Abstract
The 3’-ends of almost all eukaryotic mRNAs are formed in a two-step process, an endonucleolytic cleavage followed by polyadenylation (the addition of a poly-adenosine or poly(A) tail). These reactions take place in the pre-mRNA 3’ processing complex, a macromolecular machinery that consists of more than 20 proteins. A general framework for how the pre-mRNA 3’ processing complex assembles and functions has emerged from extensive studies over the past several decades using biochemical, genetic, computational, and structural approaches. Here we review what we have learned about this important cellular machine and discuss the remaining questions and future challenges.
Keywords: 3’ end processing, Polyadenylation, cleavage, complex assembly, function, regulation
Pre-mRNA 3’ processing is an essential step in eukaryotic mRNA biogenesis, and it directly impacts many other steps in the gene expression pathway, such as transcription termination, splicing, mRNA export, translation, and mRNA turnover1–3. 3’ processing is also a versatile mechanism for gene regulation4. For example, the efficiency of 3’ processing, the usage of alternative polyadenylation (APA) sites, and the length of poly(A) tails can all be modulated. Additionally, 3’ processing-mediated gene regulation can be global or transcript-specific. Accumulating EST data revealed that a significant portion of eukaryotic genes produce alternatively polyadenylated mRNAs5, indicating that APA may be involved in the regulation of a large set of genes6. Indeed a series of recent genomic studies showed that APA regulation is widespread and plays important roles in the immune7 and the neural systems8, oncogenesis9, stem cell differentiation10, and development11. Defects in pre-mRNA 3’ processing have been associated with a wide spectrum of human diseases12. Therefore, it is critical to understand how the pre-mRNA 3’ processing machinery functions and how its activity can be regulated.
Much progress has been made in understanding the basic molecular mechanisms and regulation of pre-mRNA 3’ processing. The core cis-elements and the majority of the basal trans-acting factors required for 3’ processing have been identified1,2,4,13. Biochemical, genetic, and structural analyses have provided insights into the functions of some 3’ processing factors13. A diverse array of mechanisms has been uncovered regarding how 3’ processing can be regulated2,4. More recently, purification of the functional human pre-mRNA 3’ processing complex has allowed compositional characterization of the entire machinery and made it possible to study its structure and dynamics throughout the cleavage and polyadenylation processes14,15. In this article, we will provide a broad overview of our current knowledge of the assembly and function of the pre-mRNA 3’ processing complex and highlight recent progress in the field. The main focus will be the mammalian pre-mRNA 3’ processing complex, but comparisons with its counterparts in yeast and plants will be provided to illustrate both the conservation and divergence of the 3’ processing system in eukaryotic evolution.
PRE-MRNA 3’ PROCESSING COMPLEX: THE COMPONENTS
Assembly of the pre-mRNA 3’ processing complex is initiated by the binding of trans-acting factors to their target cis-elements in the RNA. Unlike the spliceosome whose assembly is aided by a series of specific RNA-RNA base-pairing events16, assembly of the 3’ processing complex on the pre-mRNA is directed entirely by RNA-protein interactions1,2,4,15. As will be discussed later, the cis-elements for 3’ processing are generally short and/or degenerate (Fig. 1) and the interactions of individual 3’ processing factors with RNA are usually very weak. Therefore, an extensive network of RNA-protein and protein-protein interactions is required for the assembly of a stable 3’ processing complex (Fig. 2). Here we discuss the cis-elements and protein factors involved.
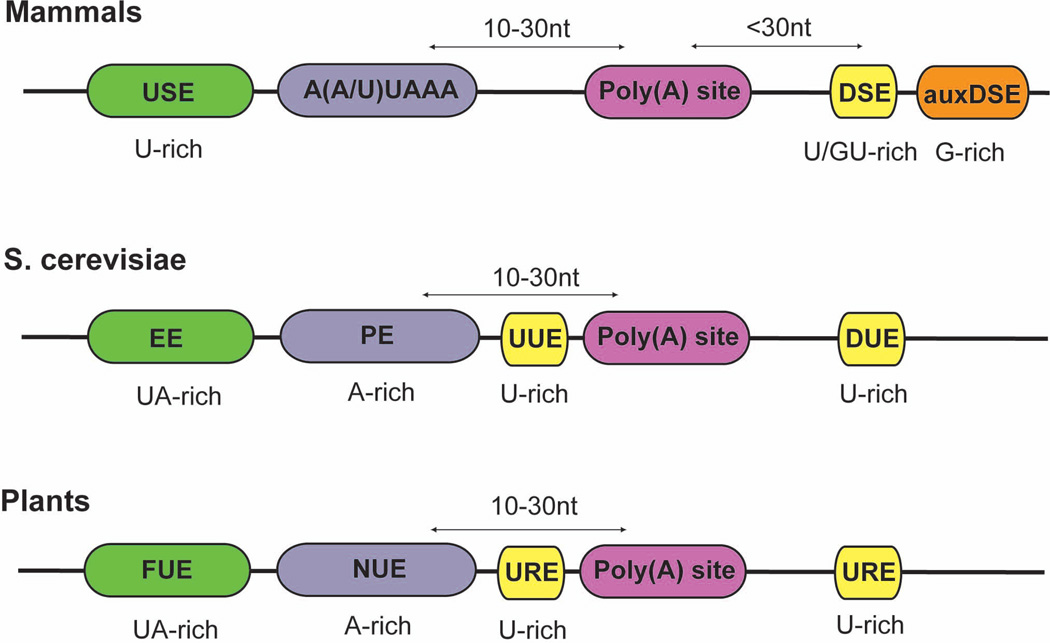
Figure 1. Comparison of the poly(A) signals from animals (canonical) the budding yeast (S. cerevisiae) and plants.
USE: upstream element; DSE: downstream element; auxDSE: auxiliary downstream element; EE, efficiencty element; PE, positioning element; UUE, upstream U-rich element; DUE, downstream U-rich element; FUE, far upstream element; NUE, near upstream element; URE, U-rich element. Same colors are used for similar cis-element to illustrate the conservation of the basic tripartite poly(A) signal across different phylogenetic groups. Adapted from reference 2.
