Abstract
The C31 subunit belongs to a complex of three subunits (C31, C34 and C82) specific to RNA polymerase (pol) III that have no counterparts in other RNA polymerases. This complex is thought to play a role in transcription initiation since it interacts with the general initiation factor TFIIIB via subunit C34. We have obtained a conditional mutation of pol III by partially deleting the acidic C-terminus of the C31 subunit. A Saccharomyces cerevisiae strain carrying this truncated C31 subunit is impaired in in vivo transcription of tRNAs and failed to grow at 37 degrees C. This conditional growth phenotype was suppressed by overexpression of the gene coding for the largest subunit of pol III (C160), suggesting an interaction between C160 and C31. The mutant pol III enzyme transcribed non-specific templates at wild-type rates in vitro, but was impaired in its capacity to transcribe tRNA genes in the presence of general initiation factors. Transcription initiation, but not termination or recycling of the enzyme, was affected in the mutant, suggesting that it could be altered on interaction with initiation factors or on the formation of the open complex. Interestingly, the C-terminal deletion was also suppressed by a high gene dosage of the DED1 gene encoding a putative helicase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Ingles C. J. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Durkovich D., Kassavetis G. A., Geiduschek E. P. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993 Feb;13(2):942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Burton N., Cavallini B., Kanno M., Moncollin V., Egly J. M. Expression in Escherichia coli: purification and properties of the yeast general transcription factor TFIID. Protein Expr Purif. 1991 Oct-Dec;2(5-6):432–441. doi: 10.1016/1046-5928(91)90105-r. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F. E., Jr, Setzer D. R. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol Cell Biol. 1992 May;12(5):2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiannilkulchai N., Moenne A., Sentenac A., Mann C. Biochemical and genetic dissection of the Saccharomyces cerevisiae RNA polymerase C53 subunit through the analysis of a mitochondrially mis-sorted mutant construct. J Biol Chem. 1992 Nov 15;267(32):23099–23107. [PubMed] [Google Scholar]
