Abstract
This article addresses the question of what information and new insights atomic force microscopy (AFM) provides that are of importance and relevance to cellular biophysical chemistry research. Three enabling aspects of AFM are discussed: (a) visualization of membrane structural features with nanometer resolution, such as microvilli, ridges, porosomes, lamellapodia, and filopodia; (b) revealing structural evolution associated with cellular signaling pathways by time-dependent and high-resolution imaging of the cellular membrane in correlation with intracellular components from simultaneous optical microscopy; and (c) qualitative and quantitative measurements of single cell mechanics by acquisition of force-deformation profiles and extraction of Young’s moduli for the membrane as well as cytoskeleton. A future prospective of AFM is also presented.
I. Introduction
The research and development of microscopy technology has always played a critical role in the history of science in general and specifically in that of cellular biology. The high magnification of the optical microscope led Schleiden and Schwann to propose the concept of the cell, which is the smallest unit of life.1 Later, this discovery was referred to as the beginning of modern cell biology. The improvement of optical microscopy is still an active area of research and technology advancement, with new dyes for confocal microscopy and higher resolution imaging being continuously reported. As light microscopy’s resolution approaches its intrinsic limit of spatial resolution due to the diffraction of light or the wavelength of visible light, researchers turn to alternative technologies to break this barrier. In the early 1930s, Ruska invented the transmission electron microscope (TEM) that extended spatial resolution beyond optical microscopy.2 TEM enables intracellular components, such as the cytoskeleton, nuclei, mitochondria, and granules, to be visualized.3–5 In addition to static structure, important insights involving cellular dynamic processes also benefited from the electron microscopy’s high resolution. A milestone example is cell secretion, as revealed by Claude, De Duve, and Palade, a dynamic process that occurs in all living cells and involves transport of intracellular products to the exterior of cells.6 Although electron microscopy enjoys high spatial resolution, the sample preparations typically require either dehydration and staining7,8 or cryogenic protocols,9 which frequently cause questions of biocompatibility or biological relevancy.
A new microscopy technique, known as atomic force microscopy (AFM), emerged in the late 1990s and offered great promise for high-resolution imaging and enabling imaging under physiological conditions.10,11 Three intrinsic advantages of this technology have attracted much attention of researchers interested in cellular biology and cellular physical chemistry. First, AFM offers high resolution. Molecular resolution, for instance, at a subnanometer level, was achieved for inorganic crystals12 as well as protein crystals.13 Applications of AFM in cellular structure characterization also proved to be very encouraging.14–16 Important structural features, such as stress fibers, were visualized. 17 Second, time-dependent imaging in near-physiological media enabled many dynamic processes to be visualized in situ and in real time, such as the identification of a new membrane structure by Jena et al., “the porosome”, first in the apical plasma membrane of pancreatic acinar cells and subsequently in neurons, where secretory vesicles specifically dock and fuse.18–20 Other examples include the activation of human platelets,21 the transport of intracellular particles,22 and the spreading of a Kupffer cell.23 Third, AFM enables physical property measurements locally and globally on a single cell. For example, measuring adhesion between an AFM tip and cellular receptor, or single molecular forces,24,25 enables the mapping of the distribution of heat shock proteins (HSP) on human umbilical venous endothelial cells (HUVECs).26 HSP is essential for cellular homeostasis and efficiently triggers cellular responses to stress conditions.27
These initial efforts of applying AFM in cellular research also met with difficulties and challenges. Although enabling subnanometer resolution in some systems, it is generally difficult to attain such resolution for living cells due to tip-induced deformation, and “soft-and-sticky” interactions between the tip and cellular surface. Time-dependent imaging is also limited by the relatively slow speed of scanning, for example, tens of minutes per frame for a 100 µm × 100 µm area. Single molecular forces require large ensembles of force curves. These issues have triggered much development in bio-AFM technology, such as advanced sample preparation for cellular imaging,28,29 advanced imaging modes for resolution improvement,30–32 and tip modification and data analysis for single molecular imaging and cellular mapping.33–35 Much of this has been reviewed previously.36–38 This article deals with recent advances and new enabling aspects of AFM in cellular physical chemistry. Specific topics include (a) membrane structural features visualized by AFM with high resolution, (b) new insights via cellular signaling processes as enabled by AFM imaging in conjunction with laser scanning confocal microscopy, and (c) single cell mechanics enabled by using modified AFM probes.
II. AFM Enables High-Resolution Imaging To Reveal Characteristic Membrane Features
Although atomic resolution has been demonstrated for crystalline surfaces using AFM,39 and molecular resolution of monolayer systems has also been attained,40,41 it is still difficult as of today to achieve as high a resolution for cells as for those model systems due to (a) the fluidic and soft-and-sticky nature of the cell membrane; (b) the complexity of the membrane composition, including multiple component lipid bilayers and membrane protein molecules; and (c) the requirements of multidimensional imaging in overall cellular morphology as well as local structures. General approaches in addressing these difficulties include (a) using aqueous media to reduce capillary forces between the AFM probe and cell;11 (b) developing advanced AFM imaging modes, such as tapping42 and contact resonance imaging (CRI),30 to conquer the soft-and-sticky nature of the potential; (c) under the guidance of optical view, using high aspect ratio probes for the overall morphology in conjunction with sharp microcantilevers for local imaging;43 and (d) combining AFM with fluorescence or confocal microscopy44–46 and utilizing protein/ligand recognition37 to reveal the chemical functionality at the cellular membrane. In comparison with existing structural characterization methods such as scanning electron microscopy (SEM) and laser scanning confocal microscopy (LSCM), AFM exhibits intrinsic advantages of imaging in near-physiological conditions, in high resolution, and enabling 3D characterization of membrane structures. Progress has been made, and new structural insights have been revealed, as exemplified below.
