Abstract
Whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a rapid method for identification of microorganisms that is increasingly used in microbiology laboratories. This identification is based on the comparison of the tested isolate mass spectrum with reference databases. Using Neisseria meningitidis as a model organism, we showed that in one of the available databases, the Andromas database, 10 of the 13 species-specific biomarkers correspond to ribosomal proteins. Remarkably, one biomarker, ribosomal protein L32, was subject to inter-strain variability. The analysis of the ribosomal protein patterns of 100 isolates for which whole genome sequences were available, confirmed the presence of inter-strain variability in the molecular weight of 29 ribosomal proteins, thus establishing a correlation between the sequence type (ST) and/or clonal complex (CC) of each strain and its ribosomal protein pattern. Since the molecular weight of three of the variable ribosomal proteins (L30, L31 and L32) was included in the spectral window observed by MALDI-TOF MS in clinical microbiology, i.e., 3640–12000 m/z, we were able by analyzing the molecular weight of these three ribosomal proteins to classify each strain in one of six subgroups, each of these subgroups corresponding to specific STs and/or CCs. Their detection by MALDI-TOF allows therefore a quick typing of N. meningitidis isolates.
Keywords: Mass spectrometry, Ribosomal proteins, Biomarkers, Neisseria meningitidis
1. Introduction
Whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) generates a spectrum based on proteins detected directly from intact microorganisms (Holland et al., 1996; Williams et al., 2003), allowing the rapid identification of bacterial isolates. This identification relies on comparison of the spectra of the sample with those of reference databases. The Andromas database was engineered using an algorithm that identifies a limited number of species-specific peaks for each entry (Carbonnelle et al., 2007; Degand et al., 2008). Briefly, to engineer the database, a set of reference isolates was chosen, and ten subcultures of each of these selected isolates, grown on different media, were analyzed. For each strain, only those peaks with a relative intensity above 0.07, and which that were constantly present in all 10 sets of data obtained for a given strain, were retained. With the Andromas database, accurate species identification is obtained if at least 68% of the species-specific peaks are present in the spectrum of the subject isolate. The failure to identify some specimens is explained by small protein variations among isolates of the same species, or by the fact that some peaks of the database cannot be observed because of the poor quality of spectra obtained from whole bacteria grown in primary culture.
In this work, we aimed at answering two questions regarding the implementation of mass spectrometry in clinical laboratories. (i) Although many studies have showed that peaks used for identification species of bacteria are ribosomal protein (Lay, 2001; Pineda et al., 2003; Ryzhov and Fenselau, 2001; Teramoto et al., 2007a, 2007b), we wanted to identify the exact nature of the biomarkers empirically used to build the Andromas database for bacterial species identification. (ii) We next aimed at identifying markers specific of strain and/or groups of strains and compare them to reliable epidemiological methods. Ribosomal proteins are good candidates for such an approach as they are universal amongst cellular life. Indeed, even though most ribosomal proteins are highly conserved within a bacterial species, some of these proteins are subject to slight variations at the strain level. As the variations of the ribosomal protein genes have been proposed for classification and typing purposes (Bennett et al., 2012; Jolley et al., 2012; Kozo, 1989; Matte-Tailliez et al., 2002; Roberts et al., 2008; Yutin et al., 2012), analysis of the ribosomal protein masses in a MALDI-TOF spectrum directly from intact bacterial cells could provide an interesting epidemiological tool for the classification of bacterial isolates to the sub-species or strain level. The use of ribosomal markers detected by MALDI-TOF would then dramatically speed up epidemiological studies in the clinical laboratory and in environmental microbiology.
For epidemiological studies, strains are routinely typed using multilocus sequence typing (MLST). They are subsequently compared by sequencing the internal fragment of seven housekeeping genes. Strains having similar sequence belong to the same sequence type (ST). STs are grouped into clonal complexes (CC) by their similarity to a central allelic profile (http://pubmlst.org/). In addition, using only ribosomal proteins, Jolley et al. (2012) showed that ribosomal multilocus sequence typing (rMLST) of Streptococcus pneumoniae had strain level resolution. Here, we examined isolates of the bacterial pathogen Neisseria meningitidis to determine the bacterial components corresponding to the species-specific peaks combining genomic and proteomic approaches (Demirev et al., 2005; Dworzanski et al., 2004; Ryzhov and Fenselau, 2001). Taking into account genomic sequence data of 100 isolates, we correlated the ribosomal protein profile of each isolates with its ST and CC. Use of these data allows classifying each isolate to a subgroup on the basis of spectra obtained in a routine clinical setting.
