Abstract
There is considerable debate about the most efficient way to interrogate rare coding variants in association studies. The options include direct genotyping of specific known coding variants in genes or, alternatively, sequencing across the entire exome to capture known as well as novel variants. Each strategy has advantages and disadvantages, but the availability of cost-efficient exome arrays has made the former appealing. Here we consider the utility of a direct genotyping chip, the Illumina HumanExome array (HE), by evaluating its content based on: 1. functionality; and 2. amenability to imputation. We explored these issues by genotyping a large, ethnically diverse cohort on the HumanOmniExpressExome array (HOEE) which combines the HE with content from the GWAS array (HOE). We find that the use of the HE is likely to be a cost-effective way of expanding GWAS, but does have some drawbacks that deserve consideration when planning studies.
Keywords: Illumina HumanExome array, expanding GWAS, genotyping rare SNPs, coding variants
INTRODUCTION
Methods to extend genome-wide association studies (GWAS) have recently become a topic of high interest. Despite a large number of notable successes in the discovery of genetic variants associated with various traits, including disease via GWAS, the variants identified to date collectively only explain a small fraction of the estimated heritability of most common, chronic diseases (Manolio et al., 2009). Unknown genetic factors, including polymorphisms that have yet to be identified through GWAS studies, likely account for the ‘missing heritability’ associated with complex traits (Yang et al., 2011; Visscher et al., 2012). One explanation for this missing heritability is that widely-used genotyping platforms for GWAS are designed to directly interrogate only common single nucleotide polymorphisms (SNPs). Therefore, rare coding variants, which have been shown to play a role in the etiology of many diseases, tend to be entirely omitted by most genotyping platforms used in GWAS as they are not in linkage disequilibrium (hence not imputable) with SNPs interrogated on these arrays (Evans et al., 2008; Sun et al., 2011). Thus, the examination of rare coding variants requires either sequencing technology or the direct genotyping of variants which have previously been identified. While the former may lead to a more comprehensive assessment of all forms of variation in coding regions, including the discovery of extremely rare and/or de novo variants, the latter provides an efficient, cost-effective alternative for interrogating a subset of known variants in coding regions (Flannick et al., 2012; Pasaniuc et al., 2012).
The value of direct genotyping of previously identified coding variants, as opposed to de novo sequencing of coding regions, is dependent on a few key issues. First, if one can identify known functionally relevant variants in coding regions it might be more expedient to focus on them in cost-effective direct genotyping studies than pursuing more costly sequencing studies that may identify many likely neutral variants. Second, if coding variants identified via sequencing are easily imputable from variants genotyped on standard GWAS platforms, then the need for directly genotyping these coding regions would be minimized and greater attention could be given to more reliable imputation strategies. Third, many coding variants, whether they are functional or amenable to imputation or not, are very rare and hence likely to be absent in many global populations. Thus, direct genotyping certain coding variants may only be useful for specific populations.
Here we assessed the potential benefits of directly genotyping rare coding variants on the Illumina Human Exome (HE) array by addressing these issues. As such, our assessment includes an examination of the functional content of variants included on the array. We also evaluated the amenability of the HE markers to imputation from the Illumina Human Omni Express (HOE). And lastly, we evaluated the allele frequency spectrum of the variants included on the HE chip. We find that, overall, the HE chip does not suffer severe drawbacks in the context of these issues, but of course is limited to assessments of known (i.e., previously identified) variants. Our analyses and results have important implications for future studies seeking to identify associations with coding variants.
MATERIAL AND METHODS
Subjects and genotyping
Participants were recruited from two southern Californian military personnel cohorts: 1. the Marine Resiliency Study (MRS), a prospective study of post-traumatic stress disorder (PTSD) involving United States Marines bound for deployment to Iraq or Afghanistan (Baker et al., 2012); and 2. a cross-sectional study of active duty service members and veterans of Operation Enduring Freedom/Operation Iraqi Freedom (OEF/OIF) (Pittman et al., 2012). The protocols for these studies were approved by the University of California-San Diego Institutional Review Board (IRB Protocols #110770, #070533, and #080851), and all subjects provided written informed consent to participate.