II.1. Characteristic Membrane Structural Features Associated with the Activation and Locomotion of Mast Cells
The morphology and nanoscopic features of cellular membranes are determined by and therefore representative of cellular signaling processes, such as cellular viability and activation status.47–49 For instance, membrane ridges appear upon antibody-mediated activation of rat basophilic leukemia cells (RBL-2H3), whereas a high density of microvilli indicates resting cells.50,51 Lamellopodia and filopodia correspond to cellular spreading and locomotion.52 Membrane blebs occur during cell apoptosis.53 Because of its capability to operate under aqueous media, AFM enables high-resolution structural characterization in correlation with cellular signaling information. This intrinsic advantage of AFM has been realized in actual case studies of cells.
The concept of using two probes for cellular imaging under optical guidance is illustrated in the configuration of combining a laser scanning confocal microscope (Fluoview 1000, Olympus America Corp.) with an atomic force microscope (MFP3D; Asylum Research Inc.).43,46 The combination allows for simultaneous measurements of topographic details, mechanical properties, and inner structures of labeled cells. Live and hydrated cells may be imaged under culture media or PBS. For inspection of overall cellular morphology, a scanning area up to 90 × 90 µm2 is taken, with a 16 µm dynamic range in the Z direction. Contact mode imaging at a constant force works well using a Micromasch CSC38 cantilever (over 20 µm in tip height) with a spring constant of 0.03 N/m. Visualization of detailed membrane features with higher resolution was achieved by changing the probe to microlevers (e.g., an Olympus Biolever (k = 0.027 N/m)) and utilizing liquid tapping mode. For both probes, the multichannel images of height, z-sensor, deflection/amplitude, and lateral/phase data were acquired simultaneously.
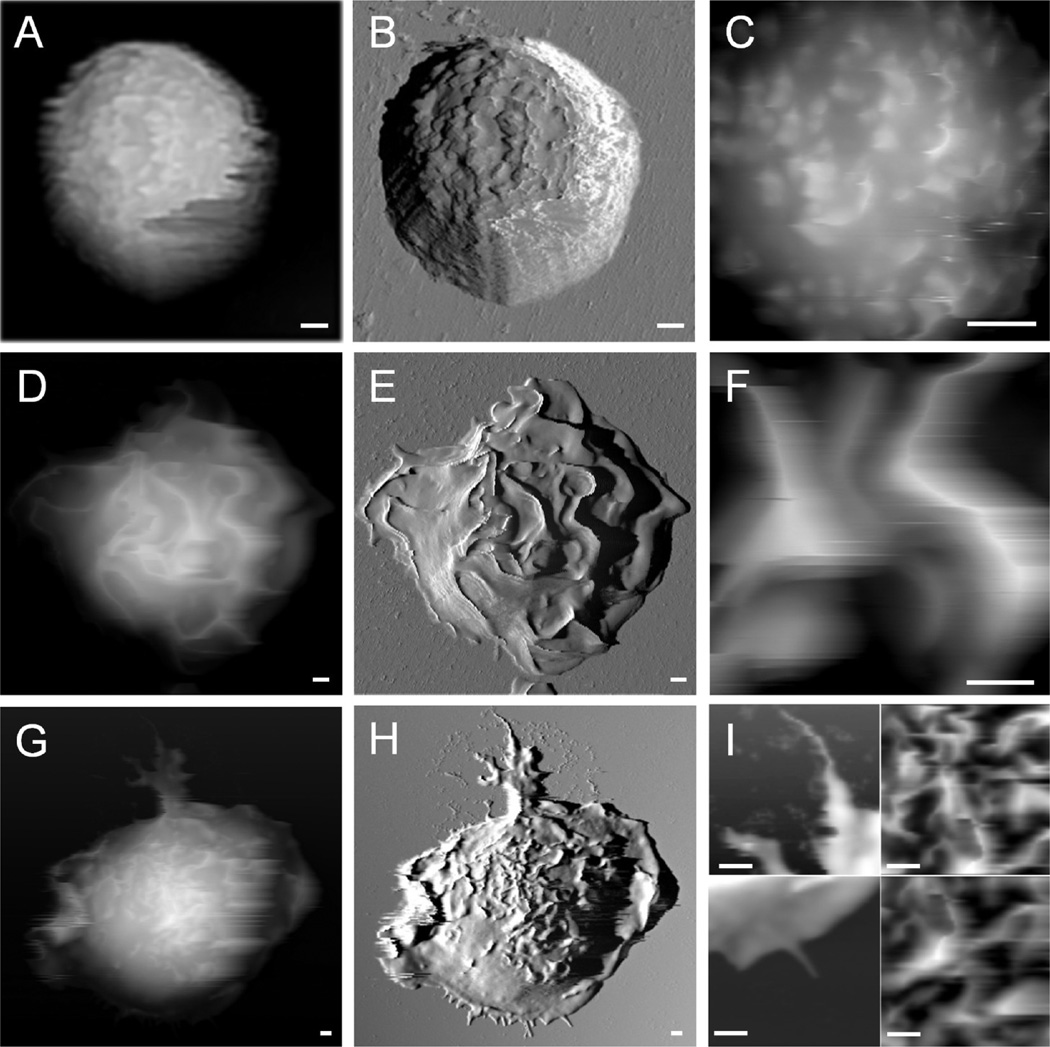
Examples of AFM images acquired in PBS for resting and stimulated bone marrow-derived mast cells (BMMCs) are shown in Figure 1, which reveals representative whole cell morphology and close-range views of mast cells at different activation status. From left to right in Figure 1, each column displays AFM topograph, deflection, and 5 × 5 µm2 zoom-in images, respectively. The resting cells exhibit a “hat” shape on the surface with a low degree of spreading. 3D measurements can be taken from designated cursor profiles: for example, the contact diameter and height are 9.0 ± 1.0 and 7.5 ± 0.5 µm, respectively, for the cell shown in Figure 1A–C. In contrast to resting mast cells, activated cells exhibit dramatically different morphology, as exemplified in Figure 1D–F. Characteristic membrane ridges appear on anti-DNP IgE-sensitized BMMCs upon activation by 2,4-dinitrophenylated bovine serum albumin (DNP-BSA) immobilized on the coverslip surface. Cell activation enhanced the cell spreading, which in turn decreased cell height. For example, the height of the cell shown in Figure 1D is 5.2 µm, 2 µm lower than the cell in Figure 1A.
