA96 thi-1 hsdR17 (rk− mk−) supE44 relA1 | Kitagawa et al. (2005) |
| MG1655 | F− λ− ilvG rfb-50 rph-1 | Blattner et al. (1997) |
| BW25113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | Baba et al. (2006) |
| MG1655 mqsR | MG1655 ΔmqsR Ω Tn5Kan-2 | Kang et al. (2004) |
| BW25113 mqsR | BW25113 ΔmqsR Ω KmR | Baba et al. (2006) |
| BW25113 mqsR mqsA | BW25113 ΔmqsR ΔmqsA Ω KmR | This study |
| BW25113 clpX | BW25113 ΔclpX Ω KmR | Baba et al. (2006) |
| BW25113 clpP | BW25113 ΔclpP Ω KmR | Baba et al. (2006) |
| BW25113 lon | BW25113 Δlon Ω KmR | Baba et al. (2006) |
| BW25113 yfjZ | BW25113 ΔyfjZ Ω KmR | Baba et al. (2006) |
| BW25113 cspD | BW25113 ΔcspD Ω KmR | Baba et al. (2006) |
| BW25113 relB | BW25113 ΔrelB Ω KmR | Baba et al. (2006) |
| BW25113 relE | BW25113 ΔrelE Ω KmR | Baba et al. (2006) |
| BW25113 hokA | BW25113 ΔhokA Ω KmR | Baba et al. (2006) |
| Plasmids | ||
| pCA24N | CmR; lacIq, pCA24N | Kitagawa et al. (2005) |
| pCA24N-mqsR | CmR; lacIq, pCA24N PT5-lac:mqsR+ | Kitagawa et al. (2005) |
| pCA24N-mqsA | CmR; lacIq, pCA24N PT5-lac:mqsA+ | Kitagawa et al. (2005) |
| pCA24N-cspD | CmR; lacIq, pCA24N PT5-lac:cspD+ | Kitagawa et al. (2005) |
| pCA24N-hokA | CmR; lacIq, pCA24N PT5-lac:hokA+ | Kitagawa et al. (2005) |
| pBS(Kan) | KmR; cloning vector | Canada et al. (2002) |
| pBS(Kan)-mqsR | KmR; pBS(Kan) Plac::mqsR+ | This study |
| pBS(Kan)-mqsA | KmR; pBS(Kan) Plac::mqsA+ | This study |
| pBS(Kan)-mqsR-mqsA | KmR; pBS(Kan) Plac::mqsR-mqsA+ | This study |
| pET28a(+)-mqsR | KmR; pET28a(+) PT7::mqsR+ | This study |
| pCA21a-mqsA | CmR; pCA21a PT7::mqsA+ | This study |
| pBAD-Myc-His C | AmpR; l-arabinose inducible expression vector | Invitrogen |
| pBAD-mqsA | AmpR; PBAD:mqsA+ in pBAD-Myc-His C | This study |
KmR, CmR and AmpR are kanamycin, chloramphenicol and ampicillin resistance, respectively.
All experiments were conducted in Luria–Bertani (LB) medium (Sambrook et al., 1989) at 37°C with the exception of LB supplemented with 0.5% glucose (LB glu) which was used to silence lac promoter transcription of toxin gene mqsR with pBS(Kan)-mqsR, and lower temperatures were used to overproduce proteins for the in vitro work. Kanamycin (50 µg ml−1) was used for pre-culturing isogenic knockout mutants and for maintaining the pBS(Kan)-based plasmids. Chloramphenicol (30 µg ml−1) was used for maintaining pCA24N-based plasmids.
