Summary
Facultative or “secondary” symbionts are common in eukaryotes, particularly insects. While not essential for host survival, they often provide significant fitness benefits [1-5]. It has been hypothesized that secondary symbionts form a “horizontal gene pool” shuttling adaptive genes among host lineages in an analogous manner to plasmids and other mobile genetic elements in bacteria [6, 7]. However, we do not know whether the distributions of symbionts across host populations reflect random acquisitions followed by vertical inheritance or whether the associations have occurred repeatedly in a manner consistent with a dynamic horizontal gene pool. Here we explore these questions using the phylogenetic and ecological distributions of secondary symbionts carried by 1,104 pea aphids, Acyrthosiphon pisum. We find that not only is horizontal transfer common, but it is also associated with aphid lineages colonizing new ecological niches, including novel plant species and climatic regions. Moreover, aphids that share the same ecologies worldwide have independently acquired related symbiont genotypes, suggesting significant involvement of symbionts in their host’s adaptation to different niches. We conclude that the secondary symbiont community forms a horizontal gene pool that influences the adaptation and distribution of their insect hosts. These findings highlight the importance of symbiotic microorganisms in the radiation of eukaryotes.
Results
We collected 1,104 pea aphids from 11 host-plant genera and 155 localities in 14 countries and assigned them to 286 haplotypes on the basis of DNA sequences from their strictly vertically inherited obligate symbiont Buchnera. Using nuclear genetic markers, we assigned most aphids (86%) to genetic clusters corresponding to populations adapted to specific host plants [8, 9].
We tested all individuals for the presence of four species of facultative (Enterobacteriaceae) symbionts known to occur regularly in pea aphids: Hamiltonella defensa, Regiella insecticola, Serratia symbiotica, and the taxon referred to as “X type” [10].
Host-Plant and Symbiont Species Associations
Symbiont species were nonrandomly distributed across aphids associated with different host plants (Table S1, part A). We confirmed known positive associations between (1) R. insecticola and aphids feeding on Trifolium spp., (2) H. defensa ad aphids from Lotus pedunculatus and Ononis spp., and (3) S. symbiotica and aphids from Cytisus scoparius and Pisum sativum. New positive associations were identified between R. insecticola and aphids from Onobrychis viciifolia and between S. symbiotica and aphids from Lotus corniculatus and Vicia cracca. The four species of symbiont were negatively correlated across aphids, with the exception of H. defensa and X type, which were positively associated.
Genetic Structure of the Facultative Symbionts
We used multilocus sequence typing (MLST) to determine the genetic structure of the four Enterobacteriaceae and asked how much variation in genetic structure could be explained by host ecology. We found host plant to be an important factor in the case of H. defensa and R. insecticola, explaining 27% and 32% of the genetic variation (p < 0.0001 and p = 0.057, respectively; analysis of molecular variance [AMOVA]; Table S1, part B), but unimportant for S. symbiotica and X type. Climate (inferred from geography) was a significant factor explaining the genetic sructure of S. symbiotica (Table S1, part B).
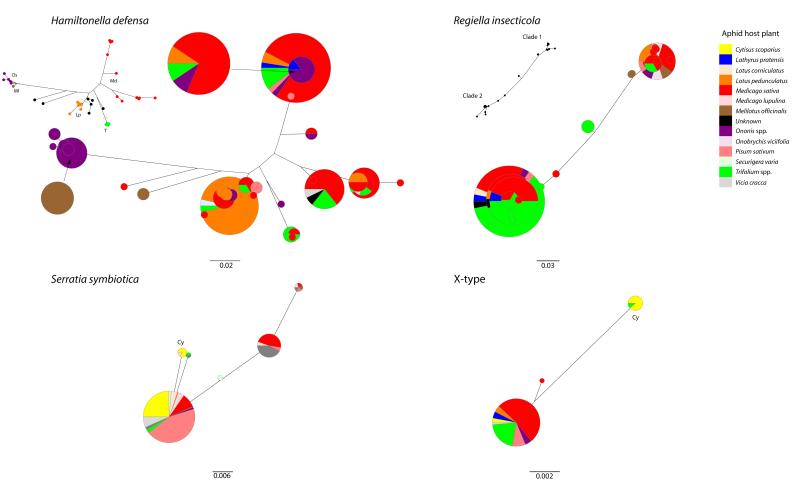
Clades of H. defensa could be identified in aphids associated with different plants (labeled on inset phylogeny in Figure 1A; see also the BaTS analysis in the Supplemental Experimental Procedures). The genetic diversity of R. insecticola is largely organized into two distinct clades (clades 1 and 2 in Figure 1B). The strong association between this symbiont and aphids feeding on Trifolium is due to bacteria from clade 2.
