Abstract
Background
Recent studies show that microRNAs (miRNAs), small non-coding RNAs that negatively regulate gene expression, may have potential for monitoring cancer status. We investigated circulating miRNAs in prostate cancer that may be associated with the progression of hormone-sensitive primary tumors to metastatic castration resistant prostate cancer (CRPC) after androgen deprivation therapy.
Methods
Using genome-wide expression profiling by TaqMan Human MicroRNA Arrays (Applied Biosystems) and/or quantitative real-time polymerase chain reaction, we compared the expression levels of miRNAs in serum samples from 28 patients of low-risk localized disease, 30 of high-risk localized disease and 26 of metastatic CRPC.
Results
we demonstrated that serum samples from patients of low-risk, localized prostate cancer and metastatic CRPC patients exhibit distinct circulating miRNA signatures. MiR-375, miR-378*, and miR-141 were significantly over-expressed in serum from CRPC patients compared to serum from low-risk localized patients, while miR-409-3p was significantly under-expressed. In prostate primary tumor samples, miR-375 and miR-141 also had significantly higher expression levels compared to those in normal prostate tissue.
Conclusions
Circulating microRNAs, particularly miR-375, miR-141, miR-378* and miR-409-3p, are differentially expressed in serum samples from prostate cancer patients. In the search for improved minimally invasive methods to follow cancer pathogenesis, the correlation of disease status with the expression patterns of circulating miRNAs may indicate the potential importance of circulating miRNAs as prognostic markers for prostate cancer progression.
Keywords: circulating microRNAs, prostate cancer, castration resistant prostate cancer
INTRODUCTION
MicroRNAs (miRNAs) are endogenous single-stranded, non-protein coding RNAs of approximately 22 nucleotides in length. MiRNAs negatively regulate gene expression by binding with imperfect complementarities to the 3’ untranslated region of their target mRNAs, reducing the stability and/or translation efficiency of target mRNAs in a sequence specific manner. MiRNAs have been estimated to modulate the expression of approximately 30% of protein coding genes in humans (1). Many studies have demonstrated distinct miRNA expression signatures in different states of health. Altered expression of miRNAs or dysfunction of miRNA pathways can affect divergent cellular processes, including cell cycle, differentiation, and proliferation, thus influencing tumorigenesis and progression.
Prostate cancer is the most frequently diagnosed male cancer and the second-leading cause of oncological mortality in men living in the United States (2). Early stage prostate cancer, called hormone-sensitive prostate cancer (HSPC), depends on androgen for growth (3). The most effective systemic treatment for HSPC has been androgen deprivation therapy (ADT), specifically by orchiectomy or luteinizing hormone-releasing hormone, with or without antiandrogens (4). Initially, the HSPC regresses, though recurrence inevitably follows and evolves to castration resistant prostate cancer (CRPC), which is usually ultimately fatal. The wide use of the prostate-specific antigen (PSA) test along with a digital rectal exam has facilitated the detection of prostate cancer in men of 50 years old or older at early disease stages. The PSA test also has been routinely used to monitor prostate cancer recurrence after therapy. However, due to the heterogeneous nature of the disease, additional prognostic markers are urgently needed for better and more accurate prediction of disease progression.
Recent studies have demonstrated that miRNAs can be exported by cells and circulate in human blood in a stable form; some of the circulating miRNAs can distinguish patients from healthy individuals (5). These findings imply possible use of circulating miRNAs as non-invasive markers for monitoring disease progression (5, 6). In prostate cancer, several reports have demonstrated that miRNAs derived from tumors of epithelial origin can be detected in blood samples and some prostate cancer related circulating miRNAs potentially correlate with risk of disease progression and some predictors of disease aggressiveness (6–9). Collectively, these studies provide evidence supporting the potential clinical use of circulating miRNAs for prognostic purposes. We explored the possibility of identifying circulating miRNA signatures for prostate cancer transition to CRPC.