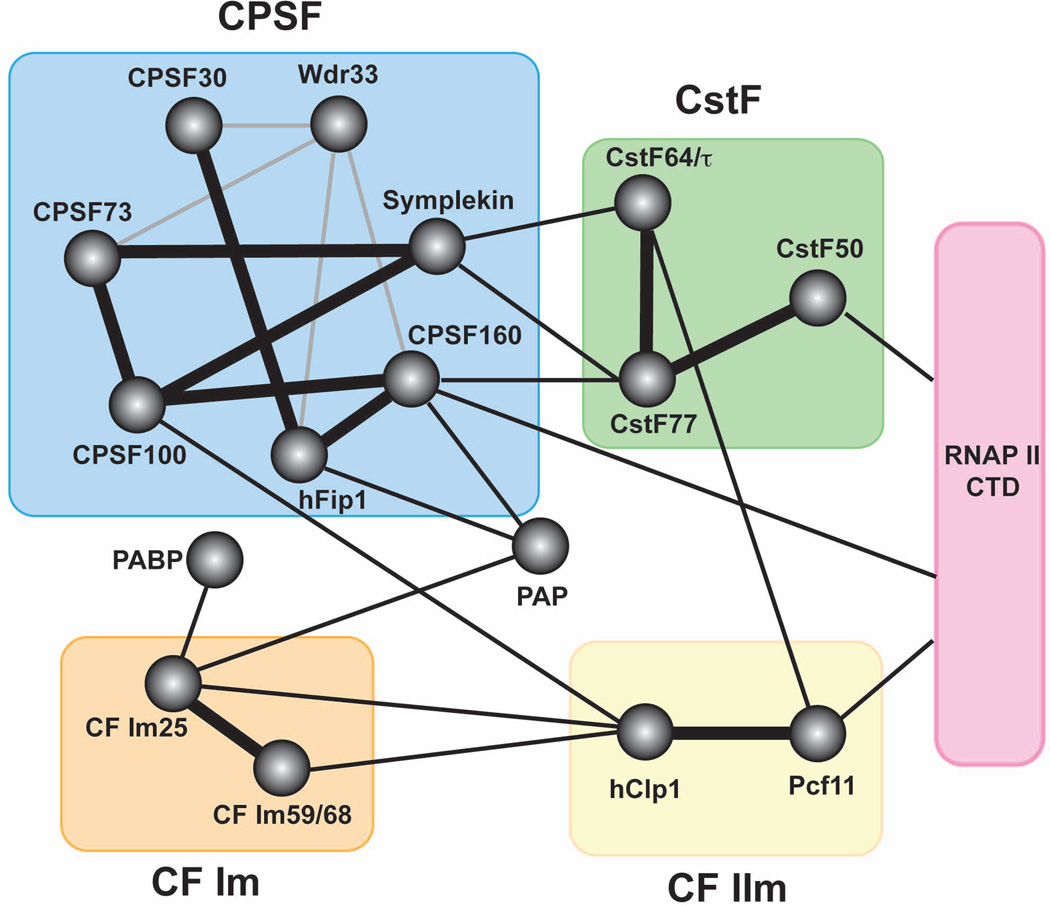
Figure 2. The protein-protein interaction network of the mammalian pre-mRNA 3’ processing complex.
Circles represent individual proteins except for RNAP II CTD (pink rectangle). Colored rectangles represent subcomplexes. Experimentally verified interactions within subcomplexes are marked with thick black lines. Experimentally verified interactions among subcomplexes are marked with thin black line. Predicted interactions based on information on homologues from other species are marked with grey lines.
Cis-elements
Assembly of the pre-mRNA 3’ processing complex is directed by multiple key cis-elements collectively called the polyadenylation signals (PAS)2. Mammalian PASes can be classified into two general types, the canonical and the noncanonical2,4,17. The main cis-elements in a canonical PAS include a highly conserved A(A/U)UAAA element located 10–30 nucleotides upstream of the cleavage/polyadenylation site (poly(A) site) and a more variable U/GU-rich downstream element (DSE) ~30 nucleotides downstream of the poly(A) site (Fig. 1). These two elements determine the general location of the poly(A) site, which is most frequently found after an adenosine residue18. Genomic studies suggest that poly(A) site selection in vivo is fairly heterogeneous and, for any given PAS, the corresponding poly(A) sites are usually clustered in a ~20nt window5,19. In addition to the A(A/U)UAAA and the DSE, functionally important upstream sequence elements (USE) and auxiliary downstream elements (auxDSE) have also been identified in many viral and cellular mRNAs2,4,20 (Fig. 1). USEs are generally U-rich while auxDSEs are mostly G-rich. These additional cis-elements help improve 3’ processing efficiency by providing binding sites for the core 3’ processing factors and/or regulatory factors.
Twenty to thirty percent of human PASes lack the A(A/U)UAAA element and it remains poorly understood how these noncanonical PASes are recognized by the 3’ processing machinery or whether they are recognized by the same 3’ processing factors that bind canonical PASes5,21. In some cases, UGUAN and related sequences have been identified as the key cis-element for directing A(A/U)UAAA-independent cleavage and polyadenylation22. Recently another type of PAS was discovered that consists of only an A-rich USE and a potent DSE23. Currently, however, it is not clear whether the noncanonical PASes share any common characteristics. Interestingly, noncanonical PASes are often associated with APA and may play important regulatory roles5.
The PASes of the budding yeast and plant pre-mRNAs are thought to be significantly different from the mammalian PASes as they lack the highly conserved A(A/U)UAAA element2,4. Instead, yeast and plant PASes include a degenerate A-rich element which includes AAUAAA-like sequences (called the Positioning Element (PE) in budding yeast and the Near Upstream Element (NUE) in plants), a UA-rich USE (called the Efficiency Element (EE) in budding yeast and the Far Upstream Element (FUE) in plants), and a U-rich element (URE) upstream and/or downstream of the poly(A) site2,4 (Fig. 1). Interestingly, these cis-elements are also shared by some mammalian noncanonical PASes22,23, indicating that PAS sequences might have been more conserved than previously thought.
Given the lack of strong, evolutionarily conserved consensus sequence in eukaryotic PASes, RNA secondary structures have long been suspected to play a role in directing the assembly of the 3’ processing complex24,25. Multiple lines of evidence support this notion. First, an in vitro selection for 3’ processing enhancers enriched motifs that are related in structure rather than sequence24. Secondly, functionally important secondary structures have been identified in the PASes of a number of viral mRNAs26,27. It remains to be seen, however, whether RNA secondary structure plays a general role in 3’ processing and if so, what specific secondary structures are required and how they are recognized.
3’ processing factors
Most, if not all, basal 3’ processing factors have been identified4,13,15 (Table 1.). Mammalian 3’ processing factors include the poly(A) polymerase (PAP), the poly(A)-binding proteins (PABPs), the RNA polymerase II large subunit (RNAP II), and four multi-subunit protein complexes, CPSF, CstF, CF Im and CF IIm1,2,13. All of these factors except for PABPs are required for cleavage, but CPSF and PAP are believed to be sufficient for the subsequent polyadenylation. PABPs are not required for either cleavage or polyadenylation, but function in activating PAP and in poly(A) tail length control28. Despite the significant divergence in the PAS sequences between yeast and mammals, most mammalian 3’ processing factors have homologues in yeast and plants2,13, indicating that the pre-mRNA 3’ processing machinery and the biochemical mechanism have been conserved.
Table 1.
Conservation of polyadenylation factors between yeast and mammals.
