Abstract
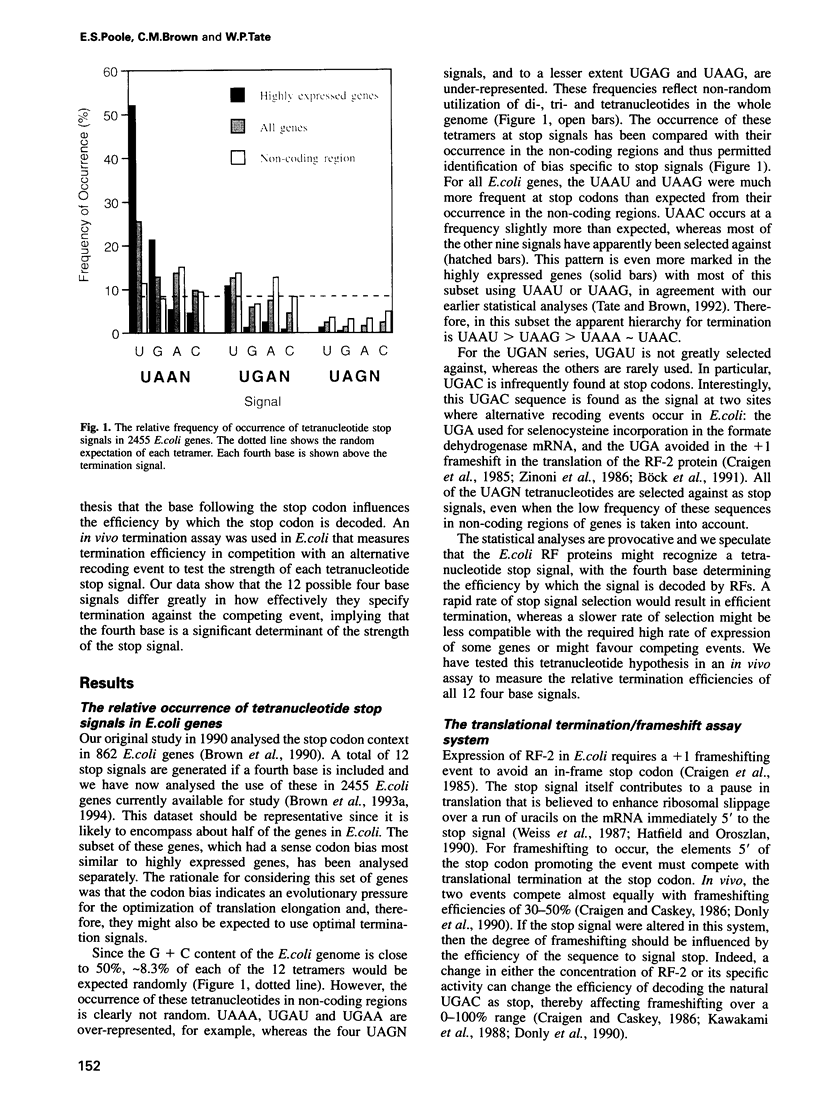
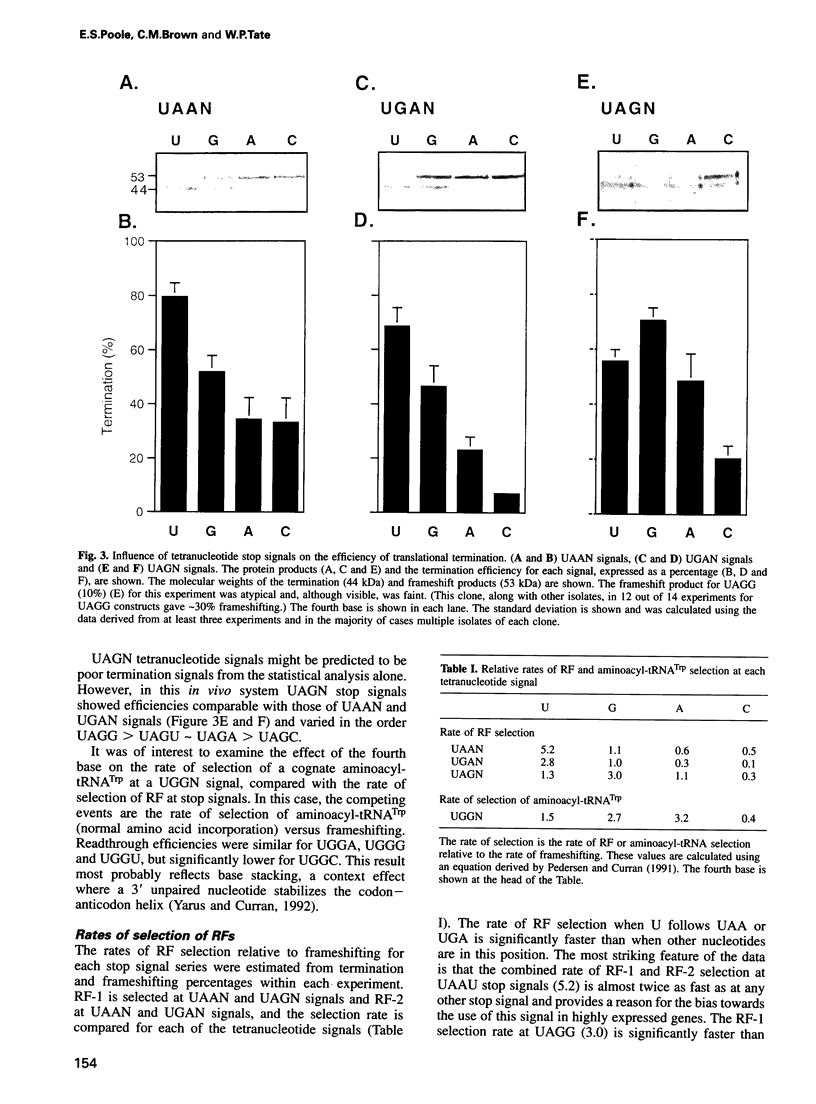
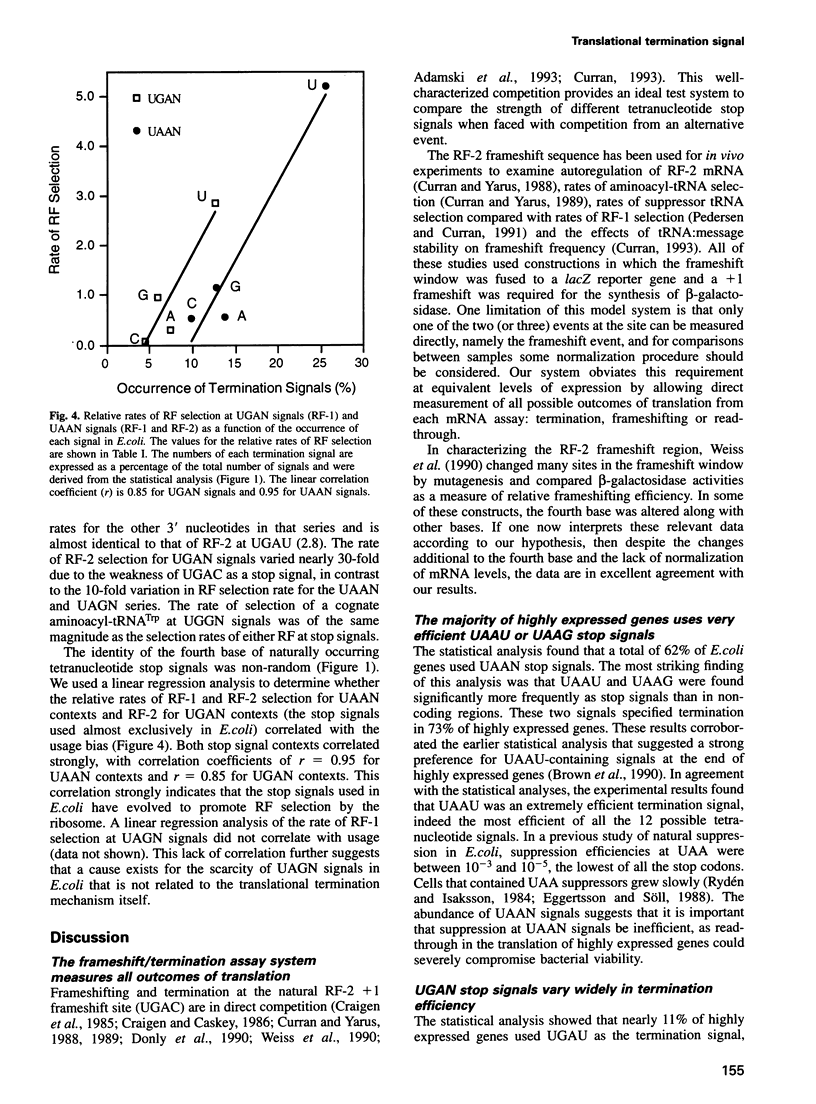
A statistical analysis of > 2000 Escherichia coli genes suggested that the base following the translational stop codon might be an important feature of the signal for termination. The strengths of each of 12 possible 'four base stop signals' (UAAN, UGAN and UAGN) were tested in an in vivo termination assay that measured termination efficiency by its direct competition with frameshifting. Termination efficiencies varied significantly depending on both the stop codon and the fourth base, ranging from 80 (UAAU) to 7% (UGAC). For both the UAAN and UGAN series, the fourth base hierarchy was U > G > A approximately C. UAG stop codons, which are used rarely in E. coli, showed efficiencies comparable with UAAN and UGAN, but differed in that the hierarchy of the fourth base was G > U approximately A > C. The rate of release factor selection varied 30-fold at UGAN stop signals, and 10-fold for both the UAAN and UAGN series; it correlated well with the frequency with which the different UAAN and UGAN signals are found at natural termination sites. The results suggest that the identity of the base following the stop codon determines the efficiency of translational termination in E. coli. They also provide a rationale for the use of the strong UAAU signal in highly expressed genes and for the occurrence of the weaker UGAC signal at several recording sites.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamski F. M., Donly B. C., Tate W. P. Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 1993 Nov 11;21(22):5074–5078. doi: 10.1093/nar/21.22.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamski F. M., McCaughan K. K., Jørgensen F., Kurland C. G., Tate W. P. The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J Mol Biol. 1994 May 6;238(3):302–308. doi: 10.1006/jmbi.1994.1293. [DOI] [PubMed] [Google Scholar]
- Arkov A. L., Korolev S. V., Kisselev L. L. Termination of translation in bacteria may be modulated via specific interaction between peptide chain release factor 2 and the last peptidyl-tRNA(Ser/Phe). Nucleic Acids Res. 1993 Jun 25;21(12):2891–2897. doi: 10.1093/nar/21.