DNA samples from 2,585 study participants were acquired, and genotyping was carried out by Illumina (http://www.illumina.com/) using the HOEE version 12v1.0. Initial allele calling was performed by Illumina in Genome Studio (http://www.illumina.com) and the overall data quality was high: sample success rate was 99.95% (9 samples failed), locus success rate was 99.86%, and genotype call rate was 99.88%. Twenty-eight replicate pairs of samples undergoing genotyping were assessed for consistency and ultimately reproducibility of the assay and agreement of genotyping calls was achieved for >99.99% over all genotypes across these 28 pairs. Additional data cleaning was performed in PLINK v1.07 (Purcell et al., 2007) and included the removal of 224 markers with heterozygous haploid genotypes on the X, Y, or mitochondrial chromosome. The final dataset included 949,469 markers genotyped in 2,548 individuals (2538 males and 10 females) with a genotyping rate greater than 99.8%.
Ancestry determination
We estimated each individual‘s degree of European, African, Native American, Central Asian, East Asian and Oceanic admixture by comparing the individual’s genotypes to allele frequencies of 10,079 SNPs in common with a large set of reference individuals (Libiger and Schork, 2013). In short, the reference sample consisted of genotype data for 2,513 individuals of known ancestry who originated from 83 populations from around the world. These data were assembled from publicly available sources including the Human Genome Diversity Project (HGDP) (Cann et al., 2002), the Population Reference (POPRES) (Nelson et al., 2008), HapMap3 (Altshuler et al., 2010), and the University of Utah dataset (Xing et al., 2009). Admixture estimates were obtained in two steps using a supervised analysis implemented in the ADMIXTURE software (Alexander et al., 2009). In the first step, we computed initial admixture estimates for all individuals associated with each world population using the entire set of reference individuals and determined the estimates’ standard errors via bootstrapping. A subset of reference individuals from populations that exhibited evidence of contributing to an individual’s ancestry based on 95% confidence intervals was then used to refine the initial admixture estimates in a subsequent supervised ADMIXTURE analysis.
Final ancestry calling was based first on self-reported race and ethnicity information, and second, within each of these main population group, genetic ancestry estimates. Subjects were placed into 5 groups: European Americans (subjects with >95% European ancestry; N=1,476), Asian Americans (>95% East Asian ancestry; N=43); African-American (subjects with >5% African ancestry and <5% Native American, Central Asian, East Asian and Oceanic ancestry; N=109), Hispanic Americans (subjects with >5% Native American and <10% African, Central Asian, East Asian and Oceanic ancestry; N=321), and Other (all others; N=599). Thus, our ancestry assignments provide initial assignments consistent with the often-used admixture program except that they have been refined by removing noise and leveraging comparisons to self-reported ancestries.
Genotype imputations
Imputations were conducted using markers available on the HOE platform. Prior to imputation, mitochondrial and unmapped SNPs were removed from each set. Markers that were individually rare (minor allele frequency MAF < 0.0002), showed a large number of missing genotypes (> 5%), or failed Hardy-Weinberg equilibrium (p < 1 × 10−6) were also removed (Supplemental Table 1). Imputations were performed using the default parameters in IMPUTE2 v2.2.2, using 1000 Genomes Phase 1 integrated variant set haplotypes for the autosomes and the interim set for the X chromosome (Howie et al., 2009). IMPUTE2 is well suited for imputations on genetically diverse and admixed populations such as that of the present study as the algorithm is robust to ancestral genetic variation within the reference panel and study datasets (Howie et al., 2011). Genomes were divided into approximately 5 Mb segments (minimum 2.5 Mb, maximum 7.5 Mb to avoid chromosome and centromere boundaries), and phasing and imputed genotypes were calculated for each. Imputed markers with low imputation quality values (Info ≤ 0.5) were dropped. GTOOL v0.7.0 was used to convert genotype probabilities into calls. Individual genotype probabilities exceeding 90% were assigned genotype calls and probabilities ≤ 90% were treated as missing genotypes. Agreement between the imputation results and markers exclusive to HOEE (i.e., HE markers) was examined by calculating the correlation coefficient, r2, between calls on a per marker level. Missing genotypes were assigned an allelic dosage representing the mean genotype at that particular locus for all calculations. Imputation was also performed based on genotype data from the HOEE platform. A comparison of the agreement between the HOE and HOEE to impute markers that were not genotyped on either platform was, likewise, conducted.