| Mammalian factor name |
Synonyms | Yeast homologue |
Function | Motifs |
|---|---|---|---|---|
| CPSF | ||||
| CPSF160 | CPSF1 | CFT1/YHH1 | RNA-binding (AAUAAA) | SFT1, CPSF_A |
| Wdr33 | WDC146 | PFS2 | WD40 repeat | |
| Symplekin | PTA1 | Scaffolding | ||
| CPSF100 | CPSF2 | CFT2/YDH1 | b-CASP | |
| CPSF73 | CPSF3 | BRR5/YSH1 | Endonuclease | b-CASP |
| hFip1 | FIP1L1 | FIP1 | RNA-binding | |
| CPSF30 | CPSF4 | YTH1 | RNA-binding | Zinc Finger |
| CstF | ||||
| CstF77 | CSTF3 | RNA14 | Dimerization | HAT repeat |
| CstF64 | CSTF2 | RNA15 | RNA-binding (DSE) | RRM |
| CstF64 t | CSTF2T | PTI1? | RNA-binding (DSE) | RRM |
| CstF50 | CSTF1 | WD40 repeat | ||
| CF Im | ||||
| CF Im 68 | CPSF6 | RNA-binding | RRM | |
| CF Im 59 | CPSF7 | RNA-binding | RRM | |
| CF Im 25 | CPSF5 | Dimerization | Nudix | |
| CF IIm | ||||
| hPcf11 | PCF11 | CID | ||
| hClp1 | HEAB | CLP1 | RNA kinase | GTPase |
| Other factors | ||||
| PAP | PAPOLA | PAP1 | Poly(A) Polymerase | RNA-binding, NTP |
| Neo-PAP | PAPOLG | Poly(A) Polymerase | RNA-binding, NTP | |
| Star-PAP | TUT1 | CID1 (S. pombe) | Poly(A) Polymerase | RNA-binding, NTP, RS |
| PABPN1 | Poly(A) tail length control | RRM | ||
| PABPC1 | PAB1 | Poly(A) binding | 4 RRMs | |
| PABPC4 | PAB1 | Poly(A) binding | 4 RRMs | |
| RNAP II large subunit | RPB1 | CTD | ||
| PP1 | PPP1CC | GLC7 | Phosphatase | PP2Ac |
| Rbbp6 | PACT | MpeI | DWNN, Zinc finger, RING | |
| Other yeast polyadenylation factors | ||||
| Unknown | HRP1 | RNA-binding | 2 RRMs | |
| Unknown | REF2 | RNA-binding | ||
| Unknown | SYC1 | CPSF73-100_C | ||
| Wdr82 | SWD2 | WD40 repeat | ||
| Ssu72 | SSU72 | CTD phosphatase | Ssu72 | |
1. CPSF
CPSF is required for both cleavage and polyadenylation. It recognizes the A(A/U)UAAA element, helps to recruit other components of the 3’ processing complex, and catalyzes cleavage1,2,13. Originally it was shown that the purified CPSF consisted of CPSF160, 100, and 73 (Ref. 29), and in some preparations also CPSF30 (Ref. 30). Later studies have added symplekin, hFip1, and Wdr33 to the list of CPSF components14,31,32. Next we will discuss the roles of the individual CPSF subunits in the context of the three aforementioned critical functions of CPSF.
CPSF in RNA binding
CPSF recognizes the A(A/U)UAAA element with remarkable specificity as any single mutation within the hexamer strongly represses processing33,34. Surprisingly, however, none of the CPSF subunits has a canonical RNA recognition motif (RRM). CPSF160 is believed to be the major CPSF subunit responsible for recognizing the A(A/U)UAAA element based on multiple lines of evidence. For example, CPSF160 can be UV-crosslinked to AAUAAA-containing RNAs33,35, and recombinant CPSF160 preferentially binds to RNAs containing AAUAAA than those with mutant hexamers36. Two fragments within the N-terminal half of CPSF160 bear limited similarities to the RRM domain sequence and were initially suspected to mediate RNA recognition36. However, a later study of Cft1p, the yeast CPSF160 homologue, attributed its RNA-binding activity to a central domain37. Although this region seems to be fairly well conserved from CFT1 to CPSF160, mutational analysis is needed to directly test the involvement of this segment in A(A/U)UAAA recognition by CPSF160.
At least two other subunits of the CPSF are involved in RNA binding, hFip1 and CPSF30 (Refs. 32,38). The C-terminal arginine-rich domain of hFip1 binds to U-rich sequences32. The RNA-binding activity of CPSF30 is mediated by its zinc finger domain and it also prefers poly(U) sequences38. Since many USEs are U-rich sequences (Fig. 1), hFip1 and CPSF30 may be involved in recognizing the USEs and other sequences, thereby providing additional RNA-protein contacts that may stabilize the specific interaction between CPSF160 and the A(A/U)UAAA element.
Protein-protein interactions of CPSF
CPSF subunits interact with one another extensively1,2,13 (Fig. 2). CPSF100, 73, and symplekin have recently been suggested to form the core of CPSF complex while other subunits join the complex as peripheral factors39. Furthermore, CPSF recruits other components of the 3’ processing complex through direct physical interactions (Fig. 2). For example, CPSF can be isolated in a pre-assembled complex with CstF, and this association is mediated by multiple factors, including CPSF160, hFip1, and symplekin31,32,36(Fig. 2). Wdr33 may also be involved in bridging CPSF and CstF as its yeast homologue Pfs2p interacts with factors that are homologous to subunits of the CPSF and CstF complexes40. These interactions allow cooperative binding of CPSF and CstF to A(A/U)UAAA and the DSE1,2,13. Following cleavage, CPSF remains bound to the A(A/U)UAAA element and anchors PAP to the RNA for polyadenylation through direct interactions between CPSF160, hFip1 and PAP32,36.
CPSF also interacts with a variety of regulatory proteins to modulate 3’ processing either globally or in a transcript-specific manner. For example, the influenza protein NS1 represses the 3’ processing of host cellular pre-mRNAs by inhibiting CPSF activity via direct interaction with CPSF30 (Ref. 41). In another example, following stress, HSF1 not only activates transcription of heat shock protein (Hsp) genes, but also promotes the 3’ processing of Hsp transcripts through its association with symplekin42. Finally, the RRM-containing floral repressor FCA interacts with CPSF through FY, the plant homologue of Wdr33, to control flowering time at least in part by modulating the 3’ processing of specific transcripts43.
CPSF in the catalysis of cleavage
CPSF73 has been identified as the endonuclease responsible for the cleavage step based on several lines of evidence44,45. First, CPSF73 can be UV-crosslinked to the cleavage site in the RNA substrate44. Secondly, CPSF73 contains a metallo-beta-lactamase domain and a β-CASP domain45. Some members of the β-CASP protein family, such as Artemis, are known endonucleases46. Thirdly, recombinant CPSF73 shows zinc-dependent endonuclease activity45. It is noted that the endonuclease activity of the recombinant CPSF73 is very weak and stimulation by other 3’ processing factor(s) may be necessary for efficient cleavage. Although CPSF100 is structurally highly similar to CPSF73, it lacks the zinc-binding motif and, as a result, does not possess endonuclease activity45. The functions of CPSF100 remain unclear.
The endonuclease activity of CPSF73 was also revealed by studies on metazoan histone pre-mRNA 3’ processing47. Directed by unique cis-elements, the 3’ processing of metazoan histone pre-mRNAs involves only a cleavage step without polyadenylation48. Recent studies suggest that some CPSF subunits, including CPSF73, 100 and symplekin, function in this process and CPSF73 functions as the endonuclease for cleavage47,49. Intriguingly, CPSF73 has been suggested to play additional role in histone pre-mRNA 3’ processing as the exonuclease for degradation of the downstream RNA50.
2. CstF
CstF recognizes the DSE and is specifically required for cleavage but apparently not for polyadenylation1,2,13. CstF is comprised of three subunits, CstF77, CstF50 and either CstF64 or its paralog CstF64 τ51,52. Recent studies suggest that CstF may function as a dimer in the 3’ processing complex53,54.
CstF in RNA binding
The RNA binding activity of CstF is mediated by CstF64/τ (Ref. 51). CstF64 contains an N-terminal RRM domain (Fig. 3), which on its own binds with high affinity to GU-rich sequences similar to those found in natural DSEs55. The intact CstF complex, however, binds to PAS-containing pre-mRNAs very weakly as shown by both UV crosslinking and gel mobility shift assays29. Stable association of CstF to DSE requires cooperative binding of CPSF to the upstream A(A/U)UAAA element29. Although the exact mechanism is not well understood, this type of obligatory cooperative binding may ensure that 3’ processing complex assembles only when both the A(A/U)UAAA and DSE are present and the spacing between the two elements is appropriate.