2. Material and methods
2.1. Strains
One hundred clinical isolates of N. meningitidis included in the 107 strains collection used to establish multilocus sequence typing (Maiden et al, 1998) was obtained from D. Caugant, WHO Collaborating Center for Reference and Research on Meningococci, Norwegian Institute of Public Health, Oslo. In addition, two previously described isolates, N. meningitidis NEM 8013 and N. meningitidis Z2491, were used. All isolates were grown on GCB (Gonococcal Medium Base) agar plates (Difco) containing Kellog’s supplements at 37 °C in 5% atmosphere for 18 h, harvested and inactivated in 70% ethanol. Pellets were conserved at −80 °C.
2.2. Proteolysis
Proteins of strains NEM 8013 and Z2491 were extracted from bacterial cells with 70% formic acid/acetonitrile (v/v) and the suspension centrifuged at 13000 g. The supernatant was then dried, and 5–100 μg of protein was dissolved in an appropriate volume of 10 mM Tris–HCl buffer, pH 7.0, containing 0.1% (w/v) SDS and 0.15% (w/v) dithioerythiol (DTT) to give a protein concentration of 1 μg/μl. The solutions were incubated at 90 °C for 60 s to reduce proteins and left to cool to room temperature. A volume of 5 μl of 0.5 M iodoacetamide (an excess) was added to the mixture, which was incubated at 37 °C for 30 min in the dark to carboaminomethylate the cysteine residues. Excess reagent and low molecular weight products were removed by ultrafiltration (Amicon, Millipore, Ireland), and the proteins was concentrated by centrifugal evaporation. Derivatised proteins were reconstituted in water to a final concentration of 1 μg/μl, and 5 μl was added to 190 μl of the appropriate reaction buffer: for endoproteinase GluC (protease V8), 50 mM sodium phosphate buffer, pH7.8, five microliters of the protease V8 (0.1 μg/μl) (ThermoScientific, USA) were added to the reaction mixture. After incubation at 37 °C for 16 h, the reactions were stopped by heating at 90 °C for 30 s. The samples were concentrated and residual acetonitrile and TFA removed under vacuum before reconstitution in H20.
2.3. De-O-glycosylation and dialysis
Proteins of strains NEM 8013 and Z2491 were extracted from bacterial cells with 70% formic acid/acetonitrile (v/v). The suspension was centrifuged at 13000 g. The resulting supernatant was dried out and the pellet resuspended in water. Twenty micrograms of extracted proteins were digested by endo-α-N-acetylgalactosaminidase (Biolabs, Great Britain). A slide-A-Lyser Dialysis Cassette 3.5K MCWO (ThermoScientific, USA) was used to desalt the two resulting suspensions before MALDI-TOF mass spectrometry.
2.4. MALDI-TOF mass spectrometry analysis
The protein suspensions (1 μl) or a swab of whole bacteria obtained from colonies was spotted onto a MALDI sample target and allowed to dry at room temperature. For each of the 102 strains, 10 MALDI-TOF MS spectra were recorded from different colonies. One microliter of matrix HCCA (saturated solution of cyano-4-hydroxycinnamic acid in acetonitrile 50%, trifluoroacetic acid 2.5%) was then added and allowed to co-crystallize with the whole bacteria. Samples were processed in the MALDI-TOF MS spectrometer (LT2-Andromas, Andromas SAS, France) with the MALDI Control software (Andromas SAS, France). Positive ions were extracted with an accelerating voltage of 20 kV in linear mode. Each spectrum was the sum of the ions obtained from 400 laser shots performed automatically on different regions of the same well. The spectra were analyzed in the 3640 to 12000 m/z range using the Andromas software.
2.5. Protein sequence analysis
The complete proteome of strains N. meningitidis NEM 8013 and Z2491 were extracted from the MicroScope Database (http://www.genoscope.cns.fr/agc/microscope/home/index.php), and the theoretical molecular weight (MW) of the protein corresponding to each open reading frame was deduced from the sequence with an online tool (http://www.bioinformatics.org/sms2/protein_mw.html). In addition, the DNA of the isolates NEM 8013 and Z2491 was extracted using Wizard® Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s instruction. The genes MNV_2186 and NMA0455 were amplified using the following primers: forward primer GGTTTGGCTTTCAGACGGTA; reverse primer GTCTTGCCGTATGGTTTCGT. Amplification was performed using 2 μl of template, 5 μl of 1× Dream Taq buffer (Fermentas, USA), 0.2 mM of each dNTP, 0.5 μM of each primer and 5 U of DreamTaq polymerase (Fermentas), qs 50 μl. Target DNA was amplified with 30 cycles (94 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min) with an initial denaturation step at 95 °C for 5 min and a final elongation step at 72 °C for 10 min. PCR products were checked on a 1% agar gel and directly sequenced using an ABI Prism automated sequencer (GATC-biotech, http://www.gatc-biotech.fr).