Figure 1. Host-Plant Associations of Secondary Symbiont Genotypes Symbiont.
phylogenies are built from six housekeeping genes, and host-plant associations are based on microsatellite assignments (genetic cluster) of the aphid. Bubble size corresponds to the number of individuals. Small inset phylogenies shows the operationally defined clades used for analyses and referred to in text. See also Table S1.
The major clades of H. defensa and R. insecticola were estimated to have diverged approximately 1.1 × 105 and 4.8 × 105 years before extant lineages of pea aphid radiated onto different host plants (see Table S1, part D, and the Supplemental Experimental Procedures for divergence estimates).
All symbionts had some degree of geographical structuring (see the Supplemental Experimental Procedures), but this was pronounced in S. symbiotica, which has a number of genotypes particularly common in arid countries such as Iran, Tunisia, and Turkey. The frequency of S. symbiotica infection is significantly higher in arid (0.67 ± 0.09 infected/haplotype) compared to temperate (0.27 ± 0.03 infected/haplotype) regions (p < 0.0001).
Transmission Dynamics of Facultative Symbionts
Using sequence data from the host and symbionts, we assessed the relative importance of vertical and horizontal transmission in explaining the associations between symbiont genotypes and aphid lineages using phylogenetic generalized linear mixed effect modeling (GLMM). We found that symbiont population structure is shaped by a combination of the two processes, both of which are strongly influenced by host ecology. Aphids are more likely to acquire by horizontal transfer specific symbiont genotypes in different ecological settings, and the majority of established vertical transmitted symbiont infections are found in aphid lineages associated with a single plant species.
We analyzed separately H. defensa and R. insecticola, the species with the highest genetic diversity. We tested (1) whether symbiont genotypes vary in the range of aphid lineages they infect and, if so, whether this variation had a (symbiont) phylogenetic basis, (2) whether closely related symbiont genotypes tended to infect closely related aphid lineages (the presence of a phylogenetic interaction due to vertical transmission), and (3) whether symbiont genotypes were associated with aphids feeding on particular plants and, if so, whether this had a phylogenetic basis (processes influenced by horizontal transmission) (analysis described in the Supplemental Experimental Procedures). A signal of vertical transmission (phylogenetic interaction) was found, explaining 13.1% (H. defensa) and 46.0% (R. insecticola) of the variation, although with wide credible intervals. The horizontal transmission of plant-specific symbiont genotypes explained 36.5% (H. defensa) and 10.3% (R. insecticola) of the variation in the symbiont-host associations. Whether this is due to a single symbiont genotype (nonphylogenetic effect) or to multiple related genotypes (phylogenetic effect) infecting aphids on the same plant species could not be determined (Table 1).
Table 1. Bayesian Phylogenetic Mixed Model Analysis of Symbiont Genotype Distributions.
| Term | Hamiltonella defensa | Regiella insecticola |
|---|---|---|
| Symbiont genotype | 0.001 (0.000–0.091) | 0.001 (0.000–0.098) |
| Symbiont phylogeny | 0.001(0.000–0.240) | 0.022 (0.000–0.276) |
| (Symbiont genotype + phylogeny) | 0.047 (0.000–0.258) | 0.006 (0.000–0.298) |
| Aphid: symbiont phylogeny | 0.131 (0.000–0.433) | 0.460 (0.159–0.703) |
| Host plant: symbiont genotype | 0.118 (0.000–0.283) | 0.113 (0.000–0.303) |
| Host plant: symbiont phylogeny | 0.173(0.000–0.483) | 0.002(0.000–0.299) |
| (Host plant: symbiont genotype + phylogeny) | 0.365 (0.162–0.603) | 0.103 (0.034–0.443) |
Results of the phylogenetic mixed model analysis indicating the five sources of variation in symbiont genotype distributions within Hamiltonella defensa and Regiella insecticola. Terms in parentheses are not unique terms but the combined effect of the preceding two (phylogenetic and nonphylogenetic) terms. Posterior modes are given with 95% credible intervals in parentheses.