Variant functional annotations
We mapped all variants to the closest gene from the UCSC Genome Browser known gene database (Fujita et al., 2011). Full details of our annotation pipeline are described in a previous publication (Torkamani et al., 2012) and the Supplemental Methods. In brief, variants were associated with all transcripts of the nearest gene(s), with functional impact predictions made independently for each transcript. If the variant fell within a known gene, its position within gene elements (e.g. exons, introns, untranslated regions, etc.) was recorded for functional impact predictions depending on the impacted gene element. All variants falling within an exon were analyzed for their impact on the amino acid sequence (e.g. synonymous, nonsynonymous, nonsense, frameshift, in-frame, intercodon etc.).
RESULTS
Characterization of the Cohort
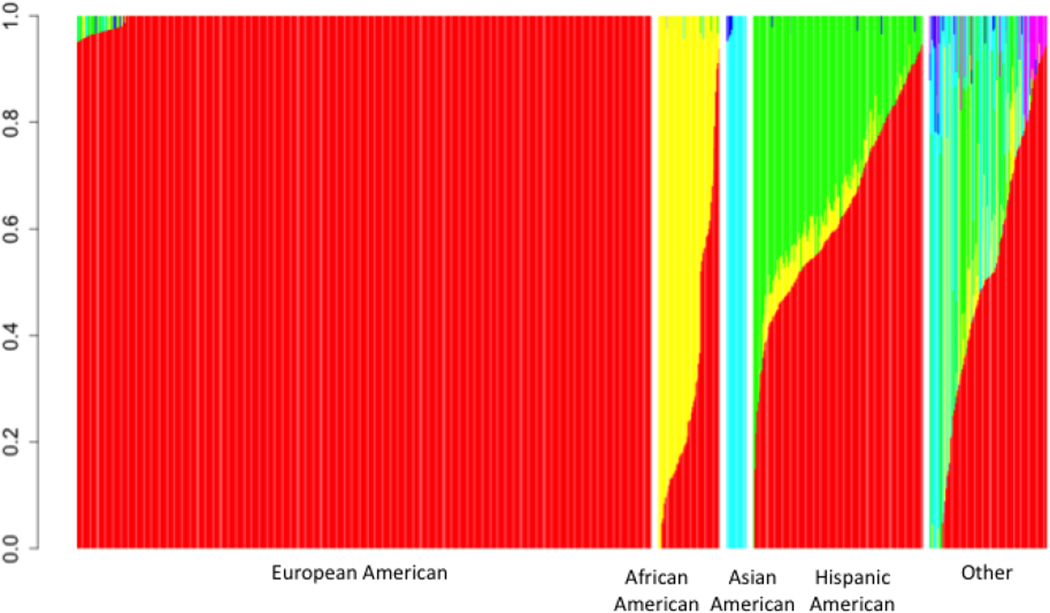
Table 1 provides a description of the cohort based on self-reported race and ethnicity information and includes the number of subjects, gender, and age of the subjects and the number of individuals removed from the study because of failed genotyping quality control (see Methods). Individual ancestry and admixture proportions were assessed within these self-reported race and ethnicity groups using genotype information (see also Methods) and a graphical representation of the ancestry/admixture among the subjects in the study is provided in Figure 1. We ultimately identified 1,476 individuals with predominantly European ancestry, 109 African-American individuals, 43 with predominantly East Asian ancestry, 321 with predominantly Hispanic American ancestry (i.e., with significant Native American admixture), and 599 with predominant ancestry from any other geoethnic population. We used these combined self-reported and genetically-determined ancestries in subsequent analyses.