Figure 1.
Characteristic membrane structural features are associated with the activation and locomotion of mast cells. Images A–C are AFM topography, deflection, and zoom-in images of a resting BMMC with its characteristic villi structures. Images D–F present an IgE-sensitized BMMC activated by DNP-BSA immobilized on the surface, showing characteristic membrane ridges. The images in G–H display an anti-DNP IgE-sensitized BMMC spreading on a DNP-BSA coated surface, with characteristic locomotion structures of lamellipodia and filopodia and characteristic activation structures of membrane ridges. Scale bars: 1 µm.
In addition to cell activation, AFM enables the capturing of transient cell membrane structures specific to cell movement. In the bottom row of Figure 1, AFM topography (Figure 1G) and the corresponding deflection (Figure 1H) images are displayed side-by-side and clearly reveal a BMMC’s movement at the moment of fixation. In locomotion, this cell uses 11 filopodia to test its way, manifested as the membrane protrusions appearing at the front end of cell movement, the leading edge. Section profiles taken on these short filopodia at the leading edge of the cell give a length range of 0.1–1 µm and a maximum height of 0.5 µm. Very likely, each filopodium represents a single integrin surface binding site. A large tail containing treelike branches and the adventurous filopodia reveal trailing and leading edges of cellular motion, respectively. The tail in the trailing edge contains a total of 5 branches, with the primary central one of 9.0 ± 0.5 µm in length and 1.10 ± 0.06 µm in height.43
The 3D quantitative characterization of cellular membrane enables the determination of the degree of activation; for example, through the density of ridges and their dimensions. Zooming in to the top of the resting cell, Figure 1C reveals microvilli and small protrusions at mesoscopic scale with apparent heights ranging from 200 to 500 nm. Upon activation, membrane ridges become prominent features, as revealed by the AFM topograph in Figure 1E,F. In Figure 1E, a total of 25 ridges are visible, with their lengths varying from 2 to 10 µm. The presence of membrane ridges on activated BMMCs is consistent with previous studies using SEM, in which the formation of membrane ridges was associated with a later stage of degranulation.51 In addition to confirming the morphologic change first revealed by SEM investigation,50,51 AFM enables 3D measurements of characteristic membrane features. For instance, the measurements of ridge height from 0.2 to 2.0 µm, as shown in Figure 1F, bring us a step closer to quantitatively correlating membrane structures with the degree of activation for mast cells. Figure 1H,I reveals characteristic membrane structures in the early stages of activation, where the ridges are visible but less prominent than those in Figure 2D–F The cell membrane was in the transition from villi to matured ridges.
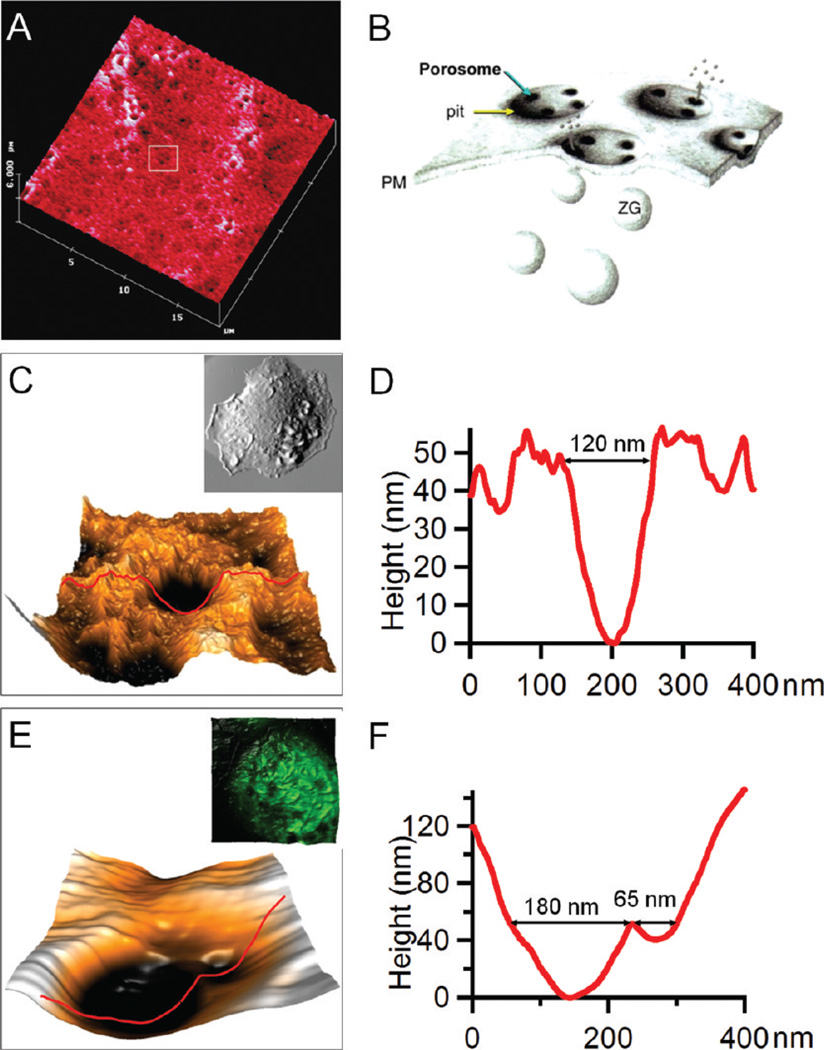
Figure 2.