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J Mol Biol. 1983 Feb 15;164(1):73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Brown C. M., Dalphin M. E., Stockwell P. A., Tate W. P. The translational termination signal database. Nucleic Acids Res. 1993 Jul 1;21(13):3119–3123. doi: 10.1093/nar/21.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., McCaughan K. K., Tate W. P. Two regions of the Escherichia coli 16S ribosomal RNA are important for decoding stop signals in polypeptide chain termination. Nucleic Acids Res. 1993 May 11;21(9):2109–2115. doi: 10.1093/nar/21.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Dalphin M. E., Tate W. P. The translational termination signal database (TransTerm) now also includes initiation contexts. Nucleic Acids Res. 1994 Sep;22(17):3620–3624. doi: 10.1093/nar/22.17.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham R. H., Sörensen P., Pagel F. T., Hijazi K. A., Mims B. H., Brechemier-Baey D., Murgola E. J. Third position base changes in codons 5' and 3' adjacent UGA codons affect UGA suppression in vivo. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):259–262. doi: 10.1016/0167-4781(90)90177-4. [DOI] [PubMed] [Google Scholar]
- Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B., Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991 Mar;5(3):515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Lee C. C., Caskey C. T. Recent advances in peptide chain termination. Mol Microbiol. 1990 Jun;4(6):861–865. doi: 10.1111/j.1365-2958.1990.tb00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. The genetic code--yesterday, today, and tomorrow. Cold Spring Harb Symp Quant Biol. 1966;31:1–9. [PubMed] [Google Scholar]
- Curran J. F. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993 Apr 25;21(8):1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989 Sep 5;209(1):65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988 Sep 5;203(1):75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Donly B. C., Edgar C. D., Adamski F. M., Tate W. P. Frameshift autoregulation in the gene for Escherichia coli release factor 2: partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 1990 Nov 25;18(22):6517–6522. doi: 10.1093/nar/18.22.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson G., Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988 Sep;52(3):354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Salser W., Epstein R. H. The influence of the reading context upon the suppression of nonsense codons. Mol Gen Genet. 