Table 1.
Descriptive Statistics for the Cohorts Studied Based on Self-Reported Race and Ethnicity.
| Measure | Number of Subjects |
Males/Females | Average Age |
# Poor Genotype QC |
|---|---|---|---|---|
| Self-Reported Race: | ||||
| Black/African American | 128 | 128/0 | 25.38 | 1 |
| American Indian/Alaska | 35 | 35/0 | 22.66 | 0 |
| Asian | 80 | 79/1 | 24.94 | 1 |
| Pacific Island/Hawaiian | 39 | 38/1 | 22.96 | 0 |
| White | 2104 | 2096/8 | 23.25 | 7 |
| Multiple Races | 125 | 125/0 | 22.50 | 0 |
| Unknown | 46 | 46/0 | 23.19 | 0 |
| Self-Reported Ethnicity: | ||||
| Non-Hispanic | 1951 | 1946/5 | 23.42 | 8 |
| Hispanic | 601 | 596/5 | 23.18 | 1 |
| Unknown | 5 | 5/0 | 22.00 | 0 |
| Total: | 2557 | 2547/10 | 23.36 | 9 |
Figure 1.
Admixture proportion of individuals included in the study. Each individual is represented by a vertical bar divided into colored segments. The size of each colored segment reflects the proportion of admixture from one of six major continental populations (red – European; Yellow – African; green – Native American; turquoise – East Asian; blue – Oceanic; magenta – Central Asian). Individuals in each ancestral category are sorted by the degree of European admixture (i.e., size of red segments).
Imputability of the HE Markers
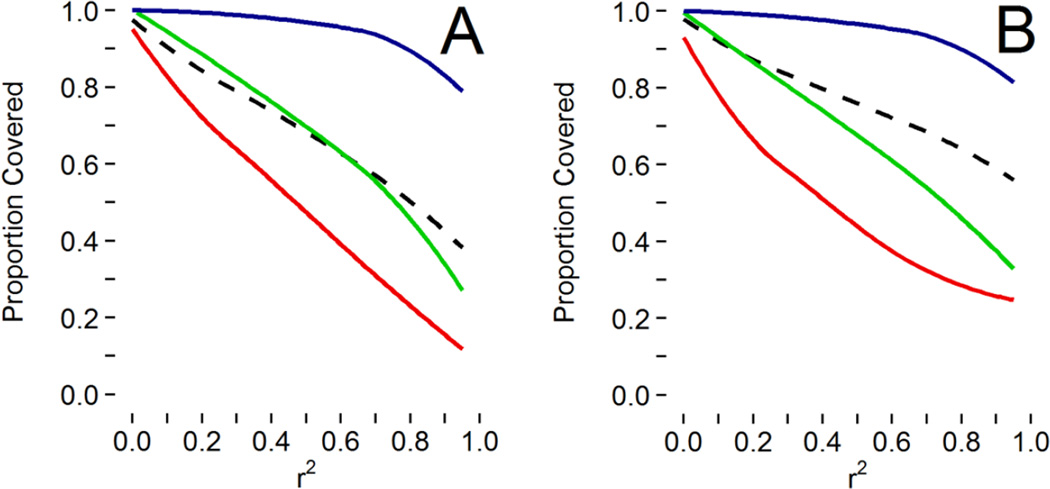
We explored the possibility that the markers which were exclusive to the HOEE array (i.e., the HE content) could be imputed from markers on the HOE array. If these markers are amenable to imputation, it would call into question the utility of the additional content on the HOEE chip. Only a modest proportion of the markers exclusive to the HOEE array were imputable from the HOE content and passed imputation quality control thresholds (N=80,205; 32.9%). Among these, markers with common variants (MAF>0.05; N=27,250) were imputed accurately across all ethnicities: 76.4% of common markers had r2>0.95 and 90.6% had r2>0.80. However, markers with moderately common (0.01≤MAF≤0.05; N=9,777) and rare (MAF<0.01; N=43,178) variants were imputed more poorly: 46.8% and 22.9% with r2>0.80, respectively. Overall, only 50.6% (N=40,620) of all imputable markers were accurately imputed across ethnicities (Figure 2a). Considering the HE included 158,878 non-monomorphic markers in this sample (among 243,783 total genotyped markers), only approximately one-quarter of variable HE content – and one-sixth of the total HE content – could be recapitulated from imputation via the HOE content. Note that we did not consider the small number of Y-chromosome (N=180) and mtDNA markers (N=245) available on the HE chip.