Porosomes captured by AFM. (A) The first example of porosomes captured by AFM on pancreatic acinar cells by B. P. Jena et al.18 (B) Schematic diagram illustrating the corresponding secretary mechanism in A. (C) A porosome captured on an activated RBL-2H3 cell. The inset in C shows the whole cell morphology of the RBL-2H3 cell on which this porosome is located. (D) The AFM section profile measurement of the porosome in C. (E) Two porosomes captured on an activated bone marrow-derived mast cell. The inset in E shows the overlay of AFM and confocal images taken on the same cell where these two porosomes were observed. (F) The AFM cursor profile measurement of the porosomes in E.
II.2. Characteristic Membrane Structural Features Associated with the Secretary Process of Cells
Using AFM, Jena and his team were the first to report the discovery of a new cellular structure, referred to as the “porosome”.18,19 Porosomes represent a characteristic structural feature at the plasma membrane present during secretary processes. Porosomes are made of macromolecular assemblies due to secretary vesicles’ docking and then fusing to release intravesicular contents to the outside media. Figure 2A gives an example of porosomes captured by AFM on pancreatic acinar cells.18 The corresponding secretion mechanism is illustrated in the schematic diagram of Figure 2B, where each “pit” at the membrane of pancreatic acinar cells contains three or four “depressions” (porosomes) and serves as the secretion site for the docking of intracellular granules. The size of these porosomes ranges from 100 to 180 nm in diameter, and their depth ranges from 15 to 30 nm.18 AFM, because of its high resolution and capability to operate in aqueous media, played a crucial role in this discovery.47,18,19 At present, porosomes have been accepted as the generic secretory machinery in cells, from exocrine cells of the pancreas, to neuroendocrine cells and neurons.54
Porosomes at the membrane of RBL-2H3 cells and BMMCs were also captured using AFM, mainly among activated cells. Figure 2C,D exemplifies the morphology and quantitative characterizations of a typical porosome captured on RBL-2H3 cells activated by antigen immobilized on the surface (whole cell shown in the inset of Figure 2C). From the AFM topograph in Figure 2C, the porosome displays a donut shape with the inner opening of 120 ± 10 nm, as measured from the section profile in Figure 2D. These donut structures and their dimensions are consistent with the previously proposed “basket” model for porosome complexes using AFM and TEM,19 where supramolecular cup-shaped lipoprotein baskets were observed at the cell plasma membrane and formed by membrane-bound secretory vesicles docking and fusing to release their vesicular contents.
BMMCs exhibit similar secretory features, as shown in Figure 2E,F, where two “depressions” were observed. These two depressions are adjacent to each other, and both are located at the valley between the protruded membrane ridges. The size of these two depressions differs, with openings of 180 ± 20 nm and 65 ± 10 nm in diameter, respectively. The arrangement and dimensions of these depressions resemble the porosomes residing in the membrane pits of secretory cells reported previously.19 More importantly, when the cell was examined using combined atomic force and confocal microscopy (inset in Figure 2E),46 these two depressions were observed to be located atop intracellular vesicles. The diameter of the intracellular vesicle is equal to the membrane height measured at the location where these two depressions reside, indicating the plasma membrane and granule membrane are in touch, and thus, these depressions are likely fusion pores (porosomes) in connection between the granule and plasma membrane.