Materials and Methods
Clinical Blood Samples
Clinical blood samples were obtained from the specimen bank of Arthur and Linda Gelb Center for Translational Research at Dana-Farber Cancer Institute (DFCI; Boston, MA). All patients had been seen at DFCI or Brigham and Women’s Hospital (Boston, MA) and gave informed consent to provide information and blood samples for research purposes. Patients with metastatic CRPC (n = 30) at the time of blood collection (between October 2003 and October 2005) were initially identified. All these patients had been treated with androgen deprivation therapy (luteinizing hormone-releasing hormone agonists or bilateral orchiectomy) and had evidence of progression of disease with either rising PSA or evidence of radiographic progression by the date of blood draw. To compare, samples with blood draw over the same period (2003–2005) were analyzed from patients with untreated localized, low-risk (n = 30) and high-risk (n = 30) disease at the time of blood draw. Degree of disease was categorized based on the D’Amico risk classification criteria (10). Specifically, low-risk patients had a tumor stage of T1c or T2a and a PSA of ≤10 ng/mL and a Gleason score of ≤ 6, and while high-risk patients had a tumor stage of T2c or a PSA of > 20 ng/mL or a Gleason score of ≥ 8.
Of the 30 CRPC and 30 low-risk samples, only 26 CRPC and 28 low-risk samples were profiled. The others samples were excluded due to having insufficient levels of RNA for profiling (verified by using qRT-PCR to measure levels of highly-expressed endogenous U6 small nuclear RNA and spiked-in synthetic Caenorhabditis elegans miRNAs (cel-miR-39, cel-miR-54, and cel-miR-238)).
Serum isolation
After blood was drawn, tubes were kept upright at room temperature for 30 minutes and then stored in a 4 °C refrigerator for a period of time ranging from 2 to 15 hours. Following centrifugation at 1000g, 4 °C for 10 minutes, serum was extracted and distributed into aliquots of 0.5 ml per 1.5 ml tube. Then the serum tubes were stored at −80°C freezer. All serum used for this study were thawed ≤ one time.
Isolation of peripheral blood mononuclear cells (PBMC)
8 ml of blood was mixed with 2 ml of HBSS and then slowly loaded onto the top of 5ml of ficoll (Ficoll-Paque™ Plus, GE Health) in a 15 ml tube. Then the tube was centrifuged for 20 minutes at 1000g at 17 °C. ~ 1.5 ml of the buffy coat residing on the top of the ficoll layer was isolated and spun down. The cell pellet isolated from the buffy coat was referred to as the PBMC.
RNA Isolation
Serum derived total RNA was isolated using mirVana™ PARIS™ Kit (Ambion) according to the protocol supplied by the manufacturer for liquid samples with modifications as reported by Mitchell et al. (6). Peripheral blood mononuclear cell (PBMC) derived total RNA was extracted with Trizol reagent (Invitrogen).
Genome-Wide MicroRNA Expression Profiling using TaqMan MicroRNA Arrays
Quality of total RNA samples was analyzed with Agilent 2100 Electrophoresis Bioanalyzer and microRNA expression was profiled utilizing TaqMan Human MicroRNA Arrays (Applied Biosystems) at the Molecular Diagnostics Core Facility of Dana-Farber Cancer Institute. Three µL neat of total RNA were used to synthesize cDNA, which was then pre-amplified by performing PCR for eighteen cycles prior to profiling. The products of the pre-amplification reaction were run on two TaqMan Low Density Array (TLDA) cards containing 384 wells each. Of the total of 728 miRNA wells, 669 probes assay for human miRNAs, nine of which are endogenous controls. Arabidopsis thaliana miRNA (ath-miR) and Mus musculus miRNA (mmu-miR) targets served as negative controls.
Measurement of miRNAs by qRT-PCR
MiRNAs were quantified by quantitative real-time polymerase chain reaction (qRT-PCR) using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) and FastStart Universal Probe Master (Roche) according to manufacturer’s protocol. All TaqMan MicroRNA Assays primer sets were purchased from Applied Biosystems and all samples were run in duplicate.
Data of clinical tumor samples
Clinical tumor sample data were obtained from a study conducted by Memorial Sloan-Kettering Cancer Center (MSKCC), in which microRNA expression of 28 normal prostate tissue samples, 99 primary prostate tumors, and 14 metastases was profiled using Agilent microRNA V2 arrays (11; http://cbio.mskcc.org/cancergenomics/prostate/data/).