Coevolution of Symbiosis and Host Niche Expansion
To explore the role of secondary symbiosis in host evolution, we treated the presence of particular symbionts as equivalent to evolutionary characters transmitted through the host phylogeny. We used ancestral state reconstruction to estimate when a symbiont became established in an aphid lineage and from this determined the number of independent acquisitions of each symbiont species (see the Supplemental Experimental Procedures). We then asked whether the rate of gains and losses of symbionts was statistically correlated with aphids colonizing new ecological niches (e.g., novel host plants).
We estimated that H. defensa and R. insecticola have independently colonized five and three major aphid clades, respectively, resulting in long-lasting associations (>2,000 years), while X type has formed only one such association (Figure S2). The situation with S. symbiotica is more complex with the bacteria found in numerous closely related aphid lineages, which likely represent many colonization and loss events.
The pea aphid’s radiation onto multiple host plants allowed us to explore the role of secondary symbionts in this process. We asked whether the rate of symbiont gain or loss is higher when aphids are associated with particular plants or whether the rate of switching to or away from feeding on a particular plant is influenced by symbiont infection. We used the program BayesDiscrete [11] to test for correlated evolution between two binary traits by comparing the log likelihood of pairs of continuous-time Markov models of trait evolution, using Bayes factor (BF) statistics (full explanation in the Supplemental Experimental Procedures). Analyses were conducted using all genotypes of a symbiont species and were then repeated for particularly well-defined clades associated with different host plants.
We found clear evidence that the pea aphid’s colonization of particular host plants is associated with infections by two of the four symbiont species. The likelihood of aphid lineages colonizing Trifolium spp., Lotus pedunculatus, Ononis spp., and Medicago sativa is significantly higher when infected with particular strains of R. insecticola and H. defensa (for full details of analysis, see Table S2). For example, the rate at which aphid lineages colonize Trifolium spp. is significantly higher when infected by R. insecticola from Clade 2 (Figure 1B), and there is a higher rate of R. insecticola loss when aphids are associated with other plant species (BF = 11.8). This relationship is weaker (BF = 2.2) when all R. insecticola genotypes are included in the analysis. We also find that pea aphids are more likely to switch to feed on L. pedunculatus and Ononis spp. if they carry H. defensa (BF = 13.9 and 16.9, respectively), and the rate of gains of these plants when uninfected is near zero, demonstrating that aphids have rarely colonized these plants without this symbiont. The same pattern was found for host-plant switches to M. sativa, but only when the analysis was restricted to one clade of H. defensa (BF = 11.7; see the Supplemental Experimental Procedures for further details).
We found no evidence that the acquisition of S. symbiotica or X type was correlated with aphid host plant. However, gains of X type are more likely in aphid lineages that already carry H. defensa (BF = 11.7). X type is sporadically distributed throughout the host phylogeny but is maintained at high frequency in a single clade of aphids, almost exclusively in coinfections with H. defensa feeding on M. sativa in Europe and North America.
Discussion
The null hypothesis of symbiont population structure assumes that acquisitions and losses occur at random on the host phylogeny. Our data clearly reject this hypothesis. In several species, we find that the acquisition of symbionts by horizontal transmission is linked to the colonization of new ecological niches by their aphid hosts. In one case, we also find that the gain of one symbiont species is associated with the presence of another. Importantly, particular symbiont genotypes are commonly found associated with aphids in the same ecological niches throughout their hosts’ global distribution. Combination of genetic data from both aphids and bacteria provides a picture of a dynamic symbiont community that is shaped by the environment of their insect hosts. Our work supports the idea that symbionts assist their host in exploiting specific ecological niches and occupying different climatic zones and is consistent with the hypothesis that symbionts form a horizontal gene pool that is actively sampled by hosts when confronting novel environmental challenge. Experimental studies are now required to test the hypotheses generated by our comparative analyses.
