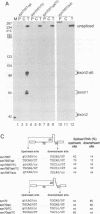
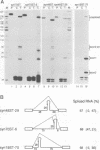
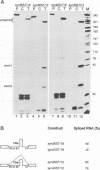
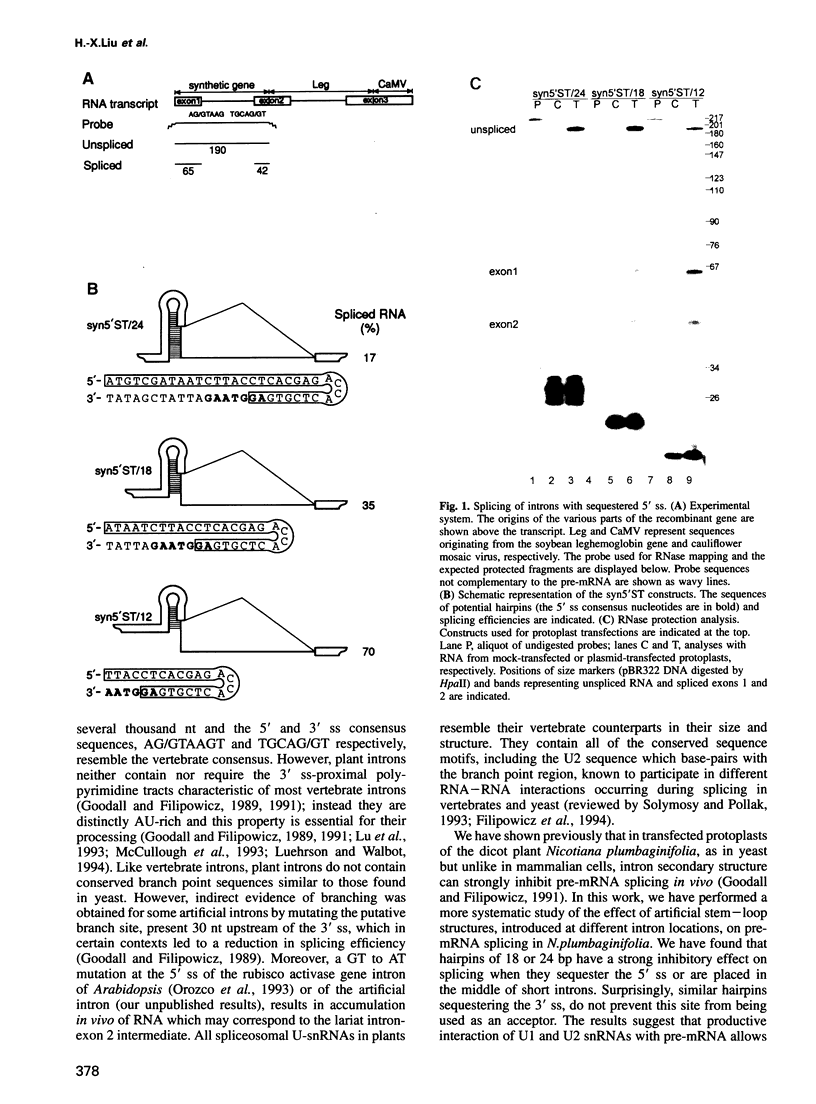
Abstract
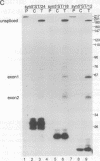
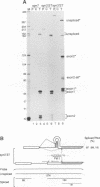
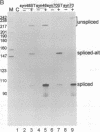
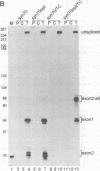
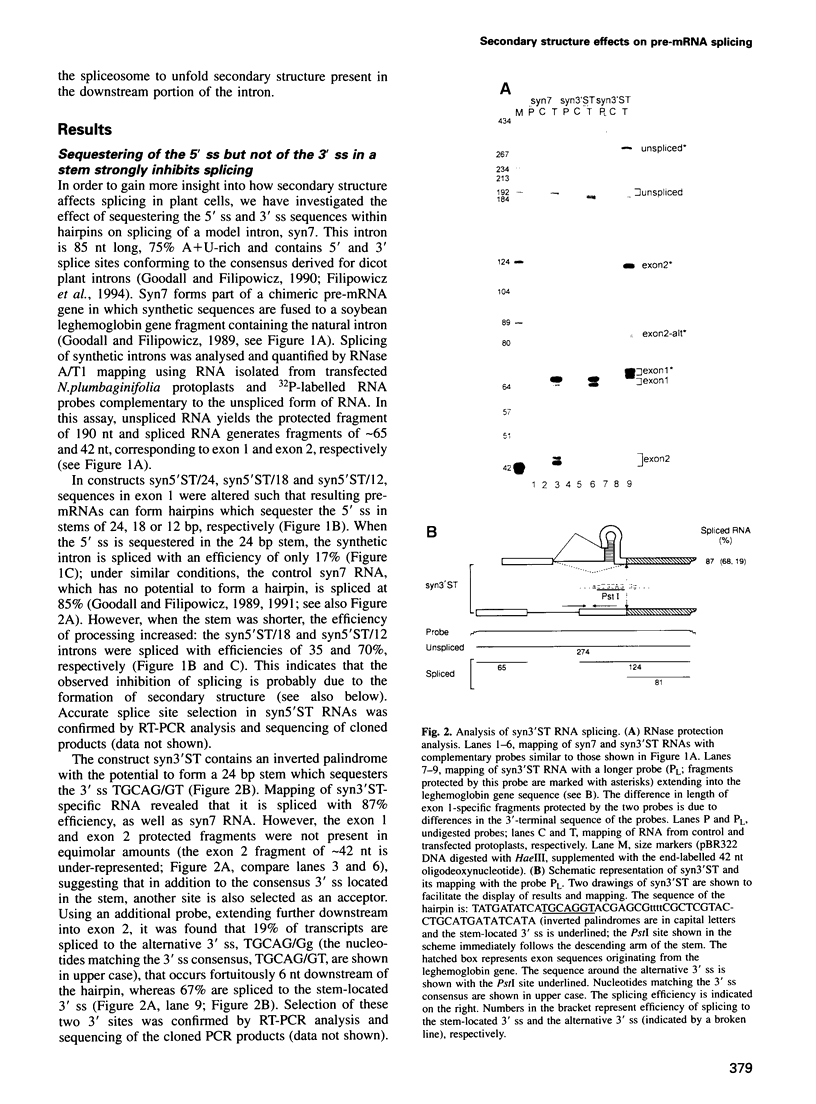
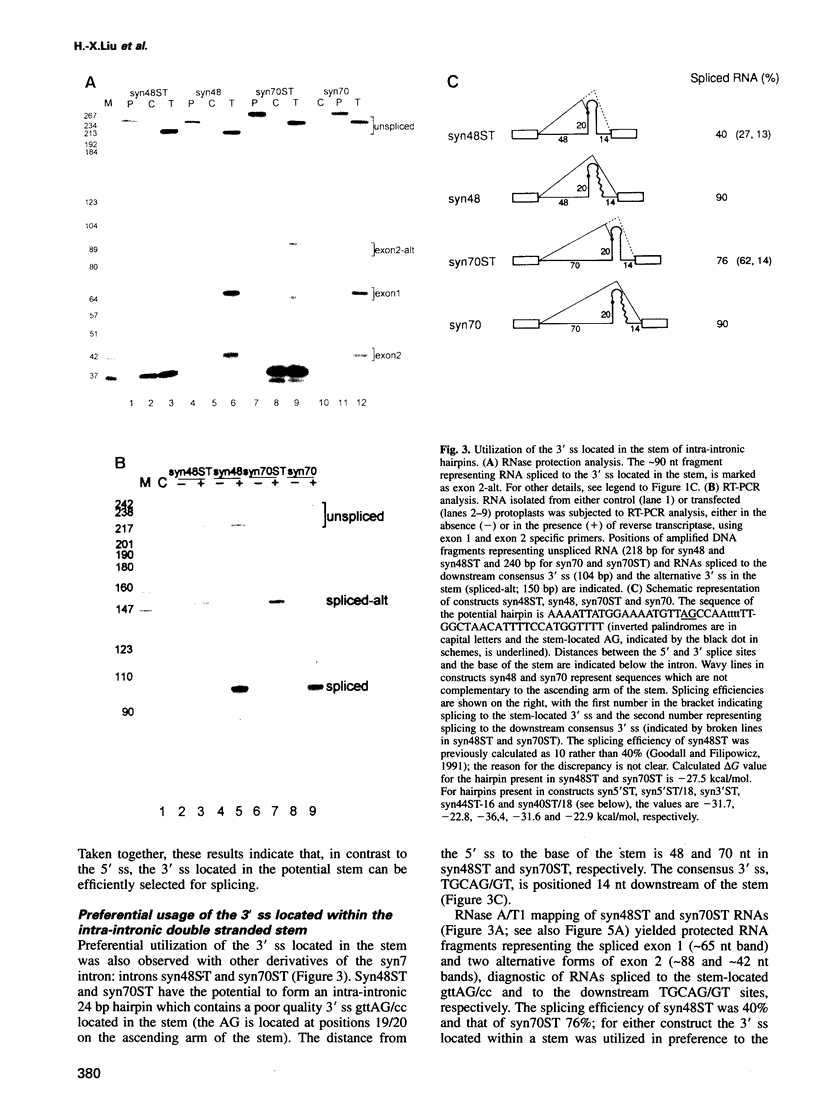
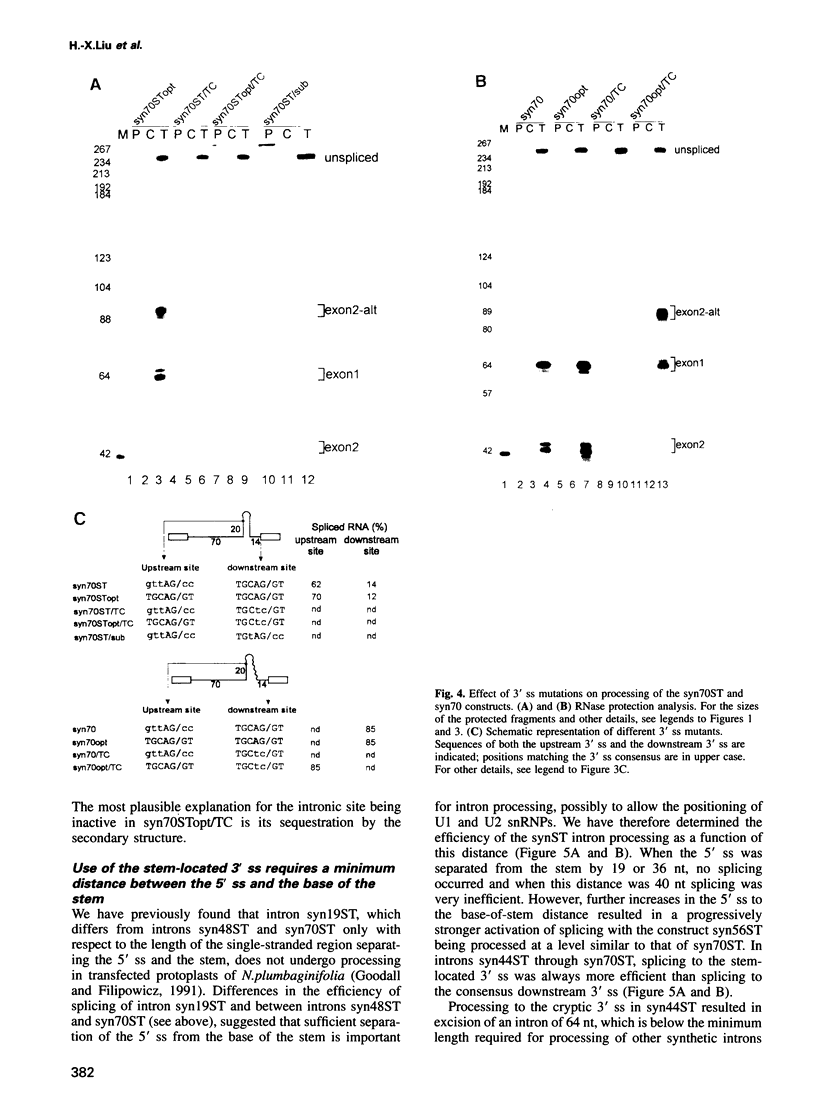
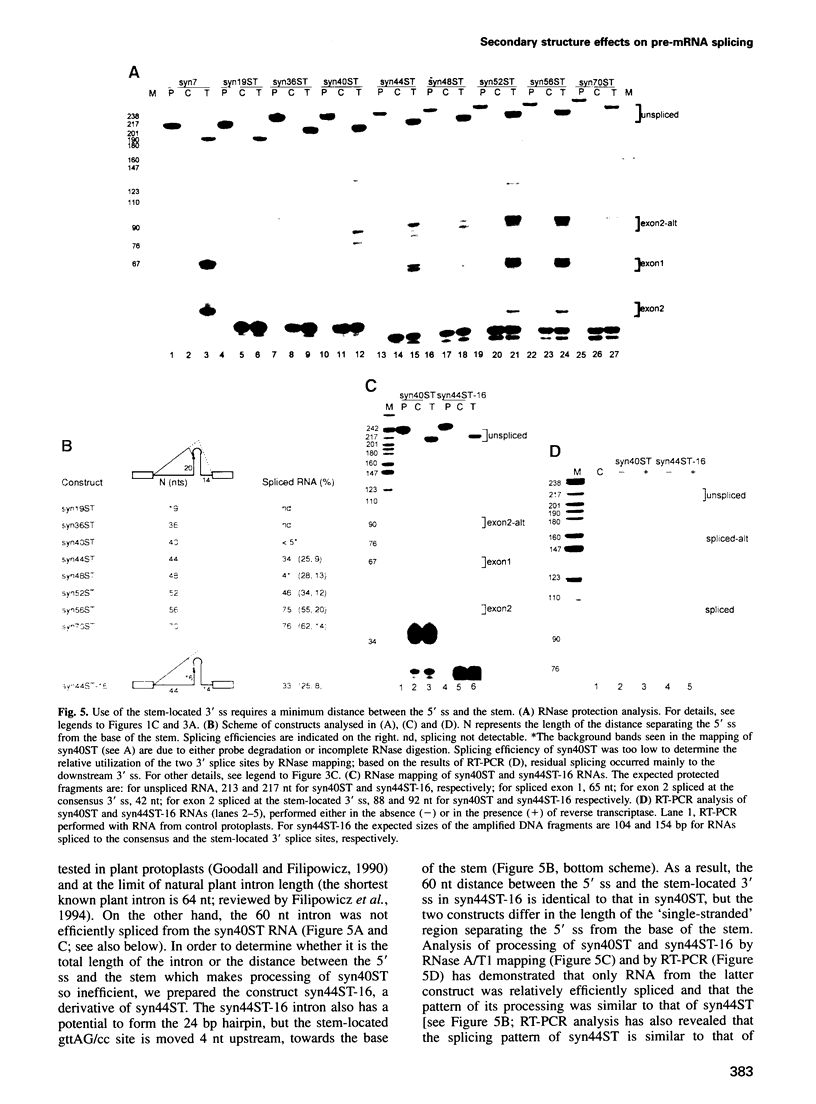
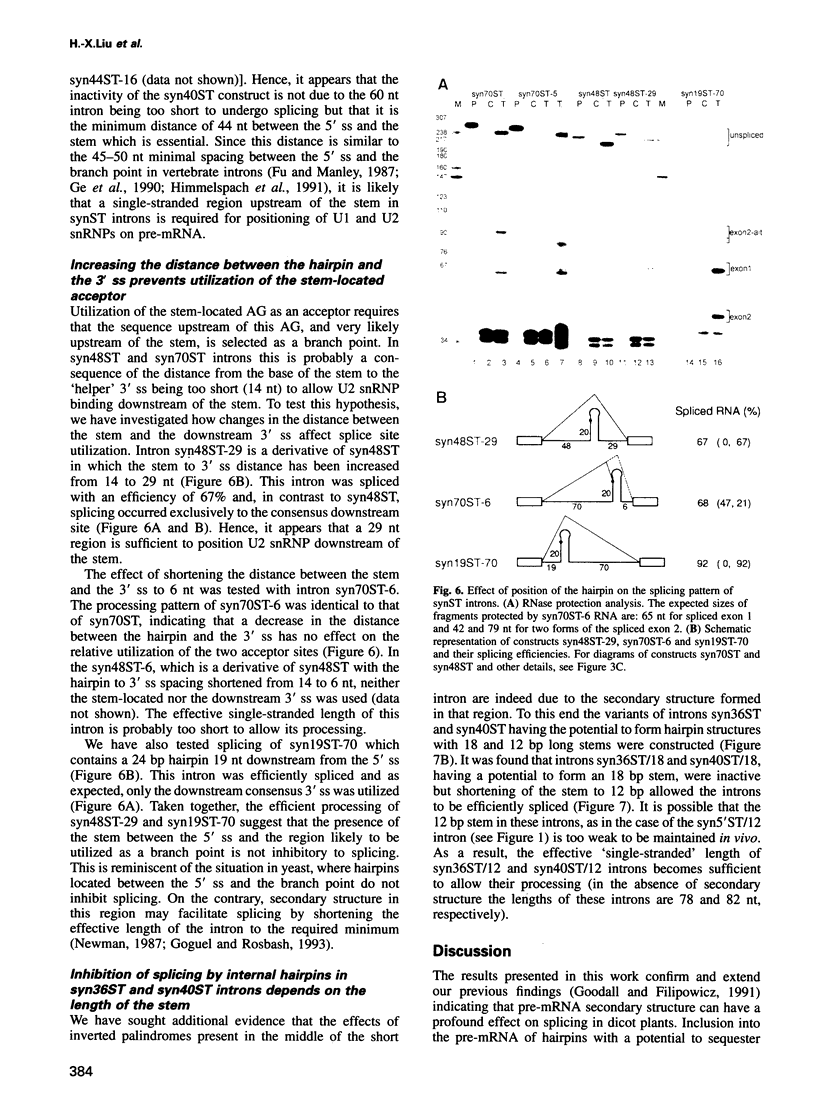
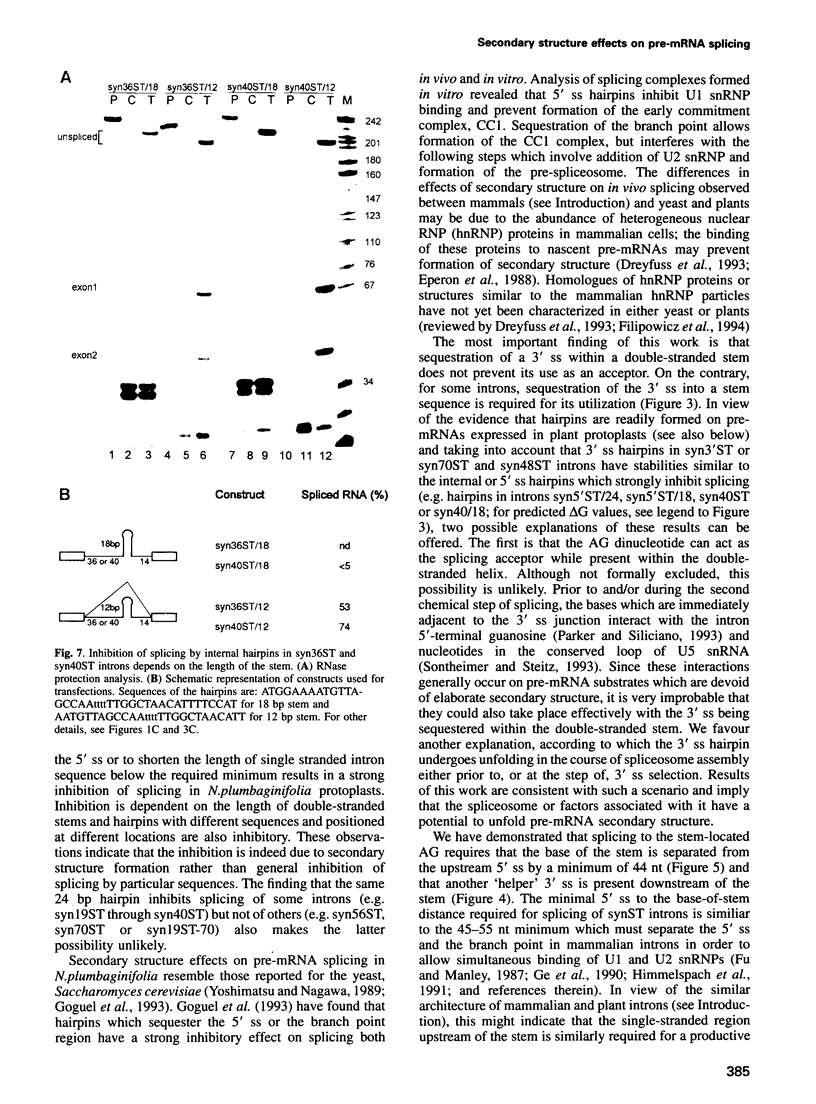
We have performed a systematic study of the effect of artificial hairpins on pre-mRNA splicing in protoplasts of a dicot plant, Nicotiana plumbaginifolia. Hairpins with a potential to form 18 or 24 bp stems strongly inhibit splicing when they sequester the 5' splice site or are placed in the middle of short introns. However, similar 24 bp hairpins sequestering the 3' splice site do not prevent this site from being used as an acceptor. Utilization of the stem-located 3' site requires