Statistical Analysis
The expression levels of each circulating miRNA are originally presented as the threshold cycle (Ct) values, defined as the fractional cycle number at which the fluorescent signal surpasses the fixed threshold in qRT-PCR. For data analysis, we used the comparative Ct method (ΔCt), normalized by subtracting the Ct value of an endogenous reference (U6-small nuclear RNA) from each miRNA (i.e., ΔCt = CtmiRNA of interest − CtU6). Data were summarized as mean and standard deviation (Std) of ΔCt value for each miRNA by disease group; fold change (calculated as the relative value 2−(ΔCT (group1)−ΔCT (group2))) was also reported. Comparisons between groups were conducted using the student t-test. The expression levels of miRNA in PBMC were analyzed similarly.
The expression levels of miRNA from prostate tissue samples were analyzed as a normalized log2 expression, with larger values representing higher expression. Comparisons between groups were conducted using the Wilcoxon rank-sum test to account for non-normal distribution, with Bonferroni correction for pair-wise comparison. The statistical analysis was performed in SAS version 9 (SAS institute, Cary, NC) and in GraphPad Prism (GraphPad Software, San Diego).
RESULTS
Patient Characteristics
This study comprises samples from three groups of patients including 28 patients of low-risk localized disease, 30 of high-risk localized disease, and 26 of metastatic CRPC. The clinical characteristics of patients are described in Table 1. In brief, patients were self-reported Caucasian men with a median age of 60 years at diagnosis. The median Gleason scores are 6 for the low-risk group, 8 for the high-risk group, and 8 for the CRPC group.
Table 1.
Clinical characteristics of the profiled population
| All cases | Localized Low-risk (n=28) |
Localized High-risk (n=30) |
Metastatic CRPC (n=26) |
|
|---|---|---|---|---|
| At diagnosis | ||||
| Age, Median[IQR] | 60 [53–69] | 56 [51–69] | 62 [60–72] | 61 [56–65] |
| PSA ng/mL, Median[IQR] | 6.9 [4.6–25.7] | 4.75 [3.1–5.7] | 8.89 [6.9–30.0] | 34.7 [6.1–81.0] |
| Biopsy Gleason Score | ||||
| 5 | 1 | 0 | 0 | 1 |
| 6 | 28 | 25 | 2 | 1 |
| 7 | 13 | 3 | 3 | 7 |
| 8 | 21 | 0 | 13 | 8 |
| 9 | 17 | 0 | 12 | 5 |
| Missing | 4 | 0 | 0 | 4 |
| Median | 7 | 6 | 8 | 8 |
| Clinical T stage | ||||
| T1 | 33 | 18 | 10 | 5 |
| T2 | 19 | 3 | 9 | 7 |
| T3 | 3 | 0 | 2 | 1 |
| Tx | 25 | 7 | 9 | 9 |
| Clinical N stage | ||||
| N0 | 34 | 7 | 22 | 5 |
| N1 | 2 | 0 | 0 | 2 |
| Nx | 44 | 21 | 8 | 15 |
| Clinical M stage | ||||
| M0 | 30 | 5 | 19 | 6 |
| M1 | 8 | 0 | 0 | 8 |
| Mx | 42 | 23 | 11 | 8 |
| At blood draw | ||||
| Year from diagnosis to blood draw, Median[IQR] | 4.8 [2.7–10.4] | |||
| Age, Median[IQR] | 71 [60–77] | |||
| PSA ng/mL, Median[IQR] | 11.8 [4.9,75.1] |
IQR, interquartile range
Identification of CRPC associated circulating microRNA
We first investigated differential expression of known circulating miRNAs isolated from serum between low-risk (n = 28) and CRPC (n = 26) patients using TaqMan Human MicroRNA Arrays. After eliminating miRNAs that were undetectable in over 90% of samples, we found a significant difference in the number of detectable miRNAs across samples between the low-risk (243.4 ± 42.6, mean ± SD) and CRPC (207.3 ± 32.2) groups (p < 0.001, t-test; Supplementary Table S1). Only 71 miRNAs were detected in every sample, but 403 miRNAs were detected in at least one sample in both groups.