2.6. nanoLC-MS/MS analysis
nanoLC-MS/MS (nanoliquid chromatography coupled to tandem mass spectrometry) was performed according to the protocol described earlier (Christie-Oleza et al., 2013). Pelleted cells (two plates) of the two N. meningitidis strains were resuspended in 300 ml of lithium dodecyl sulfate-β-mercaptoethanol sample buffer for SDS–PAGE (Invitrogen, UK) and incubated at 99 °C for 10 min prior to SDS–PAGE. The cell samples were directly loaded onto a 12% Tris–Bis NuPAGE gel (Invitrogen). SDS–PAGE was carried out using 1× 3-(N-morpholino)propanesulfonic acid solution (Invitrogen) as the running buffer. Proteins were resolved over a 6-cm migration in order to separate those proteins with a low molecular weight. A single gel band containing low-molecular-weight proteins (<20 kDa) was cut for an in-gel proteolysis with trypsin (Roche, France) following the ProteasMax protocol (Promega) as previously described (Clair et al., 2012). NanoLC-MS/MS experiments were performed using the LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher, USA) coupled to an UltiMate 3000 LC system (Dionex-LC Packings) using conditions previously described (de Groot et al., 2009). MS/MS spectra were searched against homemade protein sequence databases containing all the annotated CDS sequences of the N. meningitidis NEM 8013 or Z2491 genomes. Searches were carried out using the MASCOT 2.2.04 software (Matrix Science).
2.7. Ribosomal proteins
The molecular weights of ribosomal proteins were predicted from 100 genomes stored in the PubMLST Neisseria database using the BIGSdb export plugin (Jolley and Maiden, 2010). This uses standard BioPerl modules (Stajich et al., 2002) to translate the complete identified coding sequences and predict molecular weight based on amino acid composition.
2.8. Cluster analysis
Phylogenetic analysis based on the MALDI mass spectra was conducted by the Bionumerics software (version 5.1, Applied Maths, Kortrijk, Belgium) using a similarity coefficient curve based on Pearson correlation. The phylogenetic tree was built by the unweighted pair group method with arithmetic mean (UPGMA).
3. Results and discussion
3.1. Reproducibility of MALDI-TOF MS species-specific biomarkers
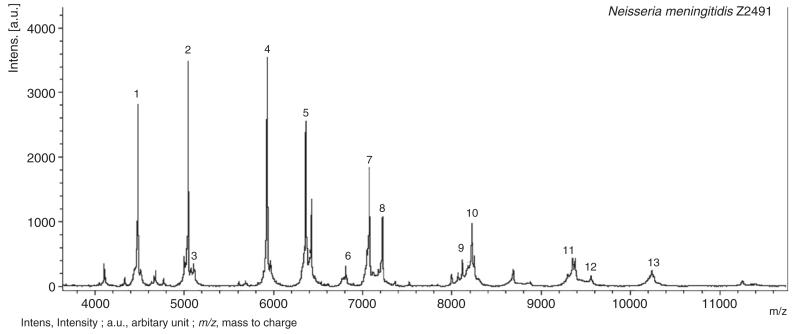
The Andromas database includes 13 species-specific peaks for N. meningitidis. A MALDI mass spectrum of the whole-cell suspensions of N. meningitidis Z2491 is shown in Fig. 1. Our first aim was to assess the reproducibility of the species-specific peaks identified in the database among colonies of a single strain and between strains of the same species. We performed 10 MALDI-TOF MS acquisition of each of the 102 isolates (Table 1). Most of the peaks were highly conserved, with 10 of 13 peaks present in over 97% of the MS acquisitions. Only peaks 5, 11 and 13, as labeled in Fig. 1, were observed in less than 97% of the MS acquisitions (78%, 89% and 90%, respectively). It should be noted out that this variability never prevented accurate species identification, as in all acquisitions the diagnostic result obtained with the Andromas software was N. meningitidis.
Fig. 1. Mass spectra of whole cell of Neisseria meningitidis Z2491.