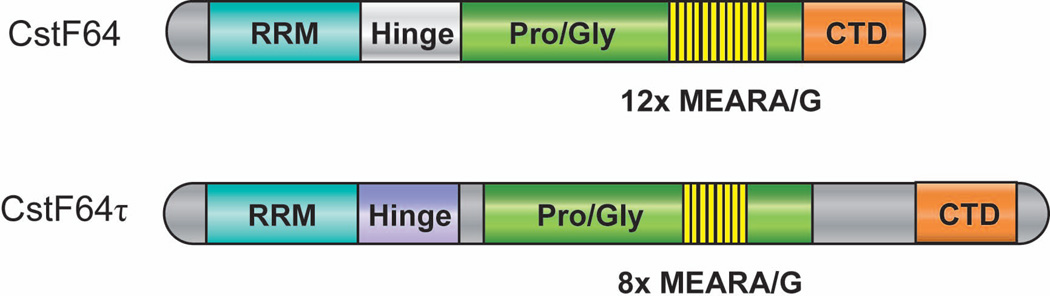
Figure 3. Domain structures of CstF 64 and CstF 64τ.
Rectangular boxes represent individual domains, which include RRM (RNA recognition motif), hinge domain, Pro/Gly (proline/glycine-rich domain), 12/8× MEARA/G (12/8 tandem copies of MEARA/G repeats), and CTD (C-terminal domain). Adapted from reference 1.
CstF64 τ shares the same domain structure as CstF64 (Fig. 3), but seems to have different RNA-binding specificity56. CstF64 τ is highly expressed in the testis and has been proposed to mediate testis-specific poly(A) site choice52. CstF64 τ knockout mice showed specific defects in spermatogenesis and male infertility57, suggesting that the CstF64 τ-mediated pre-mRNA 3’ processing plays important roles in the testis. Therefore, the relative abundance of CstF64 and related proteins may be involved in regulating tissue-specific APA. Both CstF64 and CstF64 τ seem to be evolutionarily conserved from yeast to human14 (Table 1.).
Protein-protein interactions of CstF
CstF77 is the scaffold holding the CstF complex together as it interacts with both CstF64 and CstF50 via its proline-rich domain while CstF64 and CstF50 do no interact with each other31,58 (Fig. 2). In addition, CstF77 also contains a so-called Half a TPR (HAT) domain. Crystal structures revealed that the CstF77 HAT domain forms a homodimer53,54. This is consistent with earlier observations that both CstF77 and its yeast homologue RNA14p can self-associate31,59. In addition, CstF50 may also dimerize31. Together, these results provided strong evidence that CstF functions as a dimer in the 3’ processing complex.
In addition to the aforementioned RRM domain, CstF64/τ contains a “hinge” domain, a Pro/Gly-rich region, and a highly conserved C-terminal domain (CTD) (Fig. 3). Embedded within the Pro/Gly-rich region is 12 tandem copies of MEARA/G (mouse and human) or 11 copies of LEPRG (chicken) pentapeptide repeats, but the functions of these repeats as well as the entire Pro/Gly-rich domain remain unclear1,2,13. The CstF64 “hinge” domain mediates its interactions with CstF77 and its CTD binds to Pcf11 and the transcription co-activator PC4 (Refs. 60–62). As mentioned earlier, both CstF77 and 64 interact with CPSF subunits to facilitate the cooperative binding of CPSF and CstF to pre-mRNAs (Fig. 2).
As mentioned above, CstF50 can self-associate and this interaction is mediated by its N-terminal region31. Furthermore, CstF50 contains seven WD-40 repeats, a domain known for mediating protein-protein interactions1,2,13. CstF50 binds to RNAP II CTD and this interaction has been proposed to help recruit CstF to the transcription elongation complex63. Following UV-induced DNA damage, CstF50 becomes associateed with the BRCA1-BARD1 complex and other factors and these interactions are responsible for transiently repressing pre-mRNA 3’ processing64.
3. CF Im
Similar to CstF, CF Im is involved in RNA-binding and is required only for the cleavage step1,2,13. CF Im consists of CF Im25 and one of two structurally related proteins, CF Im 59 and CF Im68. Again similar to CstF, CF Im has recently been shown to function as a dimer65,66. There are no clear homologues of CF Im in yeast (Table 1.). But it has been proposed that CF Im may be the functional equivalent of the yeast 3’ processing factor Hrp1(Ref. 22), a RNA-binding factor that does not have a clear mammalian homologue.
CF Im in RNA binding
All CF Im subunits can be UV crosslinked to RNA67, suggesting that they are all involved in RNA recognition. Recombinant CFIm25–68 binds specifically to UGUAN motif and this interaction has recently been shown to be mediated by CF Im25 (Refs. 66,68). CF Im25 contains a Nudix domain and belongs to the Nudix phosphohydrolase superfamily65. However CF Im25 differs from other Nudix hydrolases at several key residues and lacks enzymatic activity65. Instead the CF Im25 Nudix domain functions as a RNA-binding domain that specifically recognizes the UGUAN motif66. CF Im59 and 68 both have a RRM domain, but only bind to RNA in the presence of CF Im25 (Ref. 69). Currently it is unclear what RNA sequences CF Im59 and 68 recognize and what the functional significance of their RNA-binding activities is. CF Im-RNA interactions help improve the 3’ processing efficiency at canonical PASes22,70. For noncanonical PASes that lack the A(A/U)UAAA element but contain UGUAN motifs, CF Im can function as the primary RNA-binding factor for 3’ processing complex assembly and recruit other 3’ processing factors such as CPSF and PAP through direct interactions22. In keeping with the important roles of CF Im25 in PAS recognition, depletion of CF Im25 by RNAi causes significant changes in the APA profile for many transcripts71.
Protein-protein interactions of CF Im
In addition to self-association, CF Im25 also interacts with PAP and PABPN1 (Ref. 13). As mentioned earlier, both CF Im 59 and 68 have an RRM domain. Interestingly, however, the RRM of CF Im68 is required for its interaction with CF Im25 (Ref. 69). CF Im 59 and 68 also contain a C-terminal RS domain that is rich in arginine/serine dipeptide repeats. The RS domain is commonly found in the SR protein family, a large group of proteins best known as splicing regulators72. The RS domain has been shown to mediate RNA-binding as well as protein-protein interactions, including interactions with other SR proteins72. Indeed, CF Im 59 and 68 binds to a number of SR and SR-like proteins69,73, and the CF Im complex has been identified as a component of the spliceosome74. The interactions between CF Im and splicing factors play important roles in coupling splicing and 3’ processing.
4. CF IIm
CF IIm is required only for the cleavage step1,2,13, but its exact functions remain unclear. CF IIm contains at least two subunits, Pcf11 and hClp1, both conserved from yeast to human. Pcf11 homologues in yeast and Drosophila are required for both pre-mRNA 3’ processing and transcription termination75. Pcf11 binds to the RNAP II CTD in a phosphorylation-dependent manner via its CTD-interacting domain (CID domain)75. It has been proposed that RNAP II-bound Pcf11 promotes transcription termination by dismantling the elongation complex76. Additionally, the yeast Pcf11p has been shown to recruit the mRNA export adaptor Yra1p to the mRNA, thereby linking pre-mRNA 3’ processing to mRNA export77.
Although its role in pre-mRNA 3’ processing remains unknown, hClp1 has recently been identified as an RNA 5’-kinase and it functions as a component of the endonuclease complex in tRNA splicing78,79. The kinase activity of hClp1 is required for maintaining the phosphorylation of the 5’-end of the 3’ exon for the subsequent ligation78. It is still unclear, however, if the kinase activity of hClp1 is necessary for pre-mRNA 3’ processing. But the recombinant yeast Clp1 protein has no detectable kinase activity80, indicating that the kinase activity may be unique to Clp1 homologues in higher eukaryotes and this activity may not be required for pre-mRNA 3’ processing itself.