After normalizing all data to the expression level of U6 and eliminating miRNAs that were detectable in less than 10% of the samples, we found 14 out of 345 circulating miRNAs to be significantly differentially expressed between the low-risk and CRPC groups (p-values < 0.05 by t-test; Table 2). We then increased the stringency of our selection criteria. MiRNAs that had fold changes of less than 4, low expression levels (mean raw Ct values > 32), or infrequent expression (detectable in 5 or fewer samples per group) were removed, leaving four differentially expressed miRNAs: miR-375, miR-378*, miR-141, and miR-409-3p (Table 2 and Supplementary Table S2).
Table 2.
Differentially expressed circulating miRNAs between low-risk (LR) and metastatic CRPC samples.
| No. of samples detected |
Mean (Std) Raw Ct |
Mean (Std) ΔCt* |
||||||
|---|---|---|---|---|---|---|---|---|
| miRNA | LR | CRPC | LR | CRPC | LR | CRPC | Fold** change |
p-value |
| hsa-miR-375 | 26 | 25 | 26.3(1.7) | 24.2(2.1) | 5.4(2.3) | 2.4 (2.5) | +8.0 | <0.0001 |
| hsa-miR-96 | 2 | 4 | 33.5(1.1) | 31.9(1.0) | 15.1(0.6) | 9.6(1.0) | +45.3 | 0.0022 |
| hsa-miR-892b | 8 | 8 | 34.9(1.1) | 34.3(1.1) | 15(1.7) | 11.7(2.2) | +9.9 | 0.004 |
| hsa-miR-378* | 15 | 13 | 31.8(2.2) | 30.7(2.4) | 11.3(2.8) | 8.1(2.8) | +9.2 | 0.0057 |
| hsa-miR-302b | 5 | 4 | 33.1(2.7) | 28.8(0.2) | 12.9(3.6) | 7.4(0.2) | +45.3 | 0.0192 |
| hsa-miR-548c-3p | 28 | 25 | 31.6(1.9) | 31.9(1.6) | 11.4(2.5) | 9.9(1.7) | +2.8 | 0.0216 |
| hsa-miR-124 | 4 | 4 | 32.3(1.5) | 33.3(1.3) | 13.5(1.3) | 11.3(0.8) | +4.6 | 0.025 |
| hsa-miR-141 | 16 | 18 | 31.7(2.5) | 30.2(2.1) | 10.6(4.0) | 7.9(2.6) | +6.5 | 0.0276 |
| hsa-miR-875-5p | 24 | 20 | 34.1(3.2) | 32.9(2.3) | 13(3.2) | 10.8(3.3) | +4.6 | 0.0287 |
| hsa-miR-409-3p | 27 | 24 | 28.6(3.6) | 32.0(3.4) | 7.5(5.1) | 10.3(3.7) | −7.0 | 0.0297 |
| hsa-miR-548a-3p | 28 | 25 | 31.6(2.0) | 31.1(1.1) | 10.7(2.8) | 9.2(1.7) | +2.8 | 0.0302 |
| hsa-miR-623 | 13 | 15 | 33.8(2.1) | 35.6(1.4) | 12.9(2.0) | 14.3(1.4) | −2.6 | 0.0362 |
| hsa-miR-520d-5p | 28 | 26 | 27.9(2.1) | 27.5(1.7) | 6.9(2.6) | 5.6(1.9) | +2.5 | 0.0414 |
| hsa-miR-489 | 8 | 8 | 33.7(3.3) | 32.6(2.5) | 13.6(3.1) | 10.7(2.3) | +7.5 | 0.0466 |
normalized by subtracting the Ct value of U6(reference) from the Ct value of each miRNA
calculated as the relative value 2 −(ΔCT (group1) −ΔCT (group2))
Out of 669 miRNAs probed, the relative expression levels of 12 miRNAs were up-regulated (designated by (+) in fold change) in CRPC patient serum samples in comparison to the levels found in low-risk patient serum samples, while the expression level of two miRNAs were down-regulated (designated by the (−) in fold change). Bolded miRNAs were selected for further study. P-values were calculated based on the unpaired two-sided t-test, with a cut-point of 0.05.