Table 1. Presence of the Neisseria meningitidis biomarkers among the 102 tested strains.
| Peaks number | m/z (database Andromas) | % Detection of each peak among the 10 acquisitions of the 102 strains (i.e., 1020 acquisitions) | Numbers of strains which do not have the corresponding biomarker at least once, among the 10 acquisitions ofeach strain |
|---|---|---|---|
| 1 | 4486 | 99 | 1 |
| 2 | 5049 | 100 | 0 |
| 3 | 5618 | 100 | 0 |
| 4 | 5936 | 100 | 0 |
| 5 | 6343 | 78 | 22 |
| 6 | 6434 | 97 | 2 |
| 7 | 7067 | 100 | 0 |
| 8 | 7230 | 99 | 0 |
| 9 | 8126 | 98 | 1 |
| 10 | 8230 | 100 | 0 |
| 11 | 8698 | 89 | 0 |
| 12 | 9360 | 100 | 0 |
| 13 | 9393 | 90 | 0 |
m/z: mass/charge.
3.2. Identification of the nature of the species-specific biomarkers
We next assessed whether the biomarkers were proteins. Using the endoproteinase GluC, we demonstrated that all species-specific peaks were proteins as they disappeared (10) or strongly decreased (3) after enzymatic proteolysis (data not shown). In addition, we performed an enzymatic digestion with an endo-α-N-acetylgalactosaminidase that removes O-glycosylation and did not observe modification of the m/z ratio of the species-specific peaks, thus ruling out a possible O-glycosylation of these biomarkers; indeed, N-glycosylation has never been described in N. meningitidis.
The molecular weight of the biomarkers were compared to the putative proteomes of two isolates, NEM8013 and Z2491, using the MicroScope database, taking into account the possible loss of the first methionine of the protein (Gonzales and Robert-Baudouy, 1996). We observed that for 5 peaks, the theoretical masses were different from the observed masses due to the loss of the first methionine residue of the tentatively assigned proteins (Tables 2 and 3). In addition, a shotgun proteomic approach was performed using nanoLC-MS/MS in order to list the small molecular weight proteins abundantly present in cells in these physiological conditions and confirm the identities of these biomarkers (Tables 2 and 3). For isolate NEM8013 (Table 2), the bioinformatics approach allowed the identification of 11 of the 13 biomarkers; however, in 3 cases (peaks 7, 8 and 13), the genomic approach gave two possibilities. On the other hand, the proteomic approach was contributive as most protein assignments were confirmed and only one protein was finally assigned for the peaks of isolate NEM8013. Regarding strain Z2491 (Table 3), nine biomarkers were identified by the bioinformatics approach, and an additional peak (peak 12) was identified using shotgun proteomics. Comparative proteomics of both strains allows clarifying the identity of Peak 12 which corresponds to the DNA binding protein HU (NMV_1200 hup and NMA1397 for strains NEM 8013 and Z2491, respectively).
Table 2. Biomarker assignments by means of proteogenomics for the NEM 8013 strain.
| MicroScope database |
|||||
|---|---|---|---|---|---|
| Peaks number | Observed m/z | Theoretical molecular weight including PTM | Protein assignment by sequence comparison | Protein assignment by shotgun MS/MS | PTM |
| 1 | Not present in this strain | – | – | – | – |
| 2 | 5051 | 5048 | 50S protein ribosomal L34 | 50S protein ribosomal L34 | – |
| 3 | 5620 | 5617 | Hypothetical membrane associated protein (NMV_2186) | Hypothetical membrane associated protein | – |
| 4 | 5938 | 5933 | 50S ribosomal protein L33 | 50S ribosomal protein L33 | – |
| 5 | 6342 | 6338 | 50S ribosomal protein L32 | 50S ribosomal protein L32 | Methionine removed |
| 6 | 6429 | ND | ND | ND | ND |
| 7 | 7079 | 7074 | 50S ribosomal protein L29 or putative heavy-metal scavenger protein (MNV_1128) | 50S ribosomal protein L29 | – |
| 8 | 7227 | 7221 | 50S ribosomal protein L35 or truncated conserved hypothetical phage protein (NMV_1277) | 50S ribosomal protein L35 | Methionine removed |
| 9 | 8112 | 8113 | 50S ribosomal protein L31 | 50S ribosomal protein L31 | – |
| 10 | 8251 | 8249 | 30S ribosomal protein S21 | 30S ribosomal protein S21 | Methionine removed |
| 11 | 8686 | 8682 | 50S ribosomal protein L28 | 50S ribosomal protein L28 | Methionine removed |
| 12 | 9348 | 9342 | Truncated ISNme1 transposase (NMV_0585.1) | DNA-binding protein HU (NMV_1200 hup) | – |
| 13 | 9377 | 9381 | 30S ribosomal protein S20 or autotransported serine protease NA1P (pseudogene part 1) (NMV_2165.1) | 30S ribosomal protein S20 | Methionine removed |
ND: not determined; PTM: post-translational modification; m/z: mass/charge.