5. Poly(A) polymerases
PAP is perhaps the best-characterized 3’ processing factor so far1,2,13. A nucleotidyltransferase catalytic domain occupies the N-terminal half of PAP and is highly conserved (Fig. 4). A RNA-binding domain is located near the middle of the protein. RNA binding by PAP alone, however, is not sequence-specific and its recruitment to 3’ processing complex requires interactions with other 3’ processing factors including CPSF and CF Im1,2,13. The C-terminal domain (CTD) of PAP contains a bipartite nuclear localization signal (NLS) and is rich in serine and threonine residues. The CTD is a hot spot for post-translational modifications and plays important regulatory roles. First, PAP CTD is hyperphosphorylated during mitosis by the mitosis promoting factor (MPS, p34cdc2/cyclin B) and, as a result, PAP activity is strongly inhibited81. Repression of PAP activity is part of the cellular mechanism for blocking protein synthesis during mitosis. Secondly, PAP CTD binds to 14-3-3ε in a phosphorylation-dependent manner and this association inhibits PAP activity and redistributes PAP to the cytoplasm82. Thirdly, PAP CTD is acetylated by the CREB-binding protein (CBP) and acetylation inhibits its association with CF Im25 and its nuclear localization83. Finally, PAP CTD is sumoylated at multiple sites, and sumoylation stabilizes PAP, promotes its nuclear localization, but inhibits its enzymatic activity in vitro84.
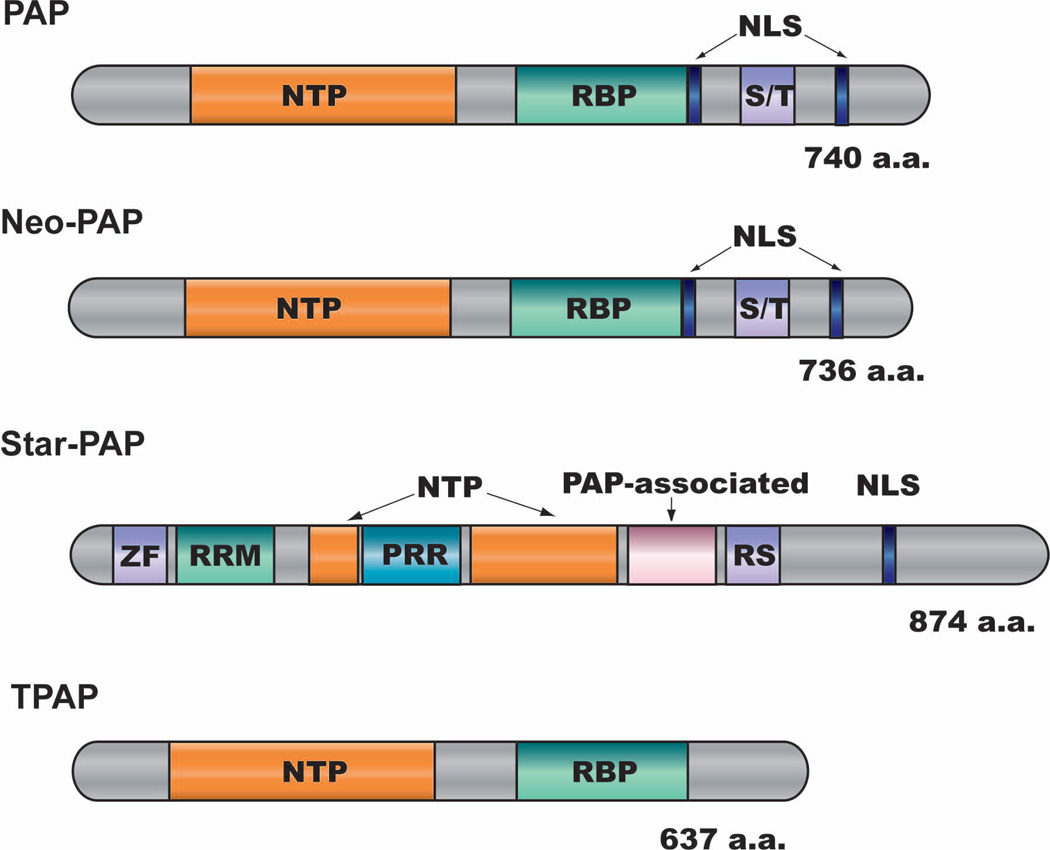
Figure 4. Domain structures of the three nuclear mammalian PAPs.
Rectangular boxes represent individual domains, which include NTP (Nucleotidyltransferase domain), RBP (PAP RNA-binding domain), NLS (nuclear localization signal), S/T (serine/threonine-rich domain), ZF (zinc finger domain), RRM (RNA recognition motif), PRR (proline-rich domain), and RS (RS domain). Adapted from reference 88.
In addition to the canonical PAP, metazoans have at least three additional nuclear poly(A) polymerases, neo-PAP, Star-PAP and TPAP (Fig. 4). Encoded by an intronless gene, TPAP is specifically expressed in the testis and can be found in both nucleus and the cytoplasm85. The functions of TPAP remain poorly understood. Neo-PAP shares the same overall domain organization with PAP and its in vitro polyadenylation activity is indistinguishable from that of the canonical PAP86,87. Neo-PAP can be incorporated into functional human 3’ processing complex in vitro14, but its in vivo functions have not been characterized.
Star-PAP has a very different domain structure (Fig. 4). It contains a zinc finger domain and a RRM near its N-terminus, a split catalytic domain in the middle, and a PAP-associated domain as well as a RS domain in the C-terminal one third88. Star-PAP was originally identified as a terminal uridylyl transferase (TUTase) specific for U6 snRNA89. The putative homologue of Star-PAP in fission yeast, Cid1, also has TUTase activity90. A later study, however, demonstrated that Star-PAP preferentially uses ATP as substrate and mainly functions as a poly(A) polymerase88. Star-PAP functions in a complex with the type I phosphatidylinositol 4-phosphate 5-kinases (PIPKIα) as well as subunits of the CPSF to control the 3’ processing of a subset of transcripts88. Interestingly, the activity of Star-PAP is directly regulated by phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P)88. Compare to S. cerevisiae which only has one nuclear PAP, the presence of multiple nuclear PAPs in metazoans may allow for more elaborate regulation of pre-mRNA 3’ processing under different conditions.