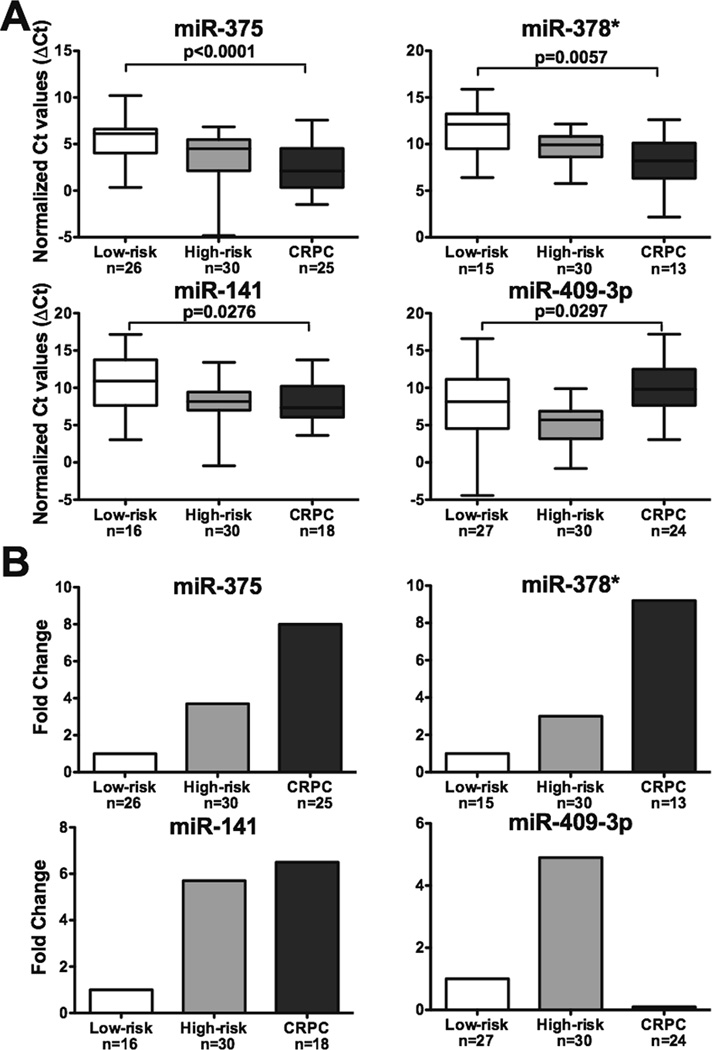
We observed that the expression levels of miR-375, miR-378*, and miR-141 are significantly up-regulated in CRPC serum samples compared to those in the low-risk group (p < 0.0001, p = 0.0057, and p = 0.0276, respectively), while the expression of miR-409-3p is down-regulated (p = 0.0297) (Fig. 1A). The increases from low-risk to CRPC for miR-375, miR-378*, and miR-141 are 8.0, 9.2, and 6.5 fold, respectively; miR-409-3p decreases 7.0 fold (Fig. 1B). Of these four miRNAs, miR-375 has the most robust expression level, which is bout 35 to 45 fold higher than that of miR-141.
Figure 1. Expression levels of miR-375, miR-378*, miR-141, and miR-409-3p in serum samples.
A. Representation of normalized expression levels of miRNAs by Ct values. Low-risk, localized (n=28) and metastatic CRPC (n=26) serum samples were profiled for miRNA expression levels. The four significantly expressed miRs were further studied in high-risk, localized (n=30) serum samples. All data were normalized to U6. Lower Ct values indicate higher expression levels. P-values were calculated comparing low-risk to CRPC using the unpaired two-sided t-test. B. Representation of normalized expression levels of miRNAs by fold change. The relative expression level of each miRNA in low-risk was arbitrarily set as 1.0.
We hypothesized that the differential expression patterns of these four serum derived circulating miRNAs may represent the result of disease progression, thus distinguishing patients of metastatic CRPC from those of low-risk, localized disease. We further determined the expression levels of these four miRNAs in serum samples derived from 30 high-risk, localized prostate cancer patients. The expression levels of circulating miR-375, miR-378*, and miR-141 in the high-risk group are between those in the low-risk and CRPC groups, thus an increase in expression level for miR-375, miR-378*, and miR-141 may correlate with progression to more advanced disease (Fig. 1). However, with miR-409-3p, we observed the expression level increases in the high-risk group but significantly declines in the CRPC group when compared to patients with low-risk disease.