Table 3. Biomarker assignments by means of proteogenomics for the Z2491 strain.
| MicroScope database |
|||||
|---|---|---|---|---|---|
| Peaks number | Observed m/z | Theoretical molecular weight including PTM | Protein assignment by sequence comparison | Protein assignment by shotgun MS/MS | PTM |
| 1 | 4488 | 4486 | 50S ribosomal protein L36 | 50S ribosomal protein L36 | – |
| 2 | 5051 | 5048 | 50S protein ribosomal L34 | 50S protein ribosomal L34 | – |
| 3 | 5620 | ND | ND | ND | ND |
| 4 | 5938 | 5933 | 50S ribosomal protein L33 | 50S ribosomal protein L33 | – |
| 5 | Not present in this strain | ||||
| 6 | 6434 | ND | ND | ND | ND |
| 7 | 7079 | 7074 | 50S ribosomal protein L29 | 50S ribosomal protein L29 | – |
| 8 | 7227 | 7221 | 50S ribosomal protein L35 | 50S ribosomal protein L35 | Methionine removed |
| 9 | 8112 | 8113 | 50S ribosomal protein L31 | 50S ribosomal protein L31 | – |
| 10 | 8251 | 8249 | 30S ribosomal protein S21 | 30S ribosomal protein S21 | Methionine removed |
| 11 | 8686 | 8682 | 50S ribosomal protein L28 | 50S ribosomal protein L28 | Methionine removed |
| 12 | 9348 | ND | ND | DNA-binding protein HU (NMA1397) | – |
| 13 | 9377 | 9381 | 30S ribosomal protein S20 | 30S ribosomal protein S20 | Methionine removed |
ND: not determined; PTM: post-translational modification; m/z: mass/charge.
A discrepancy was observed between the two strains for peak 3 (5620) that was found to be present in the spectra of the two strains. In the case of NEM8013, this peak was identified as being a hypothetical membrane protein (NMV_2186); on the other hand, this peak could not be identified by both techniques in Z2491. In addition, this peak was systematically present in the MALDI-TOF mass spectra of the 100 other strains. The homolog of NMV_2186 in NEM8013 is NMA0455 in Z2491. The comparison of the available sequences of these genes revealed only one amino-acid difference between the two strains. Resequencing of these two open reading frames confirmed the initial sequence (data not shown). The molecular weight of this protein deduced from the genome of strain Z2491 is 5650 Da, which is significantly higher than the molecular weight of peak 3 (5620). Furthermore, this peak 3, which disappeared after a protease treatment, does correspond to a protein. The most likely explanation for this is that peak 3 does not correspond to NMV_2186 and remains unidentified. Peak 6 could not be identified by both techniques in both strains, while the enzymatic digestion confirmed the protein nature of this peak. In summary, the proteins corresponding to peak 3 and peak 6 could be the consequence of post-translational modifications, other than the loss of methionine or O-glycosylation (Suh et al., 2005).
Previous studies have already shown that the vast majority of MALDI TOF peaks were proteins, after comparison with genomic (Holland et al., 1999) or protein sequence data (Ryzhov and Fenselau, 2001) for Gram-negative bacilli cell extracts. To our knowledge, this study is the first time that nanoLC-MS/MS has been used for the precise identification of species-specific peaks contained in the bacterial diagnosis databases of clinical laboratories. Altogether these data demonstrate that most of these biomarkers correspond to ribosomal proteins, as initially described by Ilina et al. (2009).
3.3. Inter-strain variability
A total of 3 biomarkers corresponding to ribosomal proteins, peaks 5, 11 and 13, were present in only 78%, 89% and 90% of the 1020 acquisitions, respectively (Table 1). In order to determine whether these relatively low frequencies were due to an intra- or inter-strain variability, the spectra of isolates that did not show one of the 13 biomarkers among the 10 acquisitions were examined. Peaks 11 and 13 were detected in the spectra of all 102 strains but only in some acquisitions, thus demonstrating that the variation of these peaks corresponded to an intra-strain variability. This variation was likely to be due to difficulties in the detection of proteins of relatively high molecular weight using MALDI-TOF mass spectrometry. Indeed, peaks of higher molecular weight are more difficult to detect than peaks corresponding to protein in the low molecular weight range (Fig. 1).
On the other hand, the only peak which was missing in a large number of strains was peak 5 (ribosomal protein L32), which was never detected in the subculture of 22 strains of the 102 strains under study and, if detected, was present in all the acquisitions of strains expressing this biomarker, showing an inter-strain variability of this protein.