1977 Mar 7;151(2):137–149. doi: 10.1007/BF00338688. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Hatfield D. L., Smith D. W., Lee B. J., Worland P. J., Oroszlan S. Structure and function of suppressor tRNAs in higher eukaryotes. Crit Rev Biochem Mol Biol. 1990;25(2):71–96. doi: 10.3109/10409239009090606. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971 Jun 14;58(2):439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Jørgensen F., Kurland C. G. Processivity errors of gene expression in Escherichia coli. J Mol Biol. 1990 Oct 20;215(4):511–521. doi: 10.1016/S0022-2836(05)80164-0. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Inada T., Nakamura Y. Conditionally lethal and recessive UGA-suppressor mutations in the prfB gene encoding peptide chain release factor 2 of Escherichia coli. J Bacteriol. 1988 Nov;170(11):5378–5381. doi: 10.1128/jb.170.11.5378-5381.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe H. V., Costello D., Wong A., Lu R. C., Wohlrab H. Mitochondrial phosphate transport. Large scale isolation and characterization of the phosphate transport protein from beef heart mitochondria. J Biol Chem. 1984 Jul 25;259(14):9115–9120. [PubMed] [Google Scholar]
- Kopelowitz J., Hampe C., Goldman R., Reches M., Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol. 1992 May 20;225(2):261–269. doi: 10.1016/0022-2836(92)90920-f. [DOI] [PubMed] [Google Scholar]
- Marshall B., Levy S. B. Prevalence of amber suppressor-containing coliforms in the natural environment. Nature. 1980 Jul 31;286(5772):524–525. doi: 10.1038/286524a0. [DOI] [PubMed] [Google Scholar]
- Martin R., Weiner M., Gallant J. Effects of release factor context at UAA codons in Escherichia coli. J Bacteriol. 1988 Oct;170(10):4714–4717. doi: 10.1128/jb.170.10.4714-4717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Bhagwat A. S. Biased DNA repair. Nature. 1992 Feb 13;355(6361):595–596. doi: 10.1038/355595b0. [DOI] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989 Sep;53(3):273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen W. T., Curran J. F. Effects of the nucleotide 3' to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J Mol Biol. 1991 May 20;219(2):231–241. doi: 10.1016/0022-2836(91)90564-m. [DOI] [PubMed] [Google Scholar]