- Chiannilkulchai N., Stalder R., Riva M., Carles C., Werner M., Sentenac A. RPC82 encodes the highly conserved, third-largest subunit of RNA polymerase C (III) from Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4433–4440. doi: 10.1128/mcb.12.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert T., Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992 Oct;6(10):1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- Costigan C., Gehrung S., Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992 Mar;12(3):1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Duimio L., Coda-Zabetta F., Sprague K. U., Ottonello S. A novel RNA polymerase III transcription factor fraction that is not required for template commitment. J Biol Chem. 1993 May 25;268(15):11199–11207. [PubMed] [Google Scholar]
- Edwards A. M., Kane C. M., Young R. A., Kornberg R. D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991 Jan 5;266(1):71–75. [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gabrielsen O. S., Marzouki N., Ruet A., Sentenac A., Fromageot P. Two polypeptide chains in yeast transcription factor tau interact with DNA. J Biol Chem. 1989 May 5;264(13):7505–7511. [PubMed] [Google Scholar]
- Gabrielsen O. S., Oyen T. B. The requirement for the A block promoter element in tRNA gene transcription in vitro depends on the ionic environment. Nucleic Acids Res. 1987 Jul 24;15(14):5699–5713. doi: 10.1093/nar/15.14.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudenus R., Mariotte S., Moenne A., Ruet A., Memet S., Buhler J. M., Sentenac A., Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988 Jul;119(3):517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Werner M., Sentenac A., Thuriaux P. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol Cell Biol. 1994 May;14(5):2905–2913. doi: 10.1128/mcb.14.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Riva M., Sentenac A., Fromageot P. Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985 Dec 5;260(28):15304–15310. [PubMed] [Google Scholar]