The structure and function of porosomes have also been reconstituted in the lipid membrane, with their compositions revealed, as well.55 The finding of porosomes as a new cellular structure and the subsequent investigations of their dynamics using AFM56 have established AFM as an important and credible new technique in cellular biology and opened a new time of “nano cellular biology”.37,47
Many other functional membrane features have also been revealed using AFM. The occurrence of heat shock proteins (HSP60) on the membrane surface of HUVECs has been investigated using antibody-modified AFM probes to enhance the tip–membrane interaction at the HSP60 present sites.26 Very similar patchy distributions of HSP60 were found for living and fixed cells, regardless of their stress status, a finding that is of much interest to the role of HSP60 in atherogenesis.26
Detergent-resistant membranes (DRMs) are yet another important and interesting membrane feature which have been isolated from cell lysates of almost all mammalian cell types.57,58 DRMs contain specific membrane proteins and are implicated in signal transduction. They are resistant to detergents due to the enrichment of sphingolipids and cholesterol. One example of DRMs is lipid rafts, existing as discrete patches in the plasma membrane of cells, with lateral dimensions ranging from tens to hundreds of nanometers.59,60 Due to its high resolution, AFM has been proposed as a promising tool to solve the complexity of DRMs. Tapping mode AFM in buffer has been performed on the surface of Triton-X 100-treated CV-1 cells, a fibroblast-like cell line originating from the kidney, and shows the unexpected large domain size (15–20 µm2) of the observed DRMs. The size range of these DRMs is about 1 order of magnitude higher than that estimated for living cells, suggesting that membrane microdomains rearrange into larger DRMs during surfactant treatment.61
The resolution and dynamics of single proteins on the plasma membrane of live cells have been achieved using scanning ion conductance microscopy (SICM).62,63 This technique, originally developed by Hansma and co-workers,64 is based on a scanned nanopipette and uses the ion current that flows between an electrode in the pipet and a bath electrode for feedback control of the pipet-sample distance. The capability to image living cells at a resolution sufficient for the identification of protein complexes, directly and without labeling, opens up a wealth of new possibilities in nanobiotechnology.
III. Combined AFM and Laser Scanning Confocal Microscopy Enable High-Resolution Imaging of Membrane in Correlation with Cytoskeleton and Intracellular Features To Reveal Cellular Signaling Pathways
The correlations among nano and mesoscopic membrane features, cytoskeleton, and intracellular functional structures enabled by combined atomic force and confocal microscopy provide new insight into cellular signaling processes, as demonstrated by the investigation of mast cell activation.46 Two pathways for the activation of mast cells have been previously reported.65–68 The antibody-mediated pathway, also known as FcεRI-dependent pathway, involves the initial binding of immunoglobulin E (IgE) to specific high-affinity IgE receptors (FcεRI) and the subsequent cross-linking of bound IgE molecules by multivalent antigens. The cross-linking process is then followed by the receptor aggregation, which further triggers the signaling cascade leading to degranulation. The FcεRI-independent pathway involves basic secretagogues, such as anaphylatoxins, neuropeptides, compound 48/80, and polylysine, etc.69 These stimuli share a common characteristic of being polycations that are able to be internalized into or penetrate halfway through the plasma membrane and stimulate pertussis toxin-sensitive G proteins.70,71
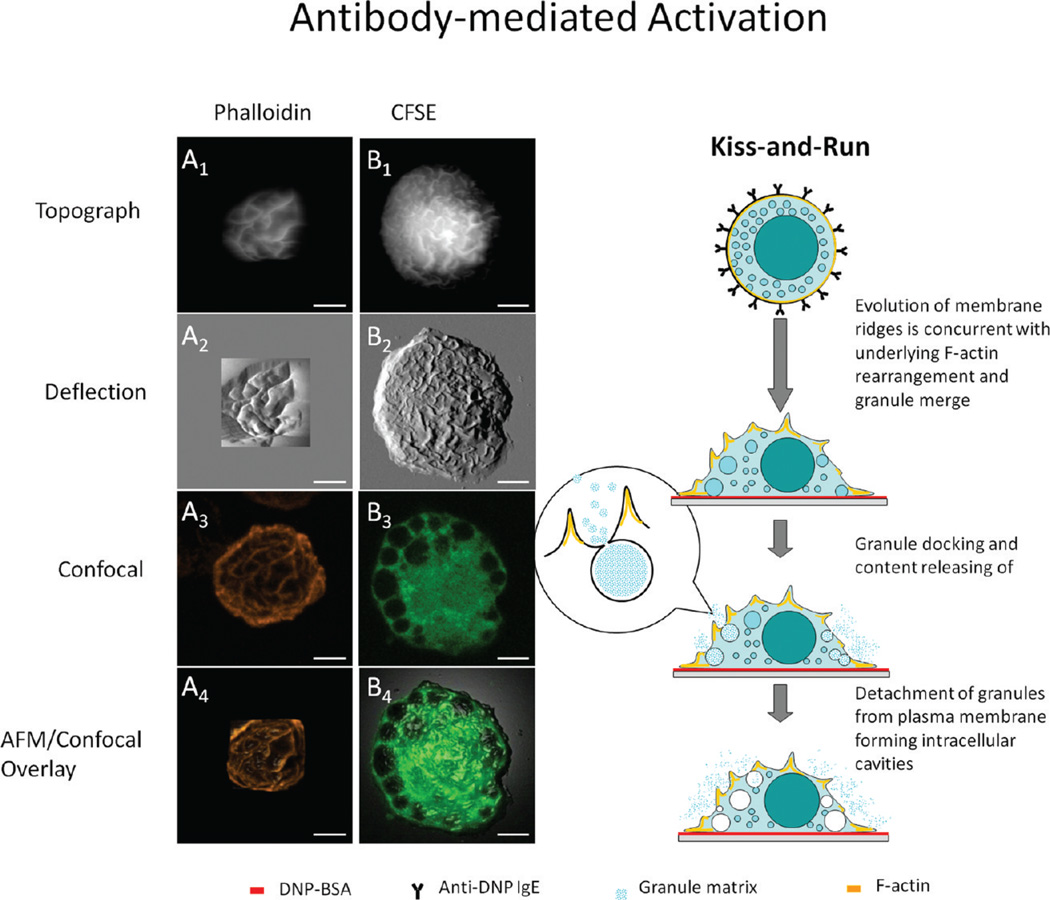
Centrifuged onto the coverslips and prior to activation, BMMCs exhibit a hat-shaped geometry with high-density microvilli present at the cellular membrane, as exemplified in Figure 1A–C. After activation via antigen, the morphology becomes dramatically different. Combined AFM and confocal imaging of BMMCs labeled with phalloidin enables the simultaneous and in situ observation of membrane morphology and cytoskeletal F-actin assemblies.46 Figure 3A1–4 presents the AFM topograph, deflection (force-error), confocal, and topograph–confocal overlay images, respectively, taken on the same cell after antibody-mediated activation. The visualization of membrane ridges shown in Figure 3A1,2 indicates the activation of this BMMC. The Z-projection images in Figure 3A3 involve only the Z slices taken on the cortical side of the cell, from which one can see the heterogeneous network arrangement of F-actin. In contrast, the resting cell shows a more homogeneous