Figure 2.
The proportion of imputable markers (N=80,205) exclusive to the HOEE (i.e. HE content) covered by imputation, based on the HOE and 1000 Genomes reference haplotypes across: a) all subjects (N=2,548); b) European Americans (N=1,476). Marker frequencies: blue – common (MAF>0.05); green – moderately common (0.01≤MAF≤0.05); red – rare (MAF<0.01); and black dashed – all.
Imputation accuracy was also assessed separately for European Americans (N=1,476, Figure 2b). We found a trend towards decreasing imputation accuracy with decreasing minor allele frequency. The proportion of markers which could be imputed accurately (r2>0.80) was 65%. The small numbers of subjects in the other ancestry groups precluded statistical comparisons.
Finally, the total number of markers that could be imputed based on the HOE and HOEE, but not present on either platform, were considered. A large number of markers were successfully imputed at an acceptable quality (i.e., information threshold greater than 0.5) on both platforms (Supplemental Table 2). The total counts and overlap between HOE and HOEE were very similar. Only slightly more markers were imputed accurately using HOEE compared to HOE (22,961,598 and 22,898,511, respectively). Markers with rare variants (MAF<0.01) accounted for roughly 54% of the approximately 23 million accurately imputed markers, while markers with common variants (MAF>0.05) accounted for 30%. In general, there was high concordance of imputed genotypes between the HOE and HOEE (Supplemental Figure S1). Approximately 17 million markers had r2>0.8. Thus, the performance of the HOE and HOEE to impute markers not present on either platform was determined to be roughly equivalent.
Functional Content for Markers Interrogated by the HE array
Of the 949,469 markers that passed genotyping QC (see Methods), the known or likely functional significance of 931,570 markers could be assessed using a suite of bioinformatics and computational procedures as described in (Torkamani et al., 2012) (see Methods). Of the 237,627 markers interrogated on the HE chip, there were 237,489 single-nucleotide variants (SNVs), 43 insertions, and 95 deletions. The classification of these markers into 9 functional groups is shown in Table 2 (left columns). Overall, 117,678 variants (49.5%) on the HE were predicted to be functional. When compared to the content on the more comprehensive HOEE array, we found that of the 122,668 HOEE functional variants, 117,678 (95.9%) were contributed by the HE. We also compared the contribution of functional content of the HE to the HOEE array after imputation (HOEEi; N = 22,961,598 markers amenable to imputation). We found that only approximately 0.7% of all variants capable of interrogation were likely to be functional (right columns of Table 2), suggesting that the HE chip is indeed substantially adding to the functional content available when using the HOE array, even after imputation. We note that some variants (N=1,143 or 0.12%) that were either interrogated on the HOEE chip or amenable to imputation were not amenable to functional prediction based on our computational procedures due to, for example, location inconsistencies in relevant databases.
Table 2.
Functional content of the variants on the Human Exome array (HE) and the Human Omni ExpressExome plus imputable marker array (HOEEi) indicating the number of variants and rate in each of nine functional classes (see Methods).