Construction of overexpression plasmids
To construct plasmids for producing MqsR, MqsA and MqsR-MqsA from a lac promoter, fragments from genomic DNA were amplified by polymerase chain reaction (PCR) using primers containing BamHI and XbaI restriction sites (Table S2) and directionally cloned into the multiple cloning site in pBS(Kan) (Canada et al., 2002). Escherichia coli TG1 (Gibson, 1984) was used as the host for plasmid construction. The sizes of the amplified fragments were 376 bp for mqsR, 396 bp for mqsA and 768 bp for mqsR-mqsA. In addition, for the nickel-enrichment DNA microarray studies with MqsA, pBAD-mqsA was constructed with primers containing XhoI and HindIII restriction sites. This vector allows mqsA to be induced using the pBAD promoter. Cloned fragments were confirmed by DNA sequencing and restriction digestion with at least different three enzymes.
To overproduce MqsR for the in vitro studies, the mqsR sequence was PCR-amplified using primer set pE-mqsR (Table S2) and ligated into the NdeI and HindIII restriction enzyme sites of pET-28a(+) (Novagen, Madison, WI) to form pET-28a(+)-mqsR with MqsR expressed from a T7 promoter. pET-28a(+) has a pBR322 replication origin, is kanamycin resistant, and MqsR contains an N-terminal hexahistidine (His6) purification tag that is cleavable by thrombin. To overproduce MqsA, the mqsA sequence was PCR-amplified using primer set pA-mqsA (Table S2) and ligated into the NdeI and HindIII restriction enzyme sites of vector pCA21a (Expression Technologies, San Diego, CA) to form pCA21a-MqsA with mqsA expressed from a T7 promoter. pCA21a has a pACYC replication origin, is chloramphenicol resistant and does not contain any expression, solubility or purification tags.
Construction of BW25113 mqsR mqsA
The whole region encoding the transcript of mqsRA except for the first 30 bp from the start codon of mqsR was deleted from the chromosome using the one-step inactivation procedure (Baba et al., 2006) with the mqsR mqsA primers (Table S2). After isolating positive colonies, deletion of the mqsR mqsA locus was verified by DNA sequencing from PCR fragments using the mqsRA (detect) primer set (Table S2).
Growth and survival assays
The toxicity of selected proteins was investigated using pBS(Kan)- and pCA24N-based expression plasmids with 1 or 2 mM IPTG added upon inoculation (for CspD and HokA, IPTG was also added at a turbidity of 0.5 at 600 nm; Fig. 3). The mqsR mqsA double mutant was cultured initially in LB glu to silence mqsR on pBS(Kan)-mqsR, then cells were grown in LB with 1 mM IPTG. Cells were diluted by 102–107 via 10-fold serial dilution steps into 0.85% NaCl solution and applied as 10 µl drops on LB agar with kanamycin or chloramphenicol to determine cell viability (Donegan et al., 1991). Two independent cultures were used for each strain.
Crystal violet biofilm assay
The biofilm formation assay was performed in 96-well polystyrene plates (Corning Costar, Cambridge, MA) (Pratt and Kolter, 1998). Briefly, each well was inoculated at an initial turbidity at 600 nm of 0.05, grown for 24 h without shaking, and the cell density (turbidity at 620 nm) and total biofilm (absorbance at 540 nm) were measured using 0.1% crystal violet staining. Normalized biofilm was calculated by dividing total biofilm by bacterial growth for each strain. Two independent cultures were used for each strain.