fluorescence contrast of F-actin.46 If these confocal images were displayed via a 2D cross-sectional view, each individual slice from the Z-stack of the activated BMMC would exhibit a broken circle, whereas that of the resting cell would display a continuous ring. This observation is consistent with a previous study by Nishida et al. using confocal microscopy72 in which the cytoskeletal F-actin was found fragmentized upon antibody-mediated activation. Our results, with the complete Z-stack, contribute further spatial distribution data on this rearrangement of F-actin at cytoskeleton. The correlation between membrane ridges observed in AFM and the F-action network seen in confocal microscopy is investigated by the overlay of the confocal and AFM images, as shown in Figure 3A4. This overlay is through the matching of at least three different landmark points selected from both AFM topographic and confocal images. The AFM topograph (Z as a function of X and Y) was displayed in 3D, and then colored according to the aligned fluorescence image using the MFP-3D software as described previously by Proksch et al.73 From the results of the overlay, it is concluded that the membrane ridges are the lipid bilayer decorating over the F-actin network underneath.46
Figure 3.
IgE-mediated degranulation pathway revealed by the combined AFM and LSCM study. From top to bottom, column A (A1–A4) presents the AFM topography, deflection, confocal z-projection, and overlay images of an anti-DNP IgE-sensitized BMMC activated by surface-immobilized DNP-BSA and labeled with fluorophore-tagged phalloidin. Note that membrane ridges are superimposed over the F-actin network underneath. Column B (B1–B4) has the same layout of images as in A and shows a BMMC activated by the same method as used in A, except the cell is now tagged with CFSE, and B3 is a single confocal slice extracted from the middle of the Z slices. Note that intracellular dye-devoid cavities are visible from the confocal image. The schematic diagram next to the right of column B illustrates the corresponding degranulation pathway derived from the combined AFM and LSCM study.
Under identical activation conditions, the fate of intracellular granules was also investigated by using carboxyfluorescein diacetate succinimidyl ester (CFSE), a cell tracing dye, to highlight the intracellular homogeneity and heterogeneity.46 The CFSE dye contains acetate groups, which yield highly fluorescent amine-reactive carboxyfluorescein succinimidyl ester upon cleavage by intracellular esterases. The succinimidyl ester group then further reacts with intracellular amine-containing proteins, lighting up the entire cell body.74 Figure 3B1,2 reveals the AFM topograph and deflection images of a BMMC labeled with CFSE, respectively. A large number of membrane ridges are clearly visible, which confirms the antigen-driven activation of this cell similar to the situation in Figure 3A1–4.50,51 Figure 3B3 reveals the presence of dye-devoid cavities inside the cell, and the diameter of these cavities varies from 0.8 to 5.0 µm. The CFSE-devoid intracellular cavities tend to gather at the periphery of the cell. Closer to the nucleous region, only two large and two small cavities were found. The lack of cavities near the central region is consistent with the fact that cavities are granules upon exocytosis due to two reasons. First, at the cell periphery, the gap between the ventral and cortical membrane is small so that granules have a higher chance to fuse with the plasma membrane. Second, during the labeling process, CFSE diffuses from the cell periphery to the center, and thus, the dye-devoid cavities must occur after granules release their contents inside. In contrast, nonactivated mast cells present a more homogeneous CFSE distribution without dye-devoid cavities.46
Taken collectively, the AFM morphology, the F-actin assembly, and intracellular granules, the antibody-mediated activation follows the so-called “kiss-and-run” process,48 as illustrated by the right schematic diagram in Figure 3. Upon cross-linking of the IgE-FcεRI complexes by surface-bound antigen, membrane ridges form due to the reorganization of cytoskeletal F-actin. In contact with the interior of the membrane, granules dock within the neighboring ridges and then open the fusion pores to release granule contents. This contact is transient, and the granules retreat to the cytoplasm afterward.48 The overall theme of this kiss-and-run model was proposed previously.75,76 New insights added from this AFM/confocal microscopy investigation include (a) revealing the origin of membrane ridges as well as the 3D characterization of their dimensions, (b) the determination of the fate of postdegranulation granules, and (c) the capture of transient fusion pores by AFM in hydrated cells.