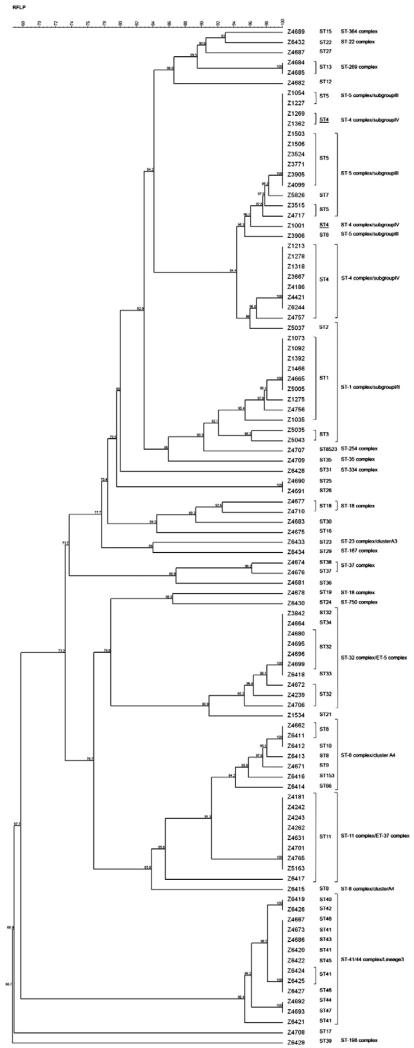
The ability to differentiate strains by MALDI-TOF has already been suggested. Arnold and Reilly (1998) used mathematical algorithm to differentiate strains of Escherichia coli on profiles obtained from cell lysates. Teramoto et al. (2007b, 2009) described with the MALDI-TOF method a phylogenetic classification using the inter-strain variability of ribosomal proteins, compared to phylogenetic tree of DNA gyrase subunit B gene. We subsequently assessed whether variability in some ribosomal proteins of N. meningitidis detected by MALDI-TOF could be used for strain grouping. We took advantage of the availability of the genome sequence of the 100 strains used in this study to analyze the molecular weight of the ribosomal proteins of these Nm strains. The ribosomal protein pattern of each strain was compared with that of the corresponding ST and CC (Fig. 2). This comparison clearly showed that there was a good correlation between the patterns of ribosomal proteins and the clonal complexes, except for ST-4 clonal complex (cc4) that includes strains having a ribosomal protein pattern identical to some strains belonging to the ST-5 clonal complex (cc5). These two clonal complexes are known to be closely related (Didelot et al., 2009).
Fig. 2. Phylogenetic tree of ribosomal proteins bases based on their molecular mass compared to ST and clonal complexes.
The correlation between the pattern of expressed ribosomal proteins and the ST showed that several different STs corresponded to the same ribosomal protein pattern (Fig. 2).
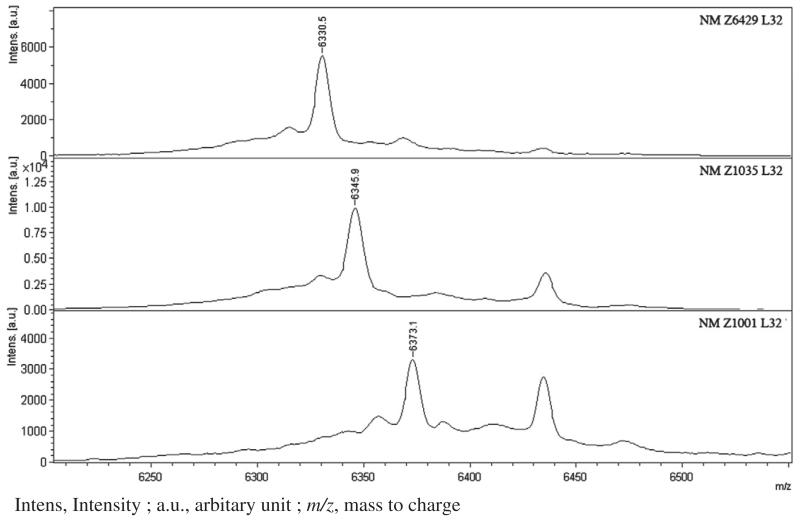
We subsequently tested the hypothesis of differentiating strains using routine MALDI-TOF MS spectra. In the spectral window employed (3640–12000 m/z), 14 ribosomal proteins could be detected. Among these, only 3 were ribosomal proteins exhibiting sequence variations, including L30, L31 and L32. The latter corresponds to peak 5. As an example, Fig. 3 shows the 3 possible variants of the L32 ribosomal protein. Table 4 indicates the STs and CCs corresponding to the different molecular weight of these 3 ribosomal proteins and the 6 resulting discriminating groups. The analysis of the molecular weights of the 3 proteins using the spectra obtained for the 102 strains allowed easily to classify each strain in one of the 6 groups, thus permitting on the basis of a routine spectrum to obtain epidemiological indications. It would be theoretically possible to determine precisely the clonal complex of each isolate, but this would require an adjustment of the spectral windows so that the molecular weight of other variable ribosomal proteins could be obtained. However in a routine setting such an adjustment may be challenging.