- James P., Hall B. D. ret1-1, a yeast mutant affecting transcription termination by RNA polymerase III. Genetics. 1990 Jun;125(2):293–303. doi: 10.1093/genetics/125.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson D. J., Beggs J. D. A suppressor of yeast spp81/ded1 mutations encodes a very similar putative ATP-dependent RNA helicase. Mol Microbiol. 1991 Apr;5(4):805–812. doi: 10.1111/j.1365-2958.1991.tb00753.x. [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., Rahe B., Pringle J., Beggs J. D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991 Feb 21;349(6311):715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Karlin S. Unusual charge configurations in transcription factors of the basic RNA polymerase II initiation complex. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5593–5597. doi: 10.1073/pnas.90.12.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Bartholomew B., Blanco J. A., Johnson T. E., Geiduschek E. P. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Blanco J. A., Johnson T. E., Geiduschek E. P. Formation of open and elongating transcription complexes by RNA polymerase III. J Mol Biol. 1992 Jul 5;226(1):47–58. doi: 10.1016/0022-2836(92)90123-2. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Joazeiro C. A., Pisano M., Geiduschek E. P., Colbert T., Hahn S., Blanco J. A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992 Dec 11;71(6):1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Hitpass L., Tsai S. Y., Weigel N. L., Allan G. F., Riley D., Rodriguez R., Schrader W. T., Tsai M. J., O'Malley B. W. The progesterone receptor stimulates cell-free transcription by enhancing the formation of a stable preinitiation complex. Cell. 1990 Jan 26;60(2):247–257. doi: 10.1016/0092-8674(90)90740-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Margottin F., Dujardin G., Gérard M., Egly J. M., Huet J., Sentenac A. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science. 1991 Jan 25;251(4992):424–426. doi: 10.1126/science.1989075. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Carraway M., Frey A. Z., Brown L., Arraj J. A. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192(1-2):288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- Moenne A., Camier S., Anderson G., Margottin F., Beggs J., Sentenac A. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 1990 Jan;9(1):271–277. doi: 10.1002/j.1460-2075.1990.tb08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
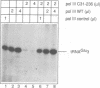
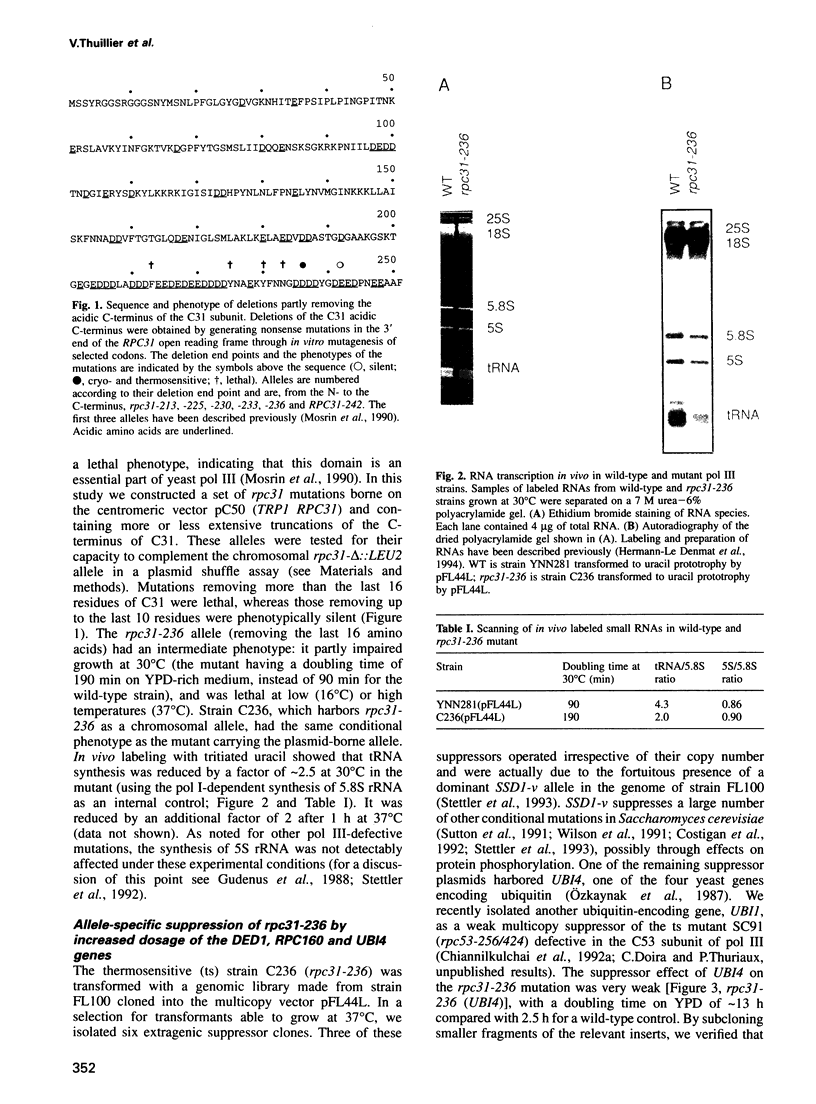