RNA isolation and whole-transcriptome analysis
Three sets of whole-transcriptome analyses were performed: (i) planktonic cells of BW25113 mqsR versus BW25113 at a turbidity of 0.5 at 600 nm, (ii) planktonic cells of BW25113/pCA24N-mqsR versus BW25113/pCA24N with 2 mM IPTG added at a turbidity of 0.5 (grown for 3 h), and (iii) planktonic cells of BW25113 mqsR/pCA24N-mqsR versus BW25113 mqsR/pCA24N grown to a turbidity of 0.5 and then 2 mM IPTG was added for 15 min (Jonas et al., 2008). Total RNA was isolated from cells as described previously (Ren et al., 2004a) using a mini bead beater (Biospec, Bartlesville, OK) and RNeasy Mini Kit (Qiagen). cDNA synthesis, fragmentation and hybridizations were as described previously (González Barrios et al., 2006). The E. coli GeneChip Genome 2.0 array (Affymetrix, Santa Clara, CA; P/N 511302) was used. Corroborating the deletion mutations and overexpression of genes, the microarray signals of the mqsR gene had very low (deletion microarray) and high (overexpression microarray) signals in their respective microarray experiments. In addition, expected signals including araA and rhaA based on the E. coli K-12 BW25113 genotype (Table 2) were also low. For the three sets of binary microarray comparisons to determine differential genes expression, if the gene with the larger transcription rate did not have a consistent transcription rate based on the 11 probe pairs (P-value less than 0.05), these genes were not used. A gene was considered differentially expressed when the P-value for comparing two chips was lower than 0.05 (to assure that the change in gene expression was statistically significant and that false positives arise less than 5%). Since the standard deviation for expression ratio for all the genes was 1.4 for condition (i), 2.0 for condition (ii) and 1.2 for condition (iii), genes were considered differentially expressed if they had greater than twofold changes for condition (i), greater than threefold for condition (ii) and greater than twofold for condition (iii) (Ren et al., 2004b). Gene functions were obtained from the Ecogene database (http://www.ecogene.org/).
Nickel-enrichment DNA microarrays
We performed two sets of nickel-enrichment DNA microarray experiments to identify promoters bound by MqsA as described previously (Zhang et al., 2008). For the first set, His-tagged MqsR was overproduced and the MqsR/MqsA complex was isolated from cells in biofilms using AG1/pCA24N-mqsR and compared with AG1/pCA24N; mqsR was induced with 2 mM IPTG and cells were grown for 24 h in 250 ml of LB medium containing 10 g glass wool (Corning Glass Works, Corning, NY). For the second set, His-tagged MqsA was overproduced and isolated from planktonic cells using BW25113/pBAD-mqsA and compared with BW25113 /pBAD-Myc-His C. l-arabinose (0.5%) was added at a turbidity of 0.8 at 600 nm in LB medium and grown for 24 h. Briefly, formaldehyde (1%) was added for 20 min with shaking at room temperature to promote cross-linking between Histagged MqsR and its affiliated MqsA which co-purified with it and DNA or between His-tagged MqsA and the DNA to which it was associated. Ni-NTA agarose gel resin (Novagen) was used to bind His-tagged MqsR/MqsA–DNA or MqsA–DNA complexes after lysing cells with a French press. The DNA fragments were labelled and DNA microarrays were performed using an E. coli Genechip antisense genome array (P/N 4011592, Affymetrix) as indicated above. The analysis of the data were performed as described previously (Zhang et al., 2008).
Electrophoretic mobility shift assays (EMSA)
To confirm binding between the promoter regions and the MqsR/MqsA (full-length MqsA, isolated as described in Supporting information) and MqsR/MqsA-N (N-terminal region of MqsA containing 1–76 aa) (Brown et al., 2009), EMSA were performed as described previously (Zhang et al., 2008). The targeted promoter regions (region 150–250 bp upstream of the start codon using primers shown in Table S2) were amplified, purified, and then labelled with biotin using the Biotin 3′-end DNA Labelling Kit (Pierce Biotechnology, Rockford, IL). After binding the protein complex (200 ng) with the biotin-labelled target promoters (6 ng), electrophoresis was conducted at 100 V at 4°C using a 6% DNA retardation gel (Invitrogen). The bound protein/DNA mixtures were transferred to a nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany) using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA), and 3′-biotin-labelled DNA was detected with the Light-Shift Chemiluminescent EMSA kit (Pierce Biotechnology).