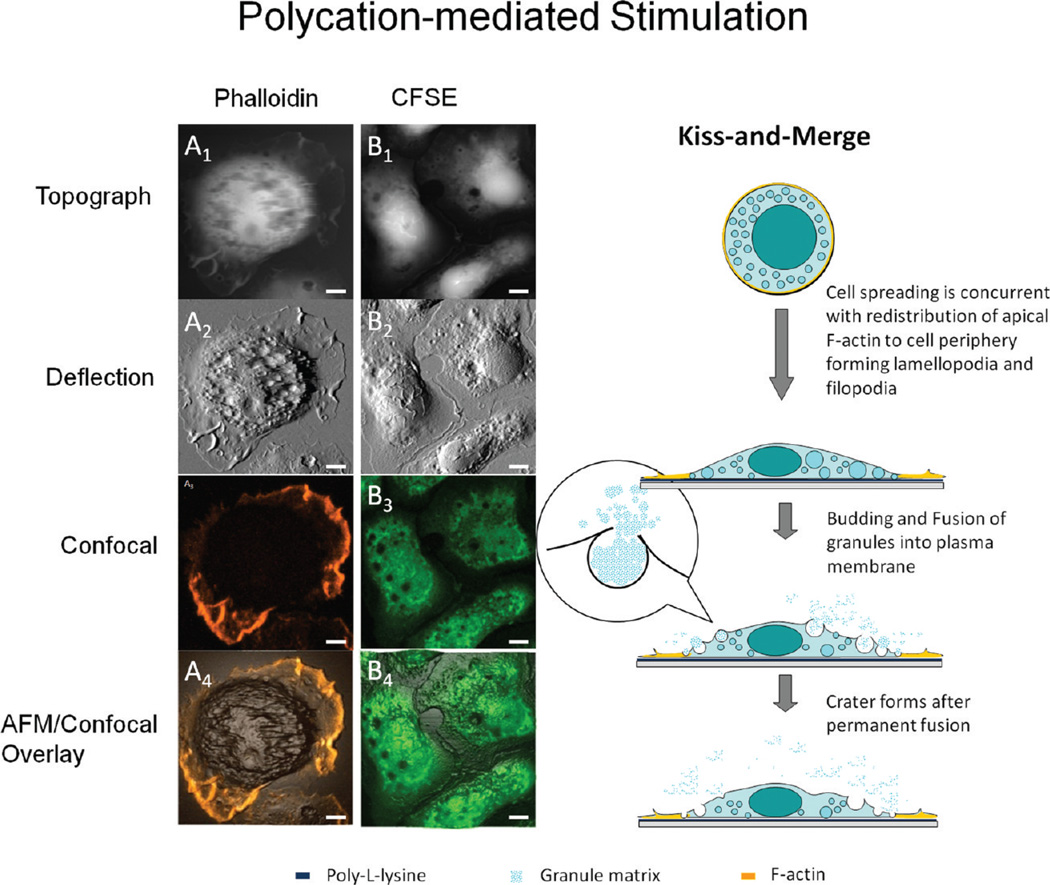
In the absence of antigen, BMMCs may be stimulated by polycations such as poly-l-lysine, as shown in Figure 4. A different degranulation behavior is revealed by the combined AFM and confocal images shown on the left of Figure 4. As shown in the AFM images (top two rows), few ridges are present at the cortical membrane, and craters become the characteristic features of BMMCs on poly-l-lysine. At locations where the AFM topographic images reveal membrane craters, the confocal contrast shows dark pits in cells stained with CFSE (Figure 4B2,3). The degree of superposition is clearly shown by the overlay image in Figure 4B4. The diameters of the dark pits in the fluorescence image range from 0.4 to 5.2 µm, consistent with the opening size of craters measured in the AFM images. Combined AFM and confocal images taken on those cells stained by phalloidin highlight the roles of cytoskeletal F-actin. Little correlation is found between F-actin distribution and formation of craters, as shown in Figure 4A1–4. Craters are mostly located at the membrane region where F-actin network is lacking. The Z-projection confocal image in Figure 4A3 shows F-actin is enriched in the lamellopodia and filopodia located at the cell edge. This observation is consistent with the prior knowledge that an F-actin network forms at the leading and trailing edges during cell locomotion.77
Figure 4.
Polycation-mediated degranulation pathway revealed by the combined AFM and LSCM study. The layout of AFM and confocal images is the same as in Figure 3, except the BMMCs are stimulated by poly-l-lysine coated on surface. Note that the main pool of actin is attracted to the cell periphery, and the dark pits observed in the CFSE fluorescence image correspond to membrane craters captured by AFM. The schematic diagram next to the right of column B illustrates a different degranulation pathway from Figure 3. Scale bars: 5 µm.
Taken collectively, we infer that the craters are formed by granules fusing into the plasma membrane where cytoskeletal F-actin network is lacking, for instance, permanent fusion. We refer to this process as “kiss-and-merge”, in contrast to the previous kiss-and-run process. The main steps are illustrated in the schematic diagram on the right of Figure 4. Upon contact with the surface-bound poly-l-lysine, cells adhere and spread. This process involves the formation of extensive lamellopodia and filopodia, as a result of cytoskeletal F-actin reassembly under the plasma membrane.77 This movement is likely faster than intracellular formation of cytoskeletal F-actin.78 Therefore, the cellular actin is enriched near the periphery and depleted at the central or bulk region of the cell. The spreading of the cell is likely to trap the granules in the “shallow regions”, where the gap between the cortical and ventral membrane is narrow. Thus, granules would merge with the plasma membrane and release matrix inside. The budding and merging of granules into the plasma membrane has been reported by previous studies using transmission electron microscopy.79 The new insights added by AFM/confocal studies include (a) the visualization and quantification of membrane craters, that is, the intermediate structures of permanent fusion; and (b) the lack of colocalization between these craters and F-actin, suggesting that the lack of F-actin participation facilitates the crater formation.