that the base of the stem is separated from the upstream 5' splice site by a minimum of approximately 45 nucleotides and that another 'helper' 3' splice site is present downstream of the stem. The results indicate that the spliceosome or factors associated with it may have a potential to unfold secondary structure present in the downstream portion of the intron, prior to or at the step of the 3' splice site selection. The finding that the helper 3' site is required for utilization of the stem-located acceptor confirms and extends previous observations, obtained with HeLa cell in vitro splicing systems, indicating that the 3' splice site may be recognized at least twice during spliceosome assembly.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Deshler J. O., Rossi J. J. Unexpected point mutations activate cryptic 3' splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991 Jul;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M. J., Piñol-Roma S., Burd C. G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Noble J., Colgan J., Manley J. L. Polyoma virus small tumor antigen pre-mRNA splicing requires cooperation between two 3' splice sites. Proc Natl Acad Sci U S A. 1990 May;87(9):3338–3342. doi: 10.1073/pnas.87.9.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goguel V., Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993 Mar 26;72(6):893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- Goguel V., Wang Y., Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol Cell Biol. 1993 Nov;13(11):6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991 Sep;10(9):2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol. 1990 May;14(5):727–733. doi: 10.1007/BF00016505. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Wiebauer K., Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Himmelspach M., Gattoni R., Gerst C., Chebli K., Stévenin J. Differential block of U small nuclear ribonucleoprotein particle interactions during in vitro splicing of adenovirus E1A transcripts containing abnormally short introns. Mol Cell Biol. 1991 Mar;11(3):1258–1269. doi: 10.1128/mcb.11.