Fig. 3. Ribosomal protein L32 in 3 differents strains of Neisseria meningitidis.
Table 4. Molecular weight of the 3 variable ribosomal proteins detected in the MALDITOF spectra (spectral window: 3640–12000 m/z), and corresponding sequence types and clonal complexes.
| Group | L30 | L31 | L32 (peak 5) | Sequence types | Clonal complexes |
|---|---|---|---|---|---|
| 1 | 6764 | 8125 | 6343 | 1,2,3 | ST-1 complex/subgroup I/II |
| 8,9,10,66,153 | ST-8 complex/Cluster A4 | ||||
| 11 | ST-11 complex/ET-37 complex | ||||
| 13 | ST-269 complex | ||||
| 15 | ST-364 complex | ||||
| 18,19,20 | ST-18 complex | ||||
| 22 | ST-22 complex | ||||
| 23 | ST-23 complex/Cluster A3 | ||||
| 24 | ST-750 complex | ||||
| 28 | ST-103 complex | ||||
| 29 | ST-167 complex | ||||
| 32,33,34 | ST-32 complex/ET-5 complex | ||||
| 37,38 | ST-37 complex | ||||
| 8523 | ST-254 complex | ||||
| 12,16,17,21,25,26,27,30,36 | Not determined | ||||
| 2 | 6764 | 8125 | 6330 | 39 | ST-198 complex |
| 3 | 6764 | 8125 | 6373 | 4 | ST-4 complex/subgroup IV |
| 5,6,7 | ST-5 complex/subgroup III | ||||
| 4 | 6817 | 8125 | 6343 | 40,41,42,43,44,45,46,47,48 | ST-41/44 complex/Lineage 3 |
| 5 | 6764 | 8141 | 6343 | 35 | ST-35 complex |
| 6 | 6764 | 8153 | 6343 | 31 | ST-334 complex |
Taken together, these data show that species-specific peaks used in the N. meningitidis database are very conserved within the species N. meningitidis, as evidenced by the very robust reproducibility of the peaks detected in the 102 strains studied in this work. We have shown, using both shotgun proteomics and genomic approaches, that the majority of these species-specific peaks correspond essentially to ribosomal proteins. The variability of some ribosomal proteins visible on the spectrum could differentiate isolates among the same species to the level of clonal complex, providing an easy and rapid epidemiological method for classifying bacterial pathogens in the clinical microbiology laboratory.
Acknowledgements
The authors would like to thank Dr. Dominique Caugant and the WHO Collaborating Center for Reference and Research on Meningococci, Norwegian Institute of Public Health, Oslo, for providing bacterial samples.
References
- Arnold RJ, Reilly JP. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 1998;12:630–636. doi: 10.1002/(SICI)1097-0231(19980529)12:10<630::AID-RCM206>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, Maiden MC. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology. 2012;158:1570–1580. doi: 10.1099/mic.0.056077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Beretti JL, Cottyn S, Quesne G, Berche P, Nassif X, Ferroni A. Rapid identification of Staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2007;45:2156–2161. doi: 10.1128/JCM.02405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza JA, Pina-Villalonga JM, Guerin P, Miotello G, Bosch R, Nogales B, Armengaud J. Shotgun nanoLC-MS/MS proteogenomics to document MALDI-TOF biomarkers for screening new members of the Ruegeria genus. Environ. Microbiol. 2013;15:133–147. doi: 10.1111/j.1462-2920.2012.02812.x. [DOI] [PubMed] [Google Scholar]
- Clair G, Armengaud J, Duport C. Restricting fermentative potential by proteome remodeling: an adaptive strategy evidenced in Bacillus cereus. Mol. Cell. Proteomics. 2012;11(M111):013102. doi: 10.1074/mcp.M111.013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot A, Dulermo R, Ortet P, Blanchard L, Guerin P, Fernandez B, Vacherie B, Dossat C, Jolivet E, Siguier P, Chandler M, Barakat M, Dedieu A, Barbe V, Heulin T, Sommer S, Achouak W, Armengaud J. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 2009;5:e1000434. doi: 10.1371/journal.pgen.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degand N, Carbonnelle E, Dauphin B, Beretti JL, Le Bourgeois M, Sermet-Gaudelus I, Segonds C, Berche P, Nassif X, Ferroni A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 2008;46:3361–3367. doi: 10.1128/JCM.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirev PA, Feldman AB, Kowalski P, Lin JS. Top-down proteomics for rapid identification of intact microorganisms. Anal. Chem. 2005;77:7455–7461. doi: 10.1021/ac051419g. [DOI] [PubMed] [Google Scholar]