Expression levels of miR-375, miR-378*, miR-141, and miR-409-3p in peripheral blood mononuclear cells
We identified differentially expressed circulating miRNA from serum samples between low-risk and CRPC patients. Three up-regulated circulating miRNAs correlate with the disease progression; however, the origin and fate of these disease specific circulating miRNAs are largely unknown. Circulating miRNAs could be exported from tumors cells or from the peripheral blood mononuclear cells (PBMC); thus, their differential expression pattern might represent the tumor progression or the host response to the tumor progression.
To determine if the differential expression patterns of miR-375, miR-378*, miR-141, and miR-409-3p in patients’ serum is correlated with their expression patterns in PBMC, qRT-PCRs were performed on total RNA isolated from PBMCs from 10 healthy blood donors, 10 low-risk, and 10 CRPC patients. The low-risk and CRPC patients are subsets of the original patients whose serum samples were used to identify the differentially expressed circulating miRNAs. We found miR-375 to have very low levels in PBMCs and did not find statistically significant differences in the expression level of the remaining three miRs in PBMCs derived from the three groups (data not shown).
Expression levels of miR-375, miR-378*, miR-141, and miR-409-3p in human prostate tumor samples
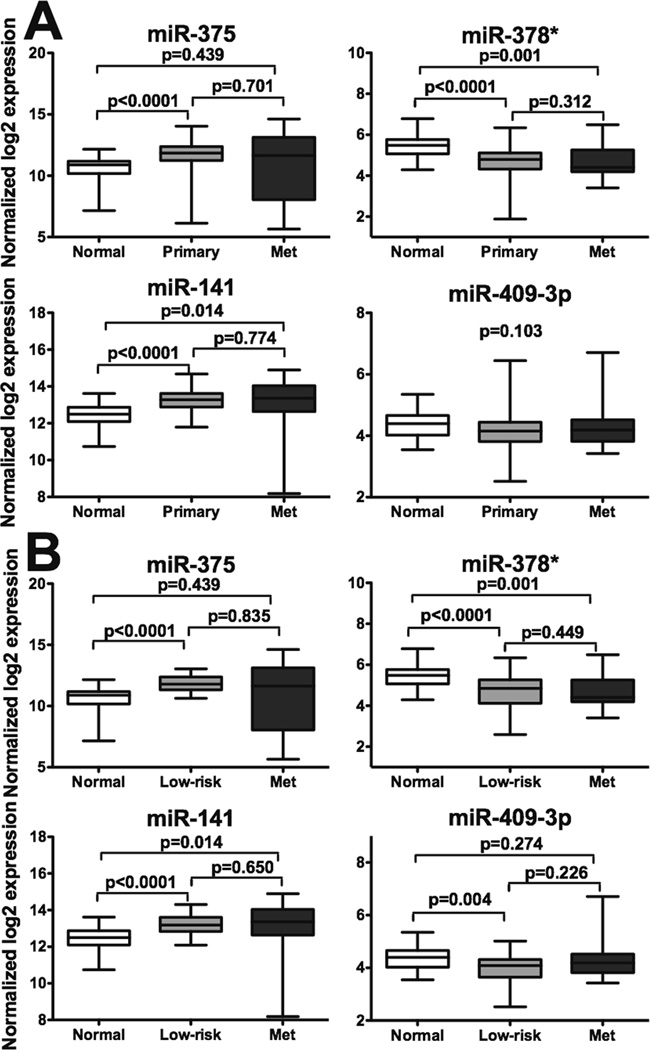
To determine if the levels of the four differentially expressed circulating miRNAs correlate with levels found in prostate tumor samples, we analyzed the miRNA-profiling data generated from the MSKCC prostate cancer cohort (11), which consists of 28 normal prostate tissue, 99 primary tumor, and 14 metastases samples. We observed that the expression of miR-375 and miR-141 is significantly increased in primary and/or metastatic tumors and that the expression of miR-378* is significantly decreased in tumor tissues, compared to those in normal tissue. No significant differences were observed between primary tumor and metastases samples. Expression of miR-409-3p is not significantly changed among different tissue samples (p = 0.103; Fig. 2A).
Figure 2. Expression levels of miR-375, miR-378*, miR-141, and miR-409-3p in human prostate tissue samples.