6. PABPs
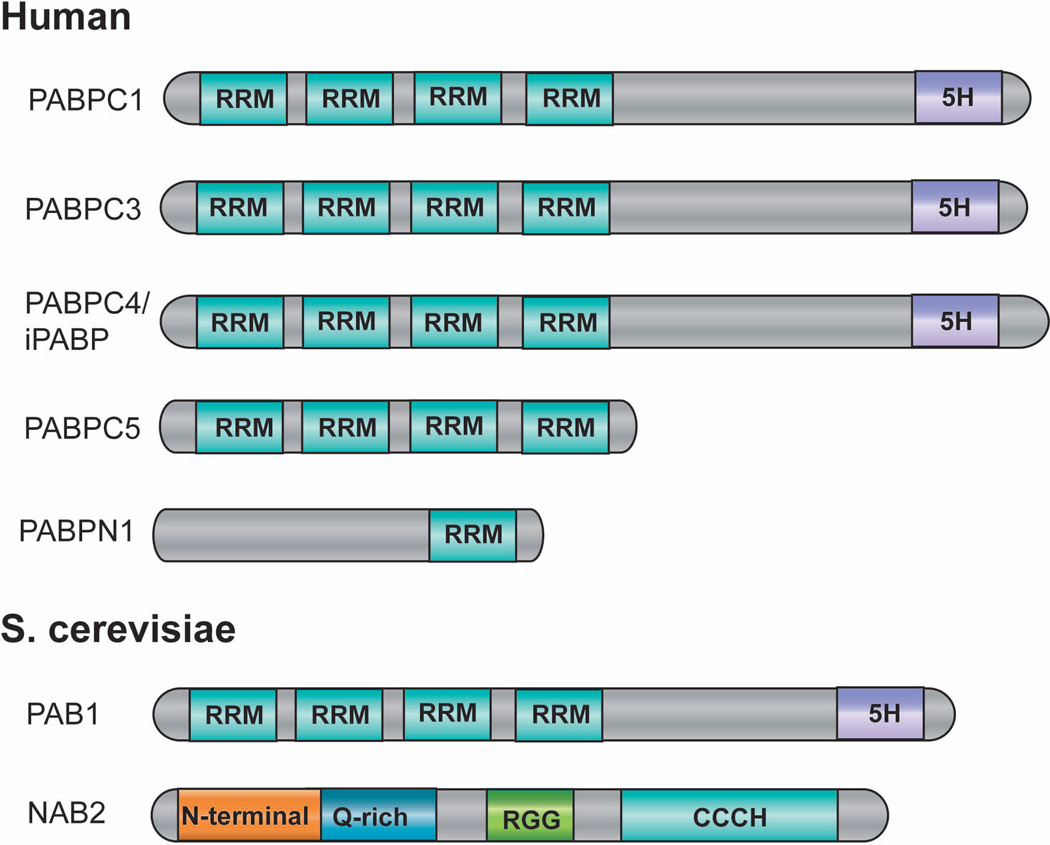
There are at least 5 PABP proteins in humans, one nuclear form (PABPN1) and 4 cytoplasmic ones (PABPC1, 3–5)28,91 (Fig. 5). The nuclear and the cytoplasmic PABPs have distinct domain structures. It has been shown that PABPN1 stimulates PAP activity and plays an important role in poly(A) tail length control92. Following cleavage, PAP, anchored to the pre-mRNA by CPSF, initiates polyadenylation in a slow and distributive reaction92. Binding of PABPN1 to the newly synthesized poly(A) tail stabilizes the polyadenylation complex and dramatically stimulates PAP activity. As a result, polyadenylation switches to a fast and processive phase. PABPN1 coats the entire length of poly(A) tail as it emerges from PAP. When the poly(A) tail reaches ~250nt in length, polyadenylation is terminated by a poorly understood but PABPN1-dependent mechanism92. PABPs have also been implicated in promoting mRNA export28,91. In the cytoplasm, PABPs play critical roles in translation by promoting 5’-3’ interaction of the mRNA and stimulating initiation28,91.
Figure 5. Domain structure of yeast and human PABPs.
Rectangular boxes represent individual domains, which include RRM (RNA recognition motif), 5H (a unique 5 conserved helices domain), RGG (RGG box involved in RNA binding), N-terminal (N-terminal domain), Q-rich (Q-rich domain) and CCCH (zinc finger domain). Adapted from reference 28.
The budding yeast has only one PABP gene (PAB1) (Fig. 5), which is the orthologue of the mammalian cytoplasmic PABPs28,91. The budding yeast does express a structurally distinct nuclear poly(A) binding protein Nab2p (Fig. 5), which is probably the functional equivalent of PABPN1. Both Nab2p and Pab1p have been implicated in poly(A) tail length control and mRNA export28,91.
7. RNAP II CTD
In addition to transcribing genes, RNAP II also plays critical roles in coordinating co-transcriptional pre-mRNA processing93–95. This function is mediated by its CTD63, a unique domain consisting of multiple repeats of the conserved heptapeptide YSPTSPS. The number of the repeat ranges from 26 in yeast to 52 in mammals. In vitro, RNAP II CTD is required for cleavage in the absence of transcription96. Although the exact functions of CTD in pre-mRNA 3’ processing are unclear, it may promote the assembly of a stable 3’ processing complex through its extensive interactions with other 3’ processing factors, including CPSF, CstF, and CF IIm63,93–95,97.
PRE-MRNA 3’ PROCESSING COMPLEX: ASSEMBLY AND DYNAMICS
In vivo, pre-mRNA 3’ processing factors are intimately connected with the transcription machinery and the 3’ processing complex assembles on pre-mRNAs co-transcriptionally93. CPSF joins the transcription machinery as early as in the preinitiation complex stage through interactions with TFIID98. CPSF, CstF, and CF Im are all associated with RNAP II during transcription elongation93,95. After transcription passes the PAS, RNAP II pauses and perhaps during this time the 3’ processing complex fully assembles on the pre-mRNA to carry out cleavage and polyadenylation93.
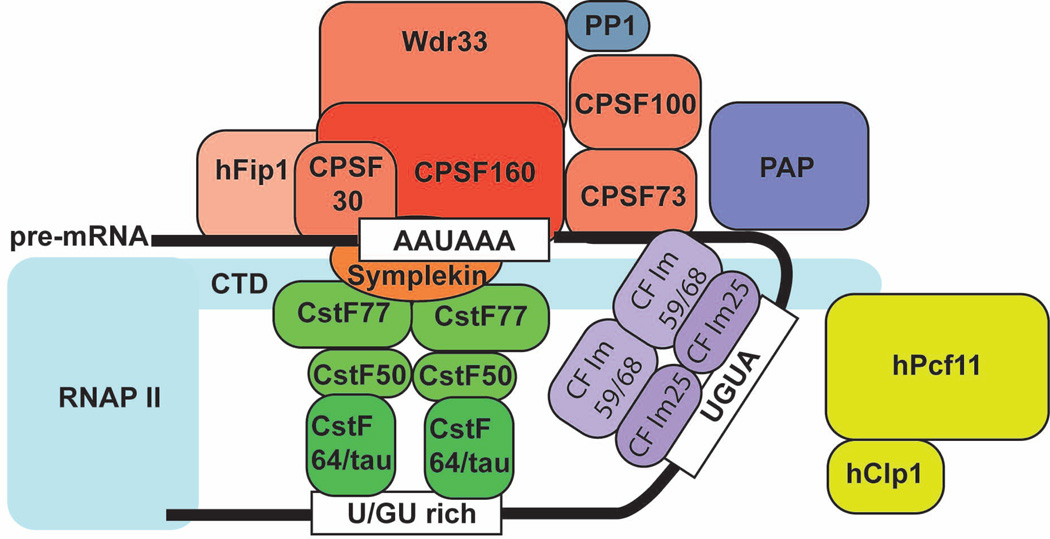
In vitro the pre-mRNA 3’ processing complex assembles in a two-step process2,15. First, CPSF, CstF and CF Im bind to the key cis-elements within PASes in a mutually stimulatory manner to form a fairly stable complex. Next CF IIm and PAP join the core complex transiently and/or weakly to form the complete 3’ processing complex (Fig. 6). In the latest model of the 3’ processing complex, CstF and CF Im both function as dimers. Dimererization of these factors may allow for more RNA-protein contacts which may in turn stablize the 3’ processing complex. It is unclear whether any structural reorganization of the assembled 3’ processing complex takes place before cleavage. As mentioned earlier, the recombinant CPSF73 has very low endonuclease activity45. For efficient cleavage, other factor(s) within the 3’ processing complex may directly stimulate CPSF73 activity by changing its conformation and/or by positioning the pre-mRNA for optimal contact with CPSF73.
Figure 6. A proposed model for the mammalian pre-mRNA 3’ processing complex.