- Phillips G. J., Arnold J., Ivarie R. The effect of codon usage on the oligonucleotide composition of the E. coli genome and identification of over- and underrepresented sequences by Markov chain analysis. Nucleic Acids Res. 1987 Mar 25;15(6):2627–2638. doi: 10.1093/nar/15.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott C. D., Kleuvers B., Göringer H. U. A rRNA-mRNA base pairing model for UGA-dependent termination. Biochimie. 1991 Jul-Aug;73(7-8):1121–1129. doi: 10.1016/0300-9084(91)90155-t. [DOI] [PubMed] [Google Scholar]
- Purohit P., Stern S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994 Aug 25;370(6491):659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Fuchs R., Higgins D. G., Stoehr P. J., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1993 Jul 1;21(13):2967–2971. doi: 10.1093/nar/21.13.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydén S. M., Isaksson L. A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193(1):38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- Salser W., Fluck M., Epstein R. The influence of the reading context upon the suppression of nonsense codons. 3. Cold Spring Harb Symp Quant Biol. 1969;34:513–520. doi: 10.1101/sqb.1969.034.01.058. [DOI] [PubMed] [Google Scholar]
- Salser W. The influence of the reading context upon the suppression of nonsense codons. Mol Gen Genet. 1969 Oct 13;105(2):125–130. doi: 10.1007/BF00445682. [DOI] [PubMed] [Google Scholar]
- Scolnick E., Tompkins R., Caskey T., Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968 Oct;61(2):768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986 Aug 26;14(16):6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W. P., Brown C. M. Translational termination: "stop" for protein synthesis or "pause" for regulation of gene expression. Biochemistry. 1992 Mar 10;31(9):2443–2450. doi: 10.1021/bi00124a001. [DOI] [PubMed] [Google Scholar]
- Tate W., Greuer B., Brimacombe R. Codon recognition in polypeptide chain termination: site directed crosslinking of termination codon to Escherichia coli release factor 2. Nucleic Acids Res. 1990 Nov 25;18(22):6537–6544. doi: 10.1093/nar/18.22.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Ribosomal frameshifting from -2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]