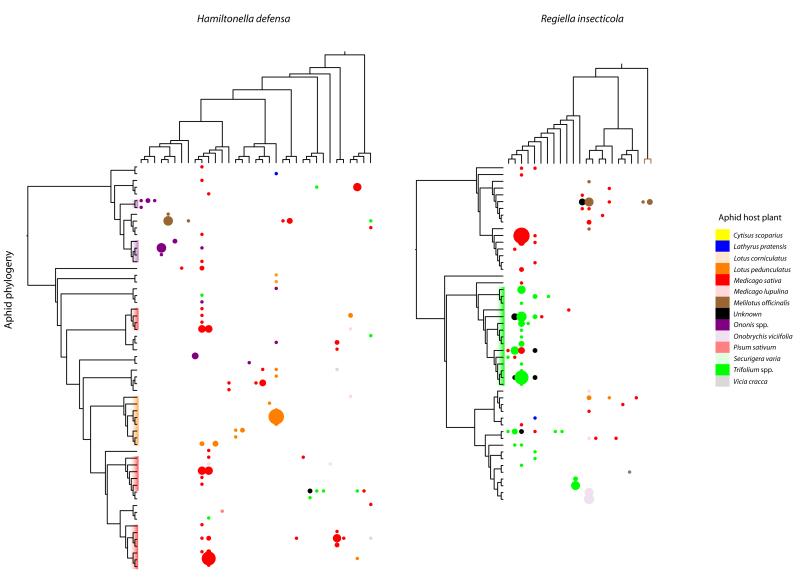
Direct evidence of horizontal transfer is observed in clades of aphids that have recently switched to feed on new host plants. For example, several clades of unrelated aphids that have switched to feed on L. pedunculatus and M. sativa are now infected by identical H. defensa genotypes (Figure 2A). Similarly, aphids that have recently switched to feed on Trifolium spp. have acquired a particular type (clade 2) of R. insecticola, which predominantly infects aphids feeding on this plant (Figure 2B). A striking feature of these symbiont associations is that we find aphids that share the same ecologies in distant geographic regions, often in very different climates, harboring the same (or closely related) bacterial genotypes. This can only be explained by relatively frequent horizontal transfer occurring in different parts of the globe, with the host’s environment affecting either the probability of symbiont infection or the persistence of different symbiont infections.
Figure 2. Interaction Matrices for Aphid, y Axis, and Symbiont Genotypes, x Axis.
Bubble size corresponds to the incidence of the genotype associations, and the color indicates the host-plant cluster of the aphid harboring the symbiont. Major aphid clades associated with Ononis spp. (purple), M. sativa (red), L. pedunculatus (orange), and Trifolium spp. (green) are highlighted on the aphid phylogeny. Aphid phylogeny is pruned to only include aphids infected by each symbiont species. See also Figure S1 for interaction matrixes on the full aphid phylogeny.
Our results provide strong support for an association between symbiont carriage and the pea aphid’s colonization of new plant species. Both R. insecticola and H. defensa populations are structured by the host plant of the aphids they infect. This could be due to differential rates of acquisition or loss on different host plants or to the symbiont providing a fitness benefit to the aphid on a particular host plant. Our results are consistent with the latter hypothesis because the analyses of correlated evolution indicate that rates of colonization of new host plants are higher when a symbiont is carried rather than when aphids on particular host plant have higher rates of gaining certain bacteria. Facultative symbionts may directly assist aphids in exploiting a new food plant, or (alternatively or in addition) they may provide indirect assistance in coping with other niche-specific challenges, such as providing protection against natural enemies associated with particular plant species. Tsuchida et al.’s [12] demonstration that transfer of R. insecticola from pea aphid to another aphid species that does not normally carry this symbiont enabled it to survive and reproduce on otherwise unsuitable Trifolium is strong support for a direct effect of symbionts on host-plant utilization. However, experiments transferring symbionts among pea aphid lineages have found mixed effects on host-plant use [12-14], and more studies are needed that include biotic and abiotic pressures experienced by aphids on different host plants in the field. Hamiltonella defensa and R. insecticola are known to confer resistance to different natural enemies [2-4], and were the selective pressures from these organisms to be host-plant specific, it may explain the associations.