qRT-PCR
qRT-PCR was performed using the StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA). After isolating RNA (Ren et al., 2004a) using RNAlater™ (Ambion, Austin, TX), 50 ng of total RNA was used for the qRT-PCR reaction using the SuperScript™ III Platinum® SYBR® Green One-Step qRT-PCR Kit (Invitrogen) or Power SYBR® Green RNA-to-CT™ 1-Step Kit (Applied Biosystems). Primers were designed using Primer3Input Software (v0.4.0) and are listed in Table S2. The housekeeping gene rrsG was used to normalize the genes expression data. The annealing temperature was 60°C for all the genes in this study. To investigate the regulation of promoters by MqsA, overnight cultures of BW25113/pBS(Kan)-mqsR and BW25113/pBS(Kan)-mqsA were cultured to a turbidity of 1 at 600 nm, then 1 mM IPTG was added for 1 h to induce mqsR and mqsA. In addition, mqsA transcript levels in the BW25113 mqsR mutant were investigated after growing to a turbidity of 0.5 at 600 nm in LB. For the MqsR/MqsA overproduction and cspD transcription experiments, overnight cultures of BW25113 mqsR mqsA/pBS(Kan)-mqsR-mqsA, BW25113 mqsR mqsA/pBS(Kan)-mqsA and BW25113 mqsR mqsA/pBS(Kan) were grown to a turbidity of 0.5 at 600 nm, and then 1 mM IPTG was added for 1 h. For the H2O2 stress and clpX, clpP, lon, cspD and mqsR transcription experiments, overnight cultures of the BW25113 wild type and BW25113 mqsR mqsA were grown to a turbidity of 1.0 at 600 nm, and then exposed to 30 mM H2O2 for 15 min (Lee et al., 2007). In addition, to examine whether MqsR/MqsA complex affects cspD transcript by H2O2 stress, overnight cultures of the BW25113 mqsR mqsA/pBS(Kan)-mqsR-mqsA and BW25113 mqsR mqsA/pBS(Kan) were inoculated into LB medium and grown to a turbidity of 1.0 at 600 nm with 1 mM IPTG for induction of the MqsR/MqsA complex, and then exposed to 30 mM H2O2 for 15 min.
Microarray accession numbers
The differential gene expression data have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Accession No. GSE14203.
Supplementary Material
Acknowledgements
This work was supported by the NIH (R01 EB003872) and the ARO (W911NF-06-1-0408). We are grateful for the Keio and ASKA strains provided by the Genome Analysis Project in Japan. W.P. is the Manning Assistant Professor of Medical Science at Brown University and T.W. is the T. Michael O’Connor Endowed Professor at Texas A&M University.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Growth curves and cell viability (cfu ml−1) for BW25113 cspD/pCA24N and BW25113 cspD/pCA24N-cspD (A), and for BW25113 hokA/pCA24N and BW25113 hokA/pCA24N-hokA (B). pCA24N-based genes were induced with 1 mM IPTG at 0 min. Data are the average of two independent cultures, and one standard deviation is shown.
Table S1. DNA binding sites for MqsR/MqsA and MqsA identified in vivo using nickel-enrichment DNA microarrays. Enrichment indicates the ratio of the MqsR/MqsA and MqsA signal relative to the empty plasmid controls, pCA24N and pBAD, respectively.