| Functional group | HE variants | Rate | HOEEi | Rate |
|---|---|---|---|---|
| Splicing Change Variants | 372 | 0.030 | 625 | 0.015 |
| Probably Damaging nscSNPs | 54,970 | 0.267 | 67,328 | 0.272 |
| Possibly Damaging nscSNPs | 39,144 | 0.190 | 46,290 | 0.187 |
| Protein motif damaging Variants | 23,304 | 0.292 | 27,283 | 0.293 |
| TFBS Disrupting Variants | 0 | 0.000 | 10 | 0.004 |
| pre-miRNA Disrupting Variants | 6 | 0.000 | 201 | 0.000 |
| miRNA-BS Disrupting Variants | 236 | 0.062 | 1,931 | 0.055 |
| ESE-BS Disrupting Variants | 17,500 | 0.117 | 27,058 | 0.117 |
| ESS-BS Disrupting Variants | 6,439 | 0.114 | 9,869 | 0.116 |
| Total Likely Functional Variants | 117,678 | 0.495 | 150,035 | 0.007 |
Overall and Functional Variant Frequencies
The majority of markers interrogated on the HE platform have very low minor allele frequencies. For example, 85% of markers exhibited minor allele frequency of 0.01 or less in our multi-ethnic cohort and similar trends were observed within each population. This observation has obvious implications on the utility of the HE in GWAS initiatives which focus on single marker tests. Assuming a small or moderate effect of variants on disease, most of the markers on the HE array will only provide sufficient power to detect associations between an allele and a disease using single marker tests if information on a very large number of case and control individuals is collected.
The mean (±s.d.) number of polymorphic markers per individual interrogated on the HE array was 15,746 (±215), and included 2,454 (±59) functional markers, 14.3 (±6.4) private markers, and 7.9 (±3.8) functional and private markers. Similar numbers were seen in the European American subgroup (total: 15,528±112; functional: 2,420±38; private: 10.1±3.8, functional & private: 5.7±2.6).
DISCUSSION
As the genetics community learns about the limitations of contemporary approaches to discovering variants that influence phenotypic expression, newer approaches will undoubtedly emerge. It is quite clear that despite the spectacular and numerous successes in identifying associated variants via GWAS initiatives focusing on common variants and linkage disequilibrium phenomena, there is a large fraction of the genetic basis of most diseases and traits that has yet to be characterized. This could be due to one or more of the following factors: (1) rarity or relatively small effect sizes of the remaining variants contributing to those conditions; (2) forms of variation not hitherto explored in as comprehensive a manner as SNPs and small indels in GWAS initiatives (e.g., copy number of variants and large structural variations); (3) complicated gene×environment interactions; (4) epigenetic factors; and, (5) other phenomena (Frazer et al., 2009; Manolio et al., 2009; Schork et al., 2009).
The contribution of rare variants to phenotypic expression is getting more and more attention given the availability of cost-efficient sequencing technologies (Bodmer and Bonilla, 2008; Frazer et al., 2009; Schork et al., 2009; Bansal et al., 2010; Gibson, 2011; Malhotra and Sebat, 2012; Pasaniuc et al., 2012). However, sequencing technologies may still be cost-prohibitive for large-scale association studies. Therefore, the genetics research community has considered the use of genotyping platforms that can interrogate previously identified variants that are not easily captured via linkage disequilibrium on standard genotyping platforms used in GWAS initiatives. Choosing the markers to be used on such arrays is crucial, but a focus on coding variants (i.e., the exome) is a logical starting point (despite the fact that coding variants tend to be rare) since it has been shown that they are likely to be functional and have been implicated in a number of diseases and phenotypes (Botstein and Risch, 2003; Jordan et al., 2010; Gorlov et al., 2011; Sunyaev, 2012). However, designing a genotyping array that would complement existing genotyping platforms is not necessarily trivial. For example, imputation strategies are gaining sophistication making it possible to avoid the use of newer assays by computationally assigning variants to individuals based on linkage disequilibrium patterns in the genome and available data sets (Marchini and Howie, 2010; Flannick et al., 2012). Thus markers interrogated on newer platforms should optimally contain those not amenable to imputation. In addition, if markers are to be chosen for direct genotyping, then it makes sense to bias them towards those likely to include functional variants. Finally, many rare variants are likely to be population-specific, including those likely to be functional (Kidd et al., 2012; Torkamani et al., 2012), making the choice of which variants to include on a genotyping array complicated. For example, a researcher may not wish to invest in a genotyping platform if many of the markers being interrogated are not likely to be found in the populations of interest.