- Mosrin C., Riva M., Beltrame M., Cassar E., Sentenac A., Thuriaux P. The RPC31 gene of Saccharomyces cerevisiae encodes a subunit of RNA polymerase C (III) with an acidic tail. Mol Cell Biol. 1990 Sep;10(9):4737–4743. doi: 10.1128/mcb.10.9.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Ozkaynak E., Finley D., Solomon M. J., Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987 May;6(5):1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafe C., Martin C., Nonet M., Podos S., Okamura S., Young R. A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990 Mar;10(3):1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
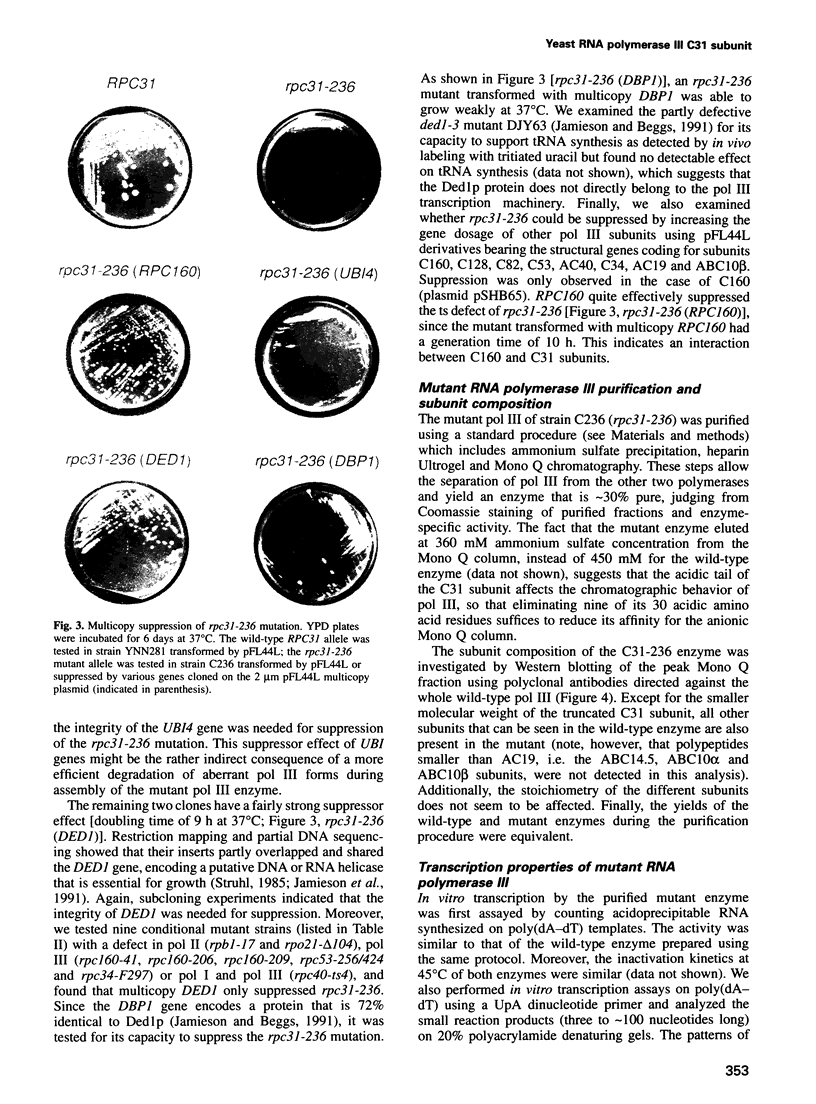
- Stettler S., Chiannilkulchai N., Hermann-Le Denmat S., Lalo D., Lacroute F., Sentenac A., Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1993 May;239(1-2):169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- Stettler S., Mariotte S., Riva M., Sentenac A., Thuriaux P. An essential and specific subunit of RNA polymerase III (C) is encoded by gene RPC34 in Saccharomyces cerevisiae. J Biol Chem. 1992 Oct 25;267(30):21390–21395. [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Immanuel D., Arndt K. T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991 Apr;11(4):2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I., Riva M., Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem. 1991 Nov 15;266(32):21971–21976. [PubMed] [Google Scholar]
- Valenzuela P., Hager G. L., Weinberg F., Rutter W. J. Molecular structure of yeast RNA polymerase III: demonstration of the tripartite transcriptive system in lower eukaryotes. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1024–1028. doi: 10.1073/pnas.73.4.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Chaussivert N., Willis I. M., Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993 Oct 5;268(28):20721–20724. [PubMed] [Google Scholar]
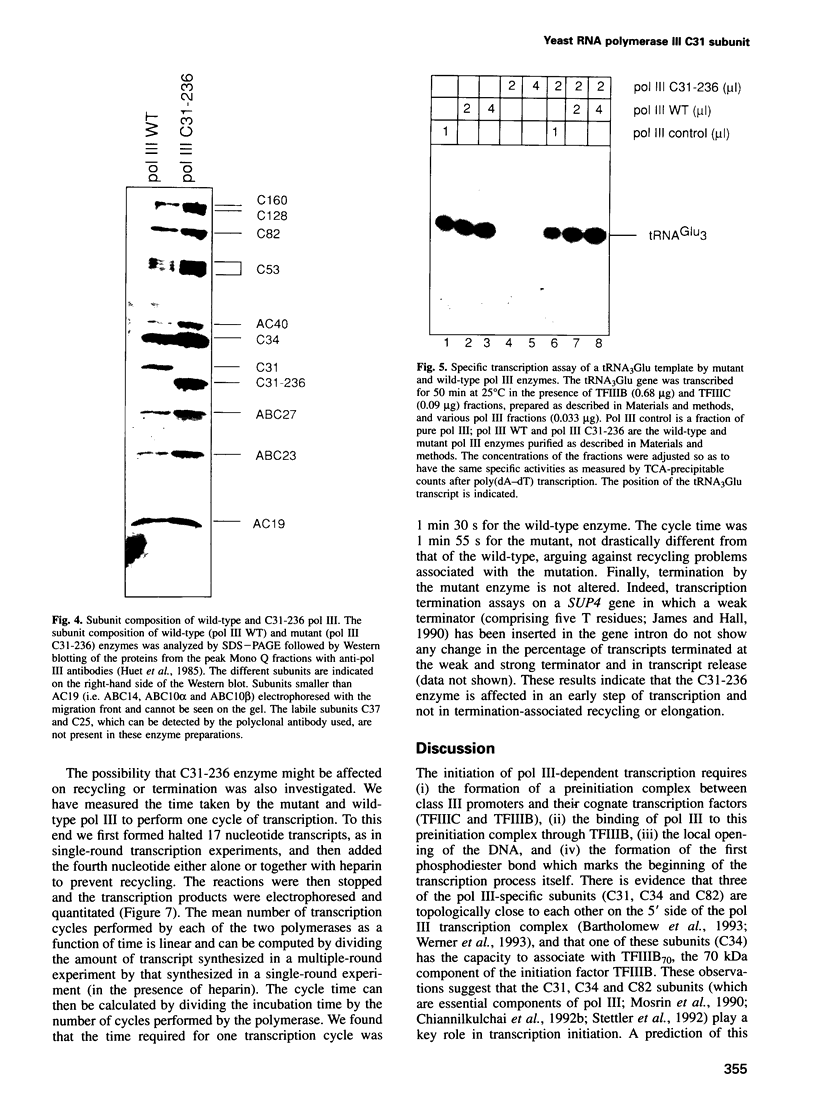
- Werner M., Hermann-Le Denmat S., Treich I., Sentenac A., Thuriaux P. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol. 1992 Mar;12(3):1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. B., Brenner A. A., White T. B., Engler M. J., Gaughran J. P., Tatchell K. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol Cell Biol. 1991 Jun;11(6):3369–3373. doi: 10.1128/mcb.11.6.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol. 1989 Jul;9(7):2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]