- Didelot X, Urwin R, Maiden MC, Falush D. Genealogical typing of Neisseria meningitidis. Microbiology. 2009;155:3176–3186. doi: 10.1099/mic.0.031534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworzanski JP, Snyder AP, Chen R, Zhang H, Wishart D, Li L. Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal. Chem. 2004;76:2355–2366. doi: 10.1021/ac0349781. [DOI] [PubMed] [Google Scholar]
- Gonzales T, Robert-Baudouy J. Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 1996;18:319–344. doi: 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- Holland RD, Wilkes JG, Rafii F, Sutherland JB, Persons CC, Voorhees KJ, Lay JO., Jr Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1996;10:1227–1232. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1227::AID-RCM659>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Holland RD, Duffy CR, Rafii F, Sutherland JB, Heinze TM, Holder CL, Voorhees KJ, Lay JO., Jr. Identification of bacterial proteins observed in MALDI TOF mass spectra from whole cells. Anal. Chem. 1999;71:3226–3230. doi: 10.1021/ac990175v. [DOI] [PubMed] [Google Scholar]
- Ilina EN, Borovskaya AD, Malakhova MM, Vereshchagin VA, Kubanova AA, Kruglov AN, Svistunova TS, Gazarian AO, Maier T, Kostrzewa M, Govorun VM. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 2009;11:75–86. doi: 10.2353/jmoldx.2009.080079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinforma. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, Wimalarathna H, Harrison OB, Sheppard SK, Cody AJ, Maiden MC. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158:1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K. Heterogeneity of ribosomal proteins among Streptomyces species and its application to identifcation. J. Gen. Microbiol. 1989;135:2635–2642. doi: 10.1099/00221287-135-10-2635. [DOI] [PubMed] [Google Scholar]
- Lay JO., Jr. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 2001;20:172–194. doi: 10.1002/mas.10003. [DOI] [PubMed] [Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matte-Tailliez O, Brochier C, Forterre P, Philippe H. Archaeal phylogeny based on ribosomal proteins. Mol. Biol. Evol. 2002;19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- Pineda FJ, Antoine MD, Demirev PA, Feldman AB, Jackman J, Longenecker M, Lin JS. Microorganism identification by matrix-assisted laser/desorption ionization mass spectrometry and model-derived ribosomal protein biomarkers. Anal. Chem. 2003;75:3817–3822. doi: 10.1021/ac034069b. [DOI] [PubMed] [Google Scholar]
- Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13953–13958. doi: 10.1073/pnas.0804861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzhov V, Fenselau C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 2001;73:746–750. doi: 10.1021/ac0008791. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MJ, Hamburg DM, Gregory ST, Dahlberg AE, Limbach PA. Extending ribosomal protein identifications to unsequenced bacterial strains using matrix-assisted laser desorption/ionization mass spectrometry. Proteomics. 2005;5:4818–4831. doi: 10.1002/pmic.200402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto K, Sato H, Sun L, Torimura M, Tao H. A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J. Proteome Res. 2007a;6:3899–3907. doi: 10.1021/pr070218l. [DOI] [PubMed] [Google Scholar]
- Teramoto K, Sato H, Sun L, Torimura M, Tao H, Yoshikawa H, Hotta Y, Hosoda A, Tamura H. Phylogenetic classification of Pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal. Chem. 2007b;79:8712–8719. doi: 10.1021/ac701905r. [DOI] [PubMed] [Google Scholar]
- Teramoto K, Kitagawa W, Sato H, Torimura M, Tamura T, Tao H. Phylogenetic analysis of Rhodococcus erythropolis based on the variation of ribosomal proteins as observed by matrix-assisted laser desorption ionization-mass spectrometry without using genome information. J. Biosci. Bioeng. 2009;108:348–353. doi: 10.1016/j.jbiosc.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Williams TL, Andrzejewski D, Lay JO, Musser SM. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 2003;14:342–351. doi: 10.1016/S1044-0305(03)00065-5. [DOI] [PubMed] [Google Scholar]
- Yutin N, Puigbo P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]