Table S2. Oligonucleotides used in this study. f indicates forward primer and r indicates reverse primer.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by 3’,5’-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almirón M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Brown BL, Grigoriu S, Kim Y, Arruda JM, Davenport A, Wood TK, et al. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5:e1000706. doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Shaw KJ. A novel family of Escherichia coli toxin–antitoxin gene pairs. J Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde PP, Davis BM, Yuan J, Waldor MK. Characterization of a higBA toxin–antitoxin locus in Vibrio cholerae. J Bacteriol. 2007;189:491–500. doi: 10.1128/JB.00909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canada KA, Iwashita S, Shim H, Wood TK. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J Bacteriol. 2002;184:344–349. doi: 10.1128/JB.184.2.344-349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny I, Gazit E. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J Biol Chem. 2004;279:8252–8261. doi: 10.1074/jbc.M308263200. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli : involvement of the yefM–yoeB toxin–antitoxin system. Mol Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, et al. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol Cell. 2009;35:154–163. doi: 10.1016/j.molcel.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Domka J, Lee J, Wood TK. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol. 2006;72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Donegan K, Matyac C, Seidler R, Porteous A. Evaluation of methods for sampling, recovery, and enumeration of bacteria applied to the phylloplane. Appl Environ Microbiol. 1991;57:51–56. doi: 10.1128/aem.57.1.51-56.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Contreras R, Zhang X-S, Kim Y, Wood TK. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS ONE. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K. Toxin–antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Bech FW, Jørgensen ST, Løbner-Olesen A, Rasmussen PB, Atlung T, et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Gultyaev AP, Franch T, Pedersen K, Mikkelsen ND. Antisense RNA-regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gibson TJ. PhD Thesis. Cambridge, UK: Cambridge University; 1984. Studies on the Epstein-Barr virus genome. [Google Scholar]
- González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- Holland A-M, Rather PN. Evidence for extracellular control of RpoS proteolysis in Escherichia coli. FEMS Microbiol Lett. 2008;286:50–59. doi: 10.1111/j.1574-6968.2008.01255.x. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors Ö. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. Systematic mutagenesis of the Escherichia coli genome. J Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wang X, Ma Q, Zhang X-S, Wood TK. Toxin–antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Page R, García-Contreras R, Palermino J-M, Zhang X-S, Doshi O, et al. Structure and function of the Escherichia coli protein YmgB: a protein critical for biofilm formation and acid-resistance. J Mol Biol. 2007;373:11–26. doi: 10.1016/j.jmb.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson RD. Hypothetical functions of toxin–antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai-Prochnow A, Webb JS, Ferrari BC, Kjelleberg S. Ecological advantages of autolysis during the development and dispersal of Pseudoalteromonas tunicata biofilms. Appl Environ Microbiol. 2006;72:5414–5420. doi: 10.1128/AEM.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiejūnaitė R, Armalytė JM, Markuckas A, Sužiedėlienė E. Escherichia coli dinJ–yafQ genes act as a toxin–antitoxin module. FEMS Microbiol Lett. 2007;268:112–119. doi: 10.1111/j.1574-6968.2006.00563.x. [DOI] [PubMed] [Google Scholar]
- Nieto C, Cherny I, Khoo SK, de Lacoba MG, Chan WT, Yeo CC, et al. The yefM–yoeB toxin–antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J Bacteriol. 2007;189:1266–1278. doi: 10.1128/JB.01130-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Borch J, Jørgensen MG, Gerdes K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol Microbiol. 2008;69:841–857. doi: 10.1111/j.1365-2958.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004a;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol Bioeng. 2004b;88:630–642. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt O, Schuenemann VJ, Hand NJ, Silhavy TJ, Martin J, Lupas AN, Djuranovic S. prlF and yhaV encode a new toxin–antitoxin system in Escherichia coli. J Mol Biol. 2007;372:894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JE, Matin A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J Mol Biol. 1991;218:129–140. doi: 10.1016/0022-2836(91)90879-b. [DOI] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky AB, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H, Kato T, Ise T, Makino K, Nakata A. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene. 1983;23:167–174. doi: 10.1016/0378-1119(83)90048-3. [DOI] [PubMed] [Google Scholar]
- Spoering AL, Vulić M, Lewis K. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol. 2006;188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephani K, Weichart D, Hengge R. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol Microbiol. 2003;49:1605–1614. doi: 10.1046/j.1365-2958.2003.03644.x. [DOI] [PubMed] [Google Scholar]
- Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin–antitoxin systems in its genome? J Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Zheng W, Crooke E, Wang YH, Inouye M. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol Microbiol. 2001;39:1572–1584. doi: 10.1046/j.1365-2958.2001.02345.x. [DOI] [PubMed] [Google Scholar]
- Zhang X-S, García-Contreras R, Wood TK. Escherichia coli transcription factor YncC (McbR) regulates colanic acid and biofilm formation by repressing expression of periplasmic protein YbiM (McbA) ISME J. 2008;2:615–631. doi: 10.1038/ismej.2008.24. [DOI] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets σs for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.