The core complex consists of CPSF bound to the A(A/U)UAAA element, CstF bound to the downstream U/GU-rich DSE as a dimer, CF Im bound to the UGUA sequences also as a dimer, and the RNAP II CTD. The nuclear PAPs (PAP/neo-PAP/star-PAP) and CF IIm associate with the core complex weakly and/or transiently.
Little is known about what exactly takes place within the 3’ processing complex after cleavage. Since the polyadenylation step can be reconstituted in vitro with CPSF and PAP1,2,13, it is possible that CstF, CF Im and CF IIm may dissociate from the complex following cleavage along with the downstream RNA. These putative compositional and/or conformational changes during the cleavage/polyadenylation transition could be controlled by post-translational modification of 3’ processing factors. For example, the yeast phosphatase Glc7p is required specifically for polyadenylation, but not for cleavage and Pta1p has been identified as a substrate99. A similar requirement for PP1, the human Glc7 homologue, has been demonstrated for mammalian polyadenylation14. It has been proposed that dephosphorylation of one or more 3’ processing factors by PP1 following cleavage may alter the RNA-protein and/or protein-protein interaction network to facilitate polyadenylation14.
Following cleavage, the A(A/U)UAAA-bound CPSF anchors PAP to the pre-mRNA to carry out polyadenylation1,2. The dynamics of the 3’ processing complex during polyadenylation has been discussed earlier (see the PABPs section). Meanwhile, the downstream RNA generated by cleavage is digested by the exoribonuclease Xrn2100,101. Xrn2 is recruited to the 3’ processing complex through interactions with the multi-functional protein p54nrb/PSF and perhaps some basal 3’ processing factors as well102. The degradation of the downstream RNA by Xrn2 is responsible, at least in part, for transcription termination as proposed by the “torpedo” model100,101.
When polyadenylation is completed, the 3’ processing complex needs to be disassembled and mature mRNAs are exported to the cytoplasm. The mechanism for 3’ processing complex disassembly remains poorly understood, but a recent study in yeast provided strong evidence that multiple mRNA export factors are required103. For example, mutations in the mRNA export receptor Mex67 leads to retention of 3’ processing factors on RNAs and inhibition of 3’ processing. For transcripts that do get processed, their poly(A) tails are unusually long. These data indicates that mRNA export factors may play an important role in the disassembly of the 3’ processing complex103. While the disassembly is taking place, some components of the 3’ processing complex remain associated with the polyadenylated mRNAs as part of the export-competent mRNP. For example, a recent study provided evidence that CF Im68 remains associated with the mRNA throughout 3’ processing and serves as an adaptor protein for export104. Similarly, another well-characterized mRNA export adaptor in yeast Yra1 is recruited to mRNA during 3’ processing through direct interaction with the cleavage factor Pcf11 and functions in the subsequent mRNA export77. Finally the multifunctional nuclear protein nucleophosmin has been shown to bind mRNA in a polyadenylation-dependent manner105, but the functional significance of this association remains unknown. The tight coupling between pre-mRNA 3’ processing and the mRNA export may ensure that only fully processed mRNAs are exported.
PRE-MRNA 3’ PROCESSING COMPLEX: REGULATION
Like other steps of the gene expression pathway, pre-mRNA 3’ processing is subject to regulation1,2,4. Many examples of 3’ processing regulation have been reported and the different modes of regulation can be classified into the following three general mechanisms.
Regulation of the basal 3’ processing factors
Changes in the protein levels, accessibility, or the activities of the basal 3’ processing factors can influence pre-mRNA 3’ processing and their effects can be global or transcript-specific.
First, changes in the protein levels of basal 3’ processing factors can regulate 3’ processing. For example, CstF64 is specifically up-regulated when primary B cells are induced to differentiate106,107. The limited CstF complexes in primary B cells preferentially bind to the high affinity distal poly(A) site of the IgM pre-mRNA, resulting in the production of the membrane-bound form of IgM. However, in activated B cells in which CstF is more abundant, 3’ processing shifts to a “first-come, first-serve” mode and a lower affinity proximal poly(A) site is now preferentially recognized, leading to synthesis of the secreted form of IgM106,107. It is highly likely that many more transcripts are regulated in a similar fashion by the changing levels of CstF64. Additionally the protein levels of CF Im subunits show developmental stage-specific variations and may cause changes in 3’ processing efficiency as well as APA patterns108.
Secondly, changes in the accessibility of the basal 3’ processing factors can regulate 3’ processing. For example, regulatory viral or cellular proteins could sequester the basal 3’ processing factors and prevent them from participating in 3’ processing. As mentioned earlier, the influenza virus NS1 protein specifically interacts with CPSF30, thereby inhibiting CPSF-RNA interaction and cleavage/polyadenylation of cellular pre-mRNAs41. As mentioned earlier, CstF50 becomes associated with the BARD1/BRCA1 complex following UV-induced DNA damage and this association leads to transient inhibition of pre-mRNA 3’ processing64.
Finally, changes in the activity of the basal 3’ processing factors can regulate 3’ processing. Multiple post-translational modifications have been shown to modulate the activities of 3’ processing factors. As mentioned earlier, hyperphosphorylation of PAP during mitosis represses PAP activity and leads to general inhibition of polyadenylation81. Other modifications could have more profound effects. For example, sumoylation affects the stability, subcellular localization, and activities of PAP84. CPSF73 and symplekin are also sumoylated and sumoylation stimulates the assembly of the 3’ processing complexes109.
Regulation of 3’ processing complex assembly by local RNA context and regulatory factors
First, 3’ processing at a specific PAS can be affected by regulatory factors bound to adjacent cis-elements. For example, 3’ processing is activated by an upstream intron110. This stimulation is mediated by multiple interactions between 3’ processing factors and splicing factors, such as the CF Im-U2AF and CPSF-U2 snRNP interactions, and their cooperative binding to the PAS and the upstream splicing cis-elements73,111. Conversely, 3’ processing is strongly inhibited by a 5’ splice site located upstream of a PAS112. This is due to inhibition of PAP activity by U1–70K and U1A of the U1 snRNP bound to the upstream 5’ splice site112. Another example of this type of regulation is provided by the 3’ processing of the HIV pre-mRNA at the 3’ LTR113. The SR protein 9G8, in a complex with CDK11 and eIF3f, binds to a specific sequence upstream of the AAUAAA hexamer. Through its interaction with CF Im bound to an adjacent UGUAN motif, 9G8 stimulates 3’ processing at the downstream PAS. Interestingly, overexpression of eIF3f or just its N-terminal 91 amino acid fragment disrupts the eIF3f-CDK11-9G8 complex and prevents efficient 3’ processing at the 3’ LTR, leading to reduced viral replication113.
Secondly, some regulatory factors could directly compete with basal 3’ processing factors for binding to specific cis-elements within the PAS, thereby inhibiting 3’ processing. For example, both the mammalian PTB (polypyrimidine tract-binding protein) and the Drosophila Sxl (sex-lethal) proteins can compete with CstF64 for binding to the DSE114,115. The outcome of this competition is determined by the affinities of these factors for the DSE and their relative abundance.
Regulation of pre-mRNA 3’ processing after complex assembly
Although regulation of pre-mRNA 3’ processing most often occurs at the earliest step, i.e. the 3’ processing complex assembly, it should be pointed out that later steps are also subject to regulation. For example, U1A protein regulates its own expression through modulating the 3’ processing of its own transcripts116. U1A proteins bind to a specific RNA structure in the 3’ UTR of its own transcripts just upstream of the PAS. The RNA-bound U1A specifically inhibits PAP activity and therefore prevents polyadenylation without affecting 3’ processing complex assembly or cleavage. This forms a negative feedback loop that helps maintain the steady state levels of U1A protein116.