A. Comparison of normalized expression levels of miRNAs in normal prostate tissue (n=28), primary prostate tumors (n=99), and metastases (n=14). B. Comparison of normalized expression levels of miRNAs in normal prostate tissue (n=28), low-risk, localized prostate tumors (n=42), and metastases (n=14). Larger log2 values represent higher expression levels. Wilcoxon sum-rank test was used to determine statistical significance, with Bonferroni correction for pair-wise comparison (p < 0.0167 as significant).
Since our study focused on low-risk, localized prostate cancer compared to metastatic CRPC, we further examined the data on 42 primary tumors of low-risk, localized prostate cancer patients according to the D’Amico risk classification, a subset of the 99 primary tumor samples of the MSKCC prostate cancer cohort. Similarly, expression of miR-375 and miR-141 is increased and expression of miR-378* is decreased in the primary tumors of low-risk, localized patients compared to those in normal tissue and no significant differences were observed between low-risk, localized tumors and metastases. We also observed a significant decrease in miR-409-3p expression in low-risk, localized tumors compared to normal tissue. Pairwise comparisons revealed statistically different expression of all four miRNAs in low-risk compared to normal samples; only miR -378* and miR-141 also demonstrate significantly different expression in metastases in comparison to those in normal prostate tissue (Fig. 2B).
DISCUSSION AND CONCLUSION
Our study demonstrates that low-risk, localized prostate cancer patients and metastatic CRPC cancer patients have distinct circulating miRNA signatures. While the number of detectable circulating miRNAs across samples decreases from low-risk to CRPC patients, the expression levels of differentially expressed miR-375, mir-378*, and miR-141 follow an increasing trend with disease progression. We speculate that the rise and fall of miR-409-3p expression from low-risk to high-risk to CRPC could be related to ADT, either a result of reduced androgen levels or other biological changes of the CRPC phenotype.
The low expression of miR-375 and the lack of a significant differential expression pattern of miR-378*, miR-141, and miR-409-3p in PBMCs from different patient groups suggest the unlikelihood that PBMC-derived miRNAs contribute to the differential expression patterns of these four circulating miRNAs between patients with low-risk, localized prostate cancer and patients with metastatic CRPC. The observation of significantly elevated expression of miR-375 and miR-141 in prostate tumor tissues relative to normal prostate tissue may suggest that the increased expression of circulating miR-375 and miR-141 in advanced prostate cancer patients may result from the selective exocytosis of these miRNAs from prostate cancer tumors into the circulation system. The expression patterns of miR-378* and miR-409-3p, compared between normal tissue and prostate tumors, did not provide any implication on the origin of the differential expression patterns of these two circulating miRNAs.
Little is known about the function of miR-378* and miR-409-3p. In breast cancer, miR-378* has been shown to have increased expression with cancer progression and to induce the Warburg effect (12). Similar to what we observed, Wach et al. also found miR-378* is down regulated in prostate tumor tissue compared to matched normal (13). Zheng et al. have found miR-409-3p suppresses radixin, a pro-metastatic gene, and that reduced levels of the miRNA are associated with metastasis in gastric cancers (14). Crucial for normal glucose homeostasis, miR-375 has an established role in regulating insulin secretion by impairing glucose-stimulated insulin exocytosis and in maintaining pancreatic cell mass (15, 16). Additionally, miR-375 potentially down-regulates Sec23A in prostate cancer cell lines, resulting in enhanced proliferation, suggesting miR-375 may have a role in promoting cell growth (17). Highly evolutionarily conserved, miR-141 is a member of the miR-200 family, which plays an essential function in epithelial-to-mesenchymal transition (18, 19). Up-regulation of miR-141 may also promote AR transcriptional activity in prostate cancer by down-regulating an AR co-repressor (20).