We explored these issues with a newly available genotyping array (the Illumina HE) designed to capture coding variants that are complementary to markers currently interrogated by other genotyping arrays. We find that as much as 49.5% of the markers interrogated by the array are likely to impact the function of genes. In addition, as only a small proportion of the HE content was amenable to imputation, we feel the addition of these markers provides an improvement over the previous GWAS array design – although it is possible that larger imputation reference panels may close this gap.
A limitation of our dataset is the unequal representation of different racial/ethnic groups with a relatively small number of Hispanics, African Americans, and subjects of other race, which precluded a detailed comparison of population-specific variants. In addition, our cohort was almost exclusively male, which effectively reduced the number of X chromosomes by half and did not allow for a comparison between gender. However, since analyses were based on the combined genomic content of the array, this should not impact our conclusions. Obviously, the choice of a genotyping platform will have to be based on the goals of a study. For example, if a study requires the accommodation of de novo, very rare, or likely population-specific variants, then the use of an array designed to interrogate variants that have been previously identified is inappropriate. However, if the goal of a study is to efficiently expand the search for likely causative variants that are ‘beneath the radar’ of standard GWAS genotyping platforms, then genotyping arrays focusing on rare variants that are likely to be functional, such as coding variants, makes sense. The design of those arrays in terms of the variants they interrogate, however, is crucial for their success.
Supplementary Material
Research Highlight.
Direct genotyping chips for coding variants provide an alternative to sequencing.
Assessed utility of Illumina HumanExome array.
Evaluation of functionality and amenability to imputation.
Exome array is cost-effective way to expand GWAS, but has some drawbacks.
ACKNOWLEDGEMENTS
CMN is supported by NIH grants 1 R01MH093500, 1 U01 MH092758, by the Marine Corps and Navy Bureau of Medicine and Surgery (BUMED). NJS is supported in part by NIH grants 5 UL1 RR025774, R21 AI085374, 5 U01 DA024417, 5 R01 HL089655, 5 R01 DA030976, 5 R01 AG035020, 1 R01 MH093500, 2 U19 AI063603, 2 U19 AG023122, 5 P01 AG027734, 1 R21 DA033813 as well as grants from Johnson and Johnson, the Veteran’s Administration, the Viterbi Foundation, the Stand-Up-to-Cancer organization, the Price Foundation and Scripps Genomic Medicine. DGB is supported to this in part by the Marine Corps and Navy Bureau of Medicine and Surgery (BUMED), VA Health Services Research and Development (VAHSR&D), VA Clinical Research and Development (VA CSR&D), NIMH R01MH093500, and the VA Center of Excellence for Stress and Mental Health (CESAMH). We acknowledge special assistance from members of VA Center of Excellence for Stress and Mental Health, VA San Diego Research and Fiscal Services, and the 1st Marine Division and Navy Medicine at 29 Palms and at Camp Pendleton.
