Abstract
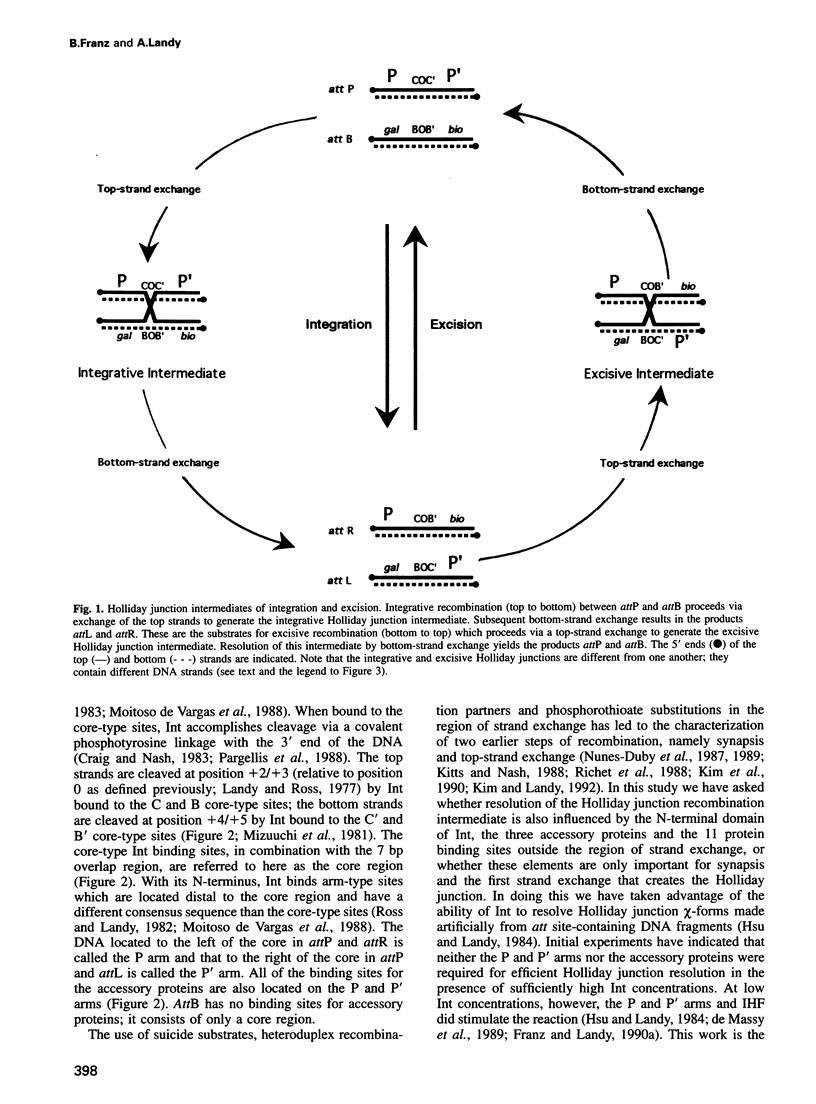
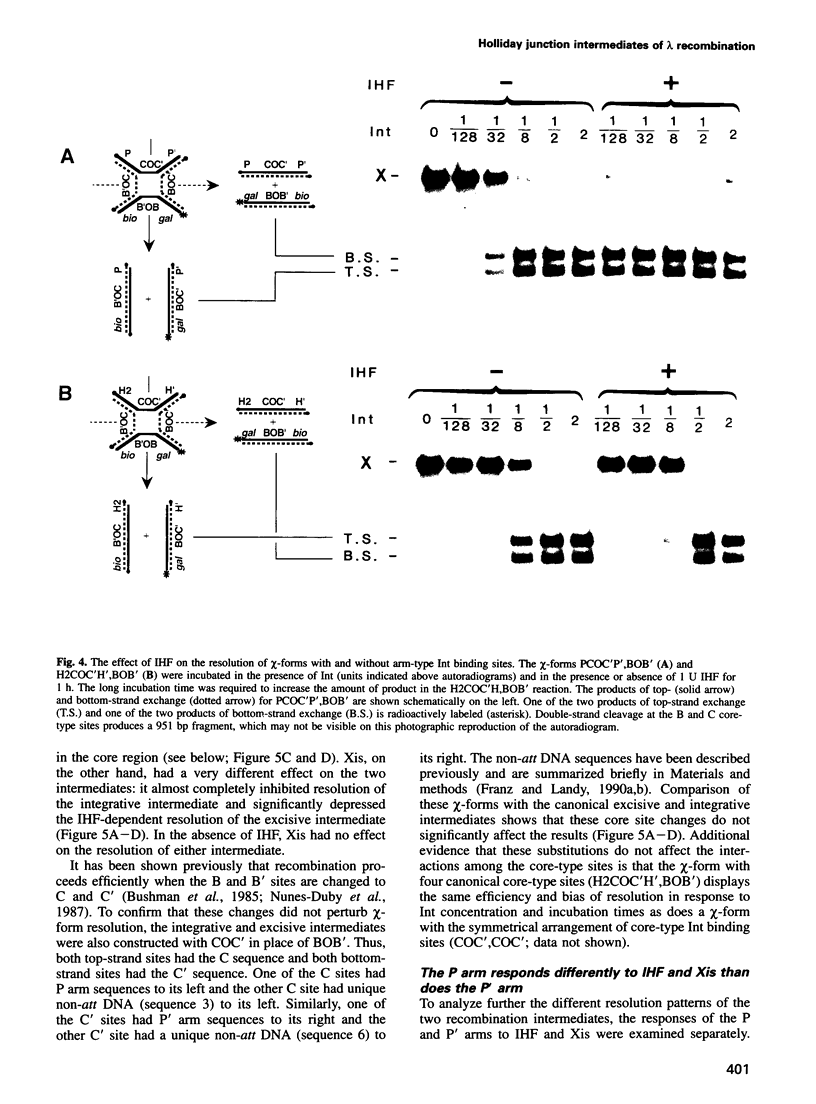
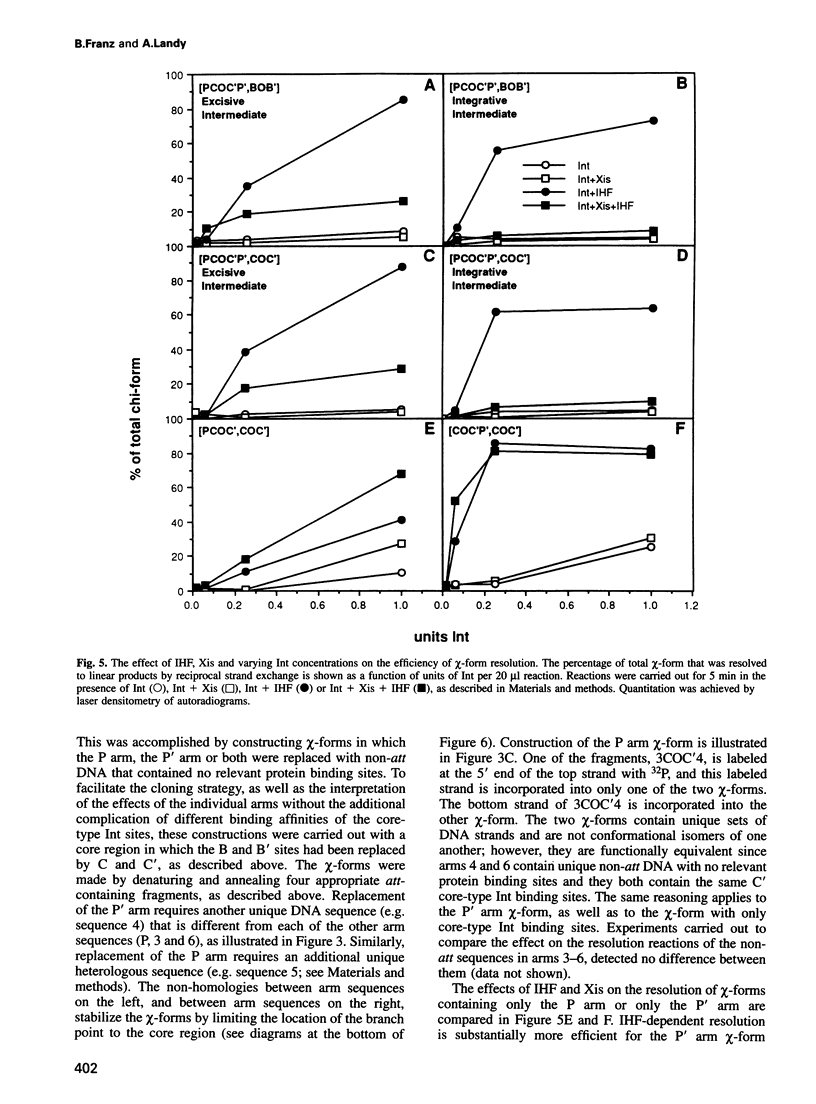
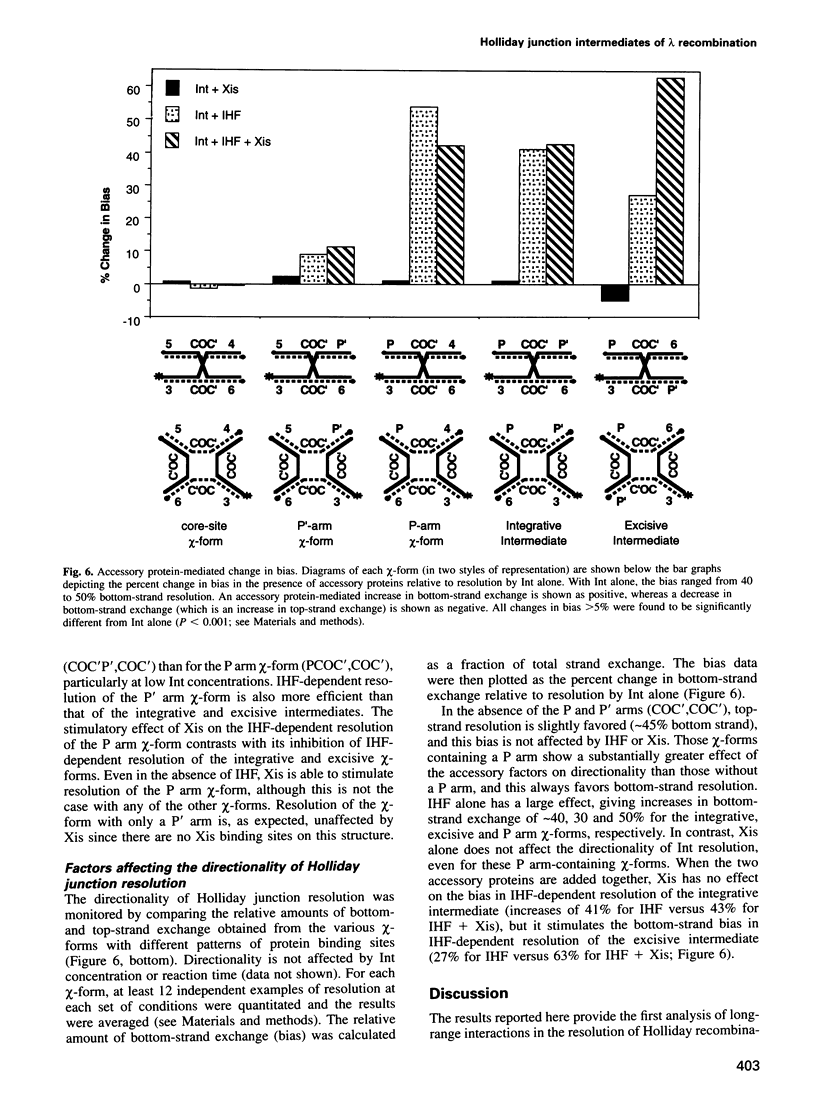
In lambda site-specific recombination, the integrative and excisive reactions proceed via two different Holliday junction intermediates, both of which are generated and resolved by a pair of sequentially ordered single strand exchanges. Factors affecting the directionality and efficiency of the second pair of strand exchanges were examined using artificial Holliday junctions (chi-forms). The integrative and excisive recombination intermediates respond differently to the accessory DNA bending proteins integration host factor and excisionase (Xis). These differences between the two recombination intermediates result from a different interaction pattern between proteins binding to the left (P arm) and right (P' arm) of the crossover region. The effect of Xis protein on the directionality of resolution, i.e. the choice of which strands are exchanged, is consistent with a role in promoting the second strand exchange during excision. Proteins binding to the left of the crossover region (P arm) primarily influence the directionality of resolution, while proteins binding to the right (P' arm) have a greater effect on the overall efficiency of resolution. Together, the effect of proteins binding to sites in the P and P' arms is to greatly enhance resolution of the two different Holliday intermediates and to favor resolution in the 'forward' direction for both integrative and excisive recombination.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Gottesman S. The form of the DNA substrate required for excisive recombination of bacteriophage lambda. J Mol Biol. 1979 Jul 5;131(3):637–649. doi: 10.1016/0022-2836(79)90012-3. [DOI] [PubMed] [Google Scholar]