Valadi et al. discovered that exosomal shuttle RNA, or mRNA and miRNA that are uniquely packaged into exosomes, can be transferred between cells; furthermore, the exosomal shuttle RNA is functional within the cells that endocytosed the exosome, indicating the possibility of lateral transfer of RNA (21). Taylor & Gercel-Taylor showed that in blood samples from ovarian cancer patients, circulating exosomes derived from tumor cells contain miRNA signatures characteristic of the biopsied tumor (22). Interestingly, exosomal miRNA were not found in samples from healthy control subjects. With their capability of regulating multiple gene targets paired with their ability to be delivered between cells, circulating miRNA may have a putative role in cell-to-cell signaling and modulating gene expression at distant targets which could affect disease progression; these exciting possibilities await exploration. The biological function and the impact of circulating miRNAs on the tumor microenvironment and tumor progression remain to be investigated. Additionally, we found some miRNAs, such as miR-205, have robust expression levels in normal prostate tissue and primary prostate tumors, but were undetectable in serum samples (data not shown), suggesting that only a subgroup of miRNAs may be selectively secreted from prostate tumors into the blood. Mechanisms of selective miRNA export from cells to the circulation system also remain to be uncovered.
The notion that serum levels of miR-141 can distinguish patients with prostate cancer from healthy controls was previously reported by Mitchell et al. while Yaman Agaoglu et al. demonstrated that plasma miR-141 only distinguishes localized/local advanced from metastatic prostate cancer patients (6, 23). Recently, several groups simultaneously observed the association of a relatively high expression level of miR-375 and miR-141 in blood and/or tumor samples of prostate cancer patients with more advanced disease compared to samples from patients with earlier stages of prostate cancer or healthy tissue (7–9, 13, 24). We again demonstrated that the elevated expression of circulating miR-375 and miR-141 can distinguish patients with metastatic CRPC from those with low-risk, localized prostate cancer. These consistent observations strongly support the potential application of circulating miR-375 and miR-141 as biomarkers for monitoring prostate cancer progression in a minimally invasive way. The present study sets the foundation for future experiments to verify the putative biomarkers with large validation cohorts and for functional studies to analyze the role of confirmed biomarkers in the cancer response to the ADT and in the transition to CRPC.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a SPORE in Prostate Cancer 2 P50 CA090381-06 (PWK).
Footnotes
Author contributions: H. N., M.Y., C.H. and S. D. performed the study. G.L. and P.K. designed and supervised the study. H. N. and W. X. did the statistical analysis of this study and all authors wrote the paper.
Competing interest statement: The authors declare no conflict of interests.
Contributor Information
Han Christine Ngoc Nguyen, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
Wanling Xie, Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
Ming Yang, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
Chen-Lin Hsieh, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
Sarah Drouin, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
Gwo-Shu Mary Lee, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
Philip W. Kantoff, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA..
REFERENCES
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:5–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Huggins C. Endocrine-induced regression of cancers. Cancer Res. 1967;27:1925–1930. [PubMed] [Google Scholar]
- 4.Ross RW, Xie W, Regan MM, Pomerantz M, Nakabayashi M, Daskivich TJ, et al. Efficacy of androgen deprivation therapy (ADT) in patients with advanced prostate cancer: association between Gleason score, prostate-specific antigen level, and prior ADT exposure with duration of ADT effect. Cancer. 2008;112:1247–1253. doi: 10.1002/cncr.23304. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Ba Y, Cai X, Yin Y, Wang K, Guo J, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 8.Selth LA, Townley S, Gillis JL, Ochnik AM, Murti K, Macfarlane RJ, et al. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. Int J Cancer. 2011 Aug 30; doi: 10.1002/ijc.26405. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 9.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 11.Taylor BS, Schultz N, Hieronymus H, Arora VK, Kaushik P, Cerami E, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, et al. miR-378(*) mediates metabolic shift in breast cancer via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130:611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 14.Zheng B, Liang L, Huang S, Zha R, Liu L, Jia D, et al. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene. 2011 Dec 19; doi: 10.1038/onc.2011.581. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 15.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, MacDonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 16.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szczyrba J, Nolte E, Wach S, Kremmer E, Stohr R, Hartmann A, et al. Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Mol Cancer Res. 2011;9:791–800. doi: 10.1158/1541-7786.MCR-10-0573. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 20.Xiao J, Gong AY, Eischeid AN, Chen D, Deng C, Young CY, et al. miR-141 modulates androgen receptor transcriptional activity in human prostate cancer cells through targeting the small heterodimer partner protein. Prostate. 2012 Feb 7; doi: 10.1002/pros.22501. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 21.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, et al. Investigation of miR-21, miR-141, miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32:583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 24.Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, Barth S, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.