Abbreviations
- HE
HumanExome array
- HOEE
HumanOmniExpressExome array
- HOE
HumanOmniExpressGWAS array
- GWAS
genome-wide association studies
- SNPs
single nucleotide polymorphisms
- MRS
the Marine Resiliency Study
- PTSD
post-traumatic stress disorder
- OEF/OIF
Operation Enduring Freedom/Operation Iraqi Freedom
- IRB
Institutional Review Board
- HGDP
Human Genome Diversity Project
- MAF
minor allele frequency
- SNVs
single-nucleotide variants
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DG, Nash WP, Litz BT, Geyer MA, Risbrough VB, Nievergelt CM, O'Connor DT, Larson GE, Schork NJ, Vasterling JJ, Hammer PS, Webb-Murphy JA. Predictors of risk and resilience for posttraumatic stress disorder among ground combat Marines: methods of the Marine Resiliency Study. Prev Chronic Dis. 2012;9:E97. doi: 10.5888/pcd9.110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Libiger O, Torkamani A, Schork N. Statistical analysis strategies for association studies involving rare variants. Nature Reviews Genetics. 2010;11:773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Evans DM, Barrett JC, Cardon LR. To what extent do scans of nonsynonymous SNPs complement denser genome-wide association studies? Eur J Hum Genet. 2008;16:718–723. doi: 10.1038/sj.ejhg.5202011. [DOI] [PubMed] [Google Scholar]
- Flannick J, Korn JM, Fontanillas P, Grant GB, Banks E, Depristo MA, Altshuler D. Efficiency and power as a function of sequence coverage, SNP array density, and imputation. PLoS Comput Biol. 2012;8:e1002604. doi: 10.1371/journal.pcbi.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov IP, Gorlova OY, Frazier ML, Spitz MR, Amos CI. Evolutionary evidence of the effect of rare variants on disease etiology. Clin Genet. 2011;79:199–206. doi: 10.1111/j.1399-0004.2010.01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DM, Ramensky VE, Sunyaev SR. Human allelic variation: perspective from protein function, structure, and evolution. Curr Opin Struct Biol. 2010;20:342–350. doi: 10.1016/j.sbi.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Gravel S, Byrnes J, Moreno-Estrada A, Musharoff S, Bryc K, Degenhardt JD, Brisbin A, Sheth V, Chen R, McLaughlin SF, Peckham HE, Omberg L, Bormann Chung CA, Stanley S, Pearlstein K, Levandowsky E, Acevedo-Acevedo S, Auton A, Keinan A, Acuna-Alonzo V, Barquera-Lozano R, Canizales-Quinteros S, Eng C, Burchard EG, Russell A, Reynolds A, Clark AG, Reese MG, Lincoln SE, Butte AJ, De La Vega FM, Bustamante CD. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libiger O, Schork NJ. A method for inferring an individual's genetic ancestry and degree of admixture associated with six major continental populations. Frontiers in Genetics. 2013;3:1–11. doi: 10.3389/fgene.2012.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Bryc K, King KS, Indap A, Boyko AR, Novembre J, Briley LP, Maruyama Y, Waterworth DM, Waeber G, Vollenweider P, Oksenberg JR, Hauser SL, Stirnadel HA, Kooner JS, Chambers JC, Jones B, Mooser V, Bustamante CD, Roses AD, Burns DK, Ehm MG, Lai EH. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Rohland N, McLaren PJ, Garimella K, Zaitlen N, Li H, Gupta N, Neale BM, Daly MJ, Sklar P, Sullivan PF, Bergen S, Moran JL, Hultman CM, Lichtenstein P, Magnusson P, Purcell SM, Haas DW, Liang L, Sunyaev S, Patterson N, de Bakker PI, Reich D, Price AL. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat Genet. 2012;44:631–635. doi: 10.1038/ng.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JO, Goldsmith AA, Lemmer JA, Kilmer MT, Baker DG. Posttraumatic stress disorder, depression, and health-related quality of life in OEF/OIF veterans. Qual Life Res. 2012;21:99–103. doi: 10.1007/s11136-011-9918-3. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Namkung J, Zhu X, Elston RC. Capability of common SNPs to tag rare variants. BMC Proc. 2011;5(Suppl 9):S88. doi: 10.1186/1753-6561-5-S9-S88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev SR. Inferring causality and functional significance of human coding DNA variants. Hum Mol Genet. 2012;21:R10–R17. doi: 10.1093/hmg/dds385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Pham P, Libiger O, Bansal V, Zhang G, Scott-Van Zeeland AA, Tewhey R, Topol EJ, Schork NJ. Clinical implications of human population differences in genome-wide rates of functional genotypes. Front Genet. 2012;3:211. doi: 10.3389/fgene.2012.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Watkins WS, Witherspoon DJ, Zhang Y, Guthery SL, Thara R, Mowry BJ, Bulayeva K, Weiss RB, Jorde LB. Fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 2009;19:815–825. doi: 10.1101/gr.085589.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.