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis S., Séraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993 Dec 24;262(5142):2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Smith J., Claude A., Lin R. J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992 Jun;11(6):2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser C. F., Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993 Dec 24;262(5142):1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lou H., McCullough A. J., Schuler M. A. 3' splice site selection in dicot plant nuclei is position dependent. Mol Cell Biol. 1993 Aug;13(8):4485–4493. doi: 10.1128/mcb.13.8.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Walbot V. Intron creation and polyadenylation in maize are directed by AU-rich RNA. Genes Dev. 1994 May 1;8(9):1117–1130. doi: 10.1101/gad.8.9.1117. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992 Nov 27;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994 May 1;8(9):1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- McCullough A. J., Lou H., Schuler M. A. Factors affecting authentic 5' splice site selection in plant nuclei. Mol Cell Biol. 1993 Mar;13(3):1323–1331. doi: 10.1128/mcb.13.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 1987 Dec 1;6(12):3833–3839. doi: 10.1002/j.1460-2075.1987.tb02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco B. M., McClung C. R., Werneke J. M., Ogren W. L. Molecular basis of the ribulose-1,5-bisphosphate carboxylase/oxygenase activase mutation in Arabidopsis thaliana is a guanine-to-adenine transition at the 5'-splice junction of intron 3. Plant Physiol. 1993 May;102(1):227–232. doi: 10.1104/pp.102.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993 Feb 18;361(6413):660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- Plumpton M., McGarvey M., Beggs J. D. A dominant negative mutation in the conserved RNA helicase motif 'SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994 Feb 15;13(4):879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991 Feb 7;349(6309):494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Chu T. T., Nadal-Ginard B. Scanning and competition between AGs are involved in 3' splice site selection in mammalian introns. Mol Cell Biol. 1993 Aug;13(8):4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer E. J., Steitz J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993 Dec 24;262(5142):1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kandels-Lewis S. 3' splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell. 1993 May 21;73(4):803–812. doi: 10.1016/0092-8674(93)90258-r. [DOI] [PubMed] [Google Scholar]
- Tanaka A., Mita S., Ohta S., Kyozuka J., Shimamoto K., Nakamura K. Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res. 1990 Dec 11;18(23):6767–6770. doi: 10.1093/nar/18.23.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T., Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989 Jun 16;244(4910):1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. The conserved dinucleotide AG of the 3' splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene. 1990 Jun 15;90(2):263–269. doi: 10.1016/0378-1119(90)90189-x. [DOI] [PubMed] [Google Scholar]