Conclusion
Despite the tremendous progress in the past decades, many key questions remain regarding the basic mechanisms of pre-mRNA 3’ processing complex assembly and function as well as its regulation. First, what are the defining features of a PAS? Although a number of key cis-elements have been identified, one or more of these elements are missing in many PASes. In the coming years, the key elements in the noncanonical PASes will undoubtedly become better defined and the mechanisms for their recognition by the 3’ processing machinery will be revealed. It will be of great interest to see whether there are any unifying features for all PASes. Secondly, what are the functions of each 3’ processing factor? Systematic structure-function analyses of individual 3’ processing factors as well as the entire 3’ processing complex will provide new insights into the molecular mechanisms of pre-mRNA 3’ processing, including RNA recognition, complex assembly, catalysis, and complex disassembly. Finally, as shown by recent studies7–11, APA is a widespread phenomenon in higher eukaryotes. It will critical to understand how 3’ processing complex assembly is regulated at alternative poly(A) sites and what is the impact of APA under physiological as well as pathological conditions. Instead of focusing on one transcript or one 3’ processing regulator at a time, it is becoming increasingly important to take a systems approach to investigate how 3’ processing is regulated at the transcriptome level. Coupling high-throughput sequencing- and microarray-based genomic analyses with biochemical characterization will be critical in this effort.
References
- 1.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutz CS. Alternative polyadenylation: a twist on mRNA 3' end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 7.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji Z, Tian B. Reprogramming of 3' untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danckwardt S, Hentze MW, Kulozik AE. 3' end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3'-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, et al. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Chan S, Martinez-Santibanez G. An up-close look at the pre-mRNA 3'-end processing complex. RNA Biol. 2009;6:522–525. doi: 10.4161/rna.6.5.9554. [DOI] [PubMed] [Google Scholar]
- 16.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Dickson AM, Wilusz J. Polyadenylation: alternative lifestyles of the A-rich (and famous?) EMBO J. 2010;29:1473–1474. doi: 10.1038/emboj.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graveley BR, Fleming ES, Gilmartin GM. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadofsky M, Alwine JC. Sequences on the 3' side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984;4:1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hans H, Alwine JC. Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol Cell Biol. 2000;20:2926–2932. doi: 10.1128/mcb.20.8.2926-2932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sittler A, Gallinaro H, Jacob M. The secondary structure of the adenovirus-2 L4 polyadenylation domain: evidence for a hairpin structure exposing the AAUAAA signal in its loop. J Mol Biol. 1995;248:525–540. doi: 10.1006/jmbi.1995.0240. [DOI] [PubMed] [Google Scholar]
- 28.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 30.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3'-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–19776. [PubMed] [Google Scholar]
- 31.Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3' processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore CL, Chen J, Whoriskey J. Two proteins crosslinked to RNA containing the adenovirus L3 poly(A) site require the AAUAAA sequence for binding. EMBO J. 1988;7:3159–3169. doi: 10.1002/j.1460-2075.1988.tb03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3'-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 37.Dichtl B, et al. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan KD, Steiniger M, Marzluff WF. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell. 2009;34:322–332. doi: 10.1016/j.molcel.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohnacker M, Barabino SM, Preker PJ, Keller W. The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3'-end-processing complex. Embo J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3'end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 42.Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
- 43.Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 44.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3' processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandel CR, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5' exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol. 2009;29:31–42. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 52.Wallace AM, et al. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci U S A. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai Y, et al. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 54.Legrand P, Pinaud N, Minvielle-Sebastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monarez RR, MacDonald CC, Dass B. Polyadenylation proteins CstF-64 and tauCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J. 2007;401:651–658. doi: 10.1042/BJ20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dass B, et al. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci U S A. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takagaki Y, Manley JL. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 59.Noble CG, Walker PA, Calder LJ, Taylor IA. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 2004;32:3364–3375. doi: 10.1093/nar/gkh664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hockert JA, Yeh HJ, MacDonald CC. The hinge domain of the cleavage stimulation factor protein CstF-64 is essential for CstF-77 interaction, nuclear localization, and polyadenylation. J Biol Chem. 285:695–704. doi: 10.1074/jbc.M109.061705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu X, et al. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3'-end processing. J Biol Chem. 2007;282:2101–2115. doi: 10.1074/jbc.M609981200. [DOI] [PubMed] [Google Scholar]
- 62.Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 63.McCracken S, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 64.Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3' end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 65.Coseno M, et al. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 2008;36:3474–3483. doi: 10.1093/nar/gkn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Q, Gilmartin GM, Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3' processing. Proc Natl Acad Sci U S A. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3' end processing of messenger RNA precursors. J Biol Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 68.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3' processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 69.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 70.Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 71.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3'-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Millevoi S, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3' end processing machineries. Embo J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 75.Proudfoot N. New perspectives on connecting messenger RNA 3' end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3'-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3' end processing factor Pcf11. Mol Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3' transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 79.Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3' end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 80.Ramirez A, Shuman S, Schwer B. Human RNA 5'-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA. 2008;14:1737–1745. doi: 10.1261/rna.1142908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 82.Kim H, Lee JH, Lee Y. Regulation of poly(A) polymerase by 14-3-3epsilon. EMBO J. 2003;22:5208–5219. doi: 10.1093/emboj/cdg486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimazu T, Horinouchi S, Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3'-end processing. J Biol Chem. 2007;282:4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- 84.Vethantham V, Rao N, Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22:499–511. doi: 10.1101/gad.1628208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee YJ, Lee Y, Chung JH. An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Lett. 2000;487:287–292. doi: 10.1016/s0014-5793(00)02367-x. [DOI] [PubMed] [Google Scholar]
- 86.Topalian SL, et al. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J Biol Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- 88.Mellman DL, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 89.Trippe R, et al. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 92.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 94.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 95.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 96.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 97.Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3' end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3' end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 99.He X, Moore C. Regulation of yeast mRNA 3' end processing by phosphorylation. Mol Cell. 2005;19:619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 100.West S, Gromak N, Proudfoot NJ. Human 5' -->3' exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 101.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 102.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3' processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qu X, et al. Assembly of an export-competent mRNP is needed for efficient release of the 3'-end processing complex after polyadenylation. Mol Cell Biol. 2009;29:5327–5338. doi: 10.1128/MCB.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruepp MD, et al. Mammalian pre-mRNA 3' end processing factor CF I m 68 functions in mRNA export. Mol Biol Cell. 2009;20:5211–5223. doi: 10.1091/mbc.E09-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palaniswamy V, Moraes KC, Wilusz CJ, Wilusz J. Nucleophosmin is selectively deposited on mRNA during polyadenylation. Nat Struct Mol Biol. 2006;13:429–435. doi: 10.1038/nsmb1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edwalds-Gilbert G, Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol Cell Biol. 1995;15:6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 108.Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78:472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- 109.Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3' processing complex. Mol Cell Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 111.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3' end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 112.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 113.Valente ST, Gilmartin GM, Venkatarama K, Arriagada G, Goff SP. HIV-1 mRNA 3' end processing is distinctively regulated by eIF3f, CDK11, and splice factor 9G8. Mol Cell. 2009;36:279–289. doi: 10.1016/j.molcel.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Castelo-Branco P, et al. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gawande B, Robida MD, Rahn A, Singh R. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 2006;25:1263–1272. doi: 10.1038/sj.emboj.7601022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gunderson SI, et al. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]