S. symbiotica population structure was most influenced by climate. Multiple unrelated aphid lineages collected in arid areas have acquired through horizontal transfer S. symbiotica genotypes specific to these locations and maintained them at significantly higher infection frequencies than in temperate regions. Serratia symbiotica is known to protect aphids from heat shock, especially genotypes collected from arid regions such as Arizona [5]. These results suggest S. symbiotica is maintained at high frequencies of infection in these regions as an adaptation to arid climates.
Conclusions
The acquisition of secondary or facultative symbionts can have important beneficial effects on their hosts’ fitness; but rather than being discrete innovations, analogous to beneficial mutations that spread to fixation, their evolutionary dynamics are much more complex. We show here that symbionts are gained and lost by host lineages at relatively high frequencies and that this is directly linked to their host’s ecology. We find evidence that the carriage of different symbionts is significantly associated with major changes in host ecology, such as the colonization of new host plants and movement into new climatic regions. Moreover, aphids that share the same ecologies worldwide have independently acquired identical or closely related symbiont genotypes. These findings are consistent with the hypothesis that symbionts play a role in determining their host’s ecological niche. They also support the idea that secondary symbionts constitute a eukaryote horizontal gene pool, a reservoir of potential adaptations, or preadaptation.
Experimental Procedures
Detection and Typing
One thousand one hundred and four aphid isolates were included in this study, approximately half of which collected as part of earlier studies on aphid-host plant interactions [8, 9] and half as new collections. Symbionts were detected using diagnostic PCR based on the 16S ribosomal RNA gene. A multilocus sequence-typing (MLST) scheme was developed for the three of four Enterobacteriaceae using six housekeeping genes, (accD, gyrB, hrpA, murE, recJ, and rpoS), based on a protocol previously developed for H. defensa (and partially for R. insecticola) [15]. Aphids were assigned to haplotypes based on the sequence of their strictly vertically transmitted primary symbiont Buchnera. We sequenced two intergenic regions (groEL-efp and cofmetE, a third region; prfC-yhgl, was used in 175 samples). Aphid phylogenies based on the primary symbiont were constructed using 365 Buchnera sequences from [16] and an additional 723 new Buchnera sequences. Nine hundred forty-five aphids were assigned to host-plant clusters using microsatellite data from 11 polymorphic nuclear loci, 525 previously analyzed assignments [8, 9], and 420 new assignments based on the same protocol as in [8]. Full details of PCR conditions and primer sequences are provided in the Supplemental Experimental Procedures and Data Set S1.
Statistical and Phylogenetic Methods
A more detailed account of all the analyses described here is in the Supplemental Experimental Procedures.
AMOVA was performed with Arlequin v.3.11 [17] for symbionts using the concatenated MLST genes and for aphids using the concatenated Buchnera, groEL-efp, and cof-metE regions. In analyses, the genetic assignment of aphids to host-plant-associated populations was used in preference to the actual collection plant (aphids can wander onto the “wrong” host plant), and geography was nested within host plant.
Recombination was found to have little impact on Enterobacteriaceae phylogenetic signals, and so the concatenated MLST gene sequences were used for subsequent analyses. Phylogenies were inferred using maximum likelihood and Bayesian methods with model fit assessed using Akaike and Bayesian information criteria.
We estimated the minimum divergence times of the major H. defensa and R. insecticola clades using the Buchnera rate of synonymous substitution, as described in Moran et al. [18]. The estimated mutation rate in [18] is one of the highest ever recorded, and it is possible that secondary symbionts, despite sharing a similar niche, have a slower rate. On the basis of this assumption, these estimates represent minimum divergence times for the symbiont clades associated with different host plants, which for the purposes of this study is sufficient for a comparison with the divergence of the corresponding pea aphid lineages (see the Supplemental Experimental Procedures for further discussion). However, these estimates should be treated as provisional until data on secondary symbiont substitution rates is available.