The correlation of membrane features with intracellular structures also reveals F-actin’s impact on the membrane morphological changes as well as local and overall deformation at nanometer scale. Upon antibody-mediated activation, the BMMC membrane exhibits a ridged morphology due to the lipid bilayers decoration over the F-actin network. If a cell membrane is thought of as like the skin of balloons, this wrinkled membrane structure is more relaxed due to cytoskeleton confinement (blow-up of Figure 3), in comparison with the stretched membrane of BMMCs spreading on poly-l-lysine (the zooming in of Figure 4).80 As such, a lack of F-actin and high lateral tension promotes permanent fusion between granules and the plasma membrane (kiss-and-merge) in the poly-l-lysine-stimulated cells, and the presence of an F-actin network, and a small lateral tension favors transient fusion (kiss-and-run) in antigen activated BMMCs.
Various other cellular signaling processes may be triggered by changing the culture conditions, such as temperature, pH, salt composition, and the concentration of effectors (regulatory molecules) and inhibitors in the media. Using single antibody-or sugar-modified AFM probes, force–distance cycles were acquired to measure the interactions of the antibodies with a protein loop protruding out of SGLT1, a Na+–glucose cotransporter in living Chinese hamster ovary (CHO) cells.24,35 SGLT1 is an important member of the ion gradient driven channels and responsible for glucose import into cells.24,35 Quantitative force measurements confirmed that this loop is one part of the extracellular domain of the transporter that was previously unknown.
Additional examples include utilizing time-dependent AFM imaging to investigate the interaction of human osteoblast-like cells (SAOS-2) and Chinese CHO cells with 2D matrixes of highly ordered, parallel arrays of collagen type I fibrils.81 It was found that cell lines expressing integrin α2β1 polarized strongly in the fibril direction. During the alignment, cells deform the matrix by reorienting individual collagen fibrils. Both the high tensile strength and pliability of collagen fibrils contribute to the anisotropic rigidity of the matrix, leading to directional cellular traction and cell polarization.81
IV. AFM Enables Determination of Single Cell Mechanics
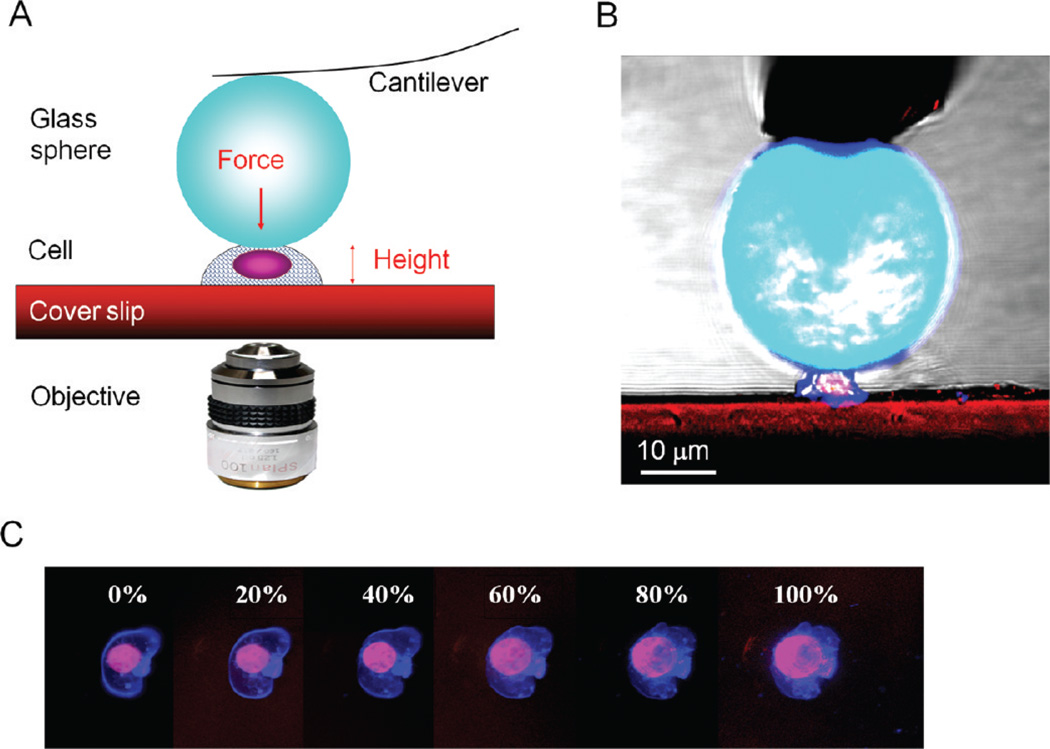
Taking advantage of the precise force measurements AFM enables, a new AFM-based methodology was developed to investigate the mechanical property of single cells, as shown schematically illustrated in Figure 5A.82 Each AFM probe is modified with a clean glass microsphere (e.g., D = 40 µm) and then used to precisely apply a compression force along the surface normal to individual cells, as shown in Figure 5B. The deflection of the cantilever depends on the force constant of the probe (cantilever) and the elastic compliance of the cell underneath. The overall force applied typically ranges from 10−5 to 10−11 N, for commercial microfabricated cantilevers. The force–deformation profile of a single cell provides qualitative and quantitative information of cell mechanics with high sensitivity and accuracy.83 High-resolution optical microscopy (e.g., confocal laser scanning microscopy) is combined with the AFM to guide the probe position and monitor cell responses to mechanical compression. During compression of the cell