- Ball C. A., Johnson R. C. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol. 1991 Jul;173(13):4032–4038. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. E., Gardner J. F., Gumport R. I. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J Mol Biol. 1985 Jan 20;181(2):187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Dunderdale H. J., West S. C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993 Sep 24;74(6):1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Bushman W., Thompson J. F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985 Nov 22;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman W., Yin S., Thio L. L., Landy A. Determinants of directionality in lambda site-specific recombination. Cell. 1984 Dec;39(3 Pt 2):699–706. doi: 10.1016/0092-8674(84)90477-x. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Franz B., Landy A. Interactions between lambda Int molecules bound to sites in the region of strand exchange are required for efficient Holliday junction resolution. J Mol Biol. 1990 Oct 20;215(4):523–535. doi: 10.1016/s0022-2836(05)80165-2. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Hoess R., Wierzbicki A., Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Molecular aspects of genetic exchange and gene conversion. Genetics. 1974 Sep;78(1):273–287. doi: 10.1093/genetics/78.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho S. H., Landy A. Dissecting the resolution reaction of lambda integrase using suicide Holliday junction substrates. EMBO J. 1994 Jun 1;13(11):2714–2724. doi: 10.1002/j.1460-2075.1994.tb06562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992 Apr 10;256(5054):198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Moitoso de Vargas L., Nunes-Düby S. E., Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990 Nov 16;63(4):773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J Mol Biol. 1988 Nov 5;204(1):95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- McCulloch R., Coggins L. W., Colloms S. D., Sherratt D. J. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994 Apr 15;13(8):1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Leon L., Huang L. C., Umlauf S. W., Cox M. M., Inman R. B. Holliday intermediates and reaction by-products in FLP protein-promoted site-specific recombination. Mol Cell Biol. 1988 Sep;8(9):3784–3796. doi: 10.1128/mcb.8.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Weisberg R., Enquist L., Mizuuchi M., Buraczynska M., Foeller C., Hsu P. L., Ross W., Landy A. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Kim S., Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989 Jun 23;244(4911):1457–1461. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Landy A. A switch in the formation of alternative DNA loops modulates lambda site-specific recombination. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):588–592. doi: 10.1073/pnas.88.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Pargellis C. A., Hasan N. M., Bushman E. W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988 Sep 23;54(7):923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integrative recombination of bacteriophage lambda DNA in vitro. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989 Nov;8(11):3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Half-att site substrates reveal the homology independence and minimal protein requirements for productive synapsis in lambda excisive recombination. Cell. 1989 Oct 6;59(1):197–206. doi: 10.1016/0092-8674(89)90881-7. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988 Jan 15;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A. Bacteriophage lambda int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983 May;33(1):261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Snyder U. K., Landy A. Helical-repeat dependence of integrative recombination of bacteriophage lambda: role of the P1 and H1 protein binding sites. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6323–6327. doi: 10.1073/pnas.85.17.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., de Vargas L. M., Skinner S. E., Landy A. Protein-protein interactions in a higher-order structure direct lambda site-specific recombination. J Mol Biol. 1987 Jun 5;195(3):481–493. doi: 10.1016/0022-2836(87)90177-x. [DOI] [PubMed] [Google Scholar]
- Weisberg R. A., Enquist L. W., Foeller C., Landy A. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J Mol Biol. 1983 Oct 25;170(2):319–342. doi: 10.1016/s0022-2836(83)80151-x. [DOI] [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- de Massy B., Dorgai L., Weisberg R. A. Mutations of the phage lambda attachment site alter the directionality of resolution of Holliday structures. EMBO J. 1989 May;8(5):1591–1599. doi: 10.1002/j.1460-2075.1989.tb03543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]