Ancestral state reconstruction was carried out use the program Mesquite [19]. Aphid haplotypes (with those represented by single individuals omitted) were assigned to different states (symbiont presence, host-plant cluster) using relaxed and conservative rules. For the relaxed assignment, if one or more individuals belonging to an aphid haplotype was infected, the haplotype was considered positive for the symbiont, and in the conservative assignment a haplotype was considered positive if >25% of the individuals were infected. When symbiosis was analyzed as a trait of the aphid (e.g., in ancestral state reconstructions and when correlated evolution was studied), analyses were performed with both the conservative and relaxed methods of assignment for comparison. So that phylogenetic uncertainty could be accounted for, reconstructions were summarized over 5,000 trees from the posterior distribution. Using the same 5,000 trees, correlated evolution of symbiont and host-plant states was analyzed using BayesDiscrete in BayesTraits [11]. Bayesian phylogenetic mixed model analysis was carried out using techniques described in Hadfield and Nakagawa (J.D.H., unpublished data) and implemented as an R library.
Supplementary Material
Acknowledgments
We thank Annabel Morley and Hannah Halford for laboratory assistance, Kevin Foster and Stu West for editing, and four anonymous referees for comments. We also thank Andrew Meade for assistance with BayesDiscrete, Dave Walsh and Ailsa McLean for comments, and Bob Foottit for providing aphid samples. This research was supported by NERC grant NE/G017638/1.
Footnotes
Accession Numbers
The GenBank accession numbers for the MLST gene sequences determined in this study are KF575178 to KF575321.
References
- 1.Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- 2.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scarborough CL, Ferrari J, Godfray HCJ. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 4.Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 2013;16:214–218. doi: 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 5.Russell JA, Moran NA. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc. Biol. Sci. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaenike J. Population genetics of beneficial heritable symbionts. Trends Ecol. Evol. 2012;27:226–232. doi: 10.1016/j.tree.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl. Acad. Sci. USA. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peccoud J, Ollivier A, Plantegenest M, Simon J-C. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl. Acad. Sci. USA. 2009;106:7495–7500. doi: 10.1073/pnas.0811117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari J, West JA, Via S, Godfray HCJ. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution. 2012;66:375–390.9. doi: 10.1111/j.1558-5646.2011.01436.x. [DOI] [PubMed] [Google Scholar]
- 10.Guay J-F, Boudreault S, Michaud D, Cloutier C. Impact of environmental stress on aphid clonal resistance to parasitoids: Role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect Physiol. 2009;55:919–926. doi: 10.1016/j.jinsphys.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 12.Tsuchida T, Koga R, Matsumoto S, Fukatsu T. Interspecific symbiont transfection confers a novel ecological trait to the recipient insect. Biol. Lett. 2011;7:245–248. doi: 10.1098/rsbl.2010.0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean AHC, van Asch M, Ferrari J, Godfray HCJ. Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc. Biol. Sci. 2011;278:760–766. doi: 10.1098/rspb.2010.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari J, Scarborough CL, Godfray HCJ. Genetic variation in the effect of a facultative symbiont on host-plant use by pea aphids. Oecologia. 2007;153:323–329. doi: 10.1007/s00442-007-0730-2. [DOI] [PubMed] [Google Scholar]
- 15.Degnan PH, Moran NA. Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol. Ecol. 2008;17:916–929. doi: 10.1111/j.1365-294X.2007.03616.x. [DOI] [PubMed] [Google Scholar]
- 16.Peccoud J, Simon J-C, McLaughlin HJ, Moran NA. Post-Pleistocene radiation of the pea aphid complex revealed by rapidly evolving endosymbionts. Proc. Natl. Acad. Sci. USA. 2009;106:16315–16320. doi: 10.1073/pnas.0905129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 18.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 19.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011 http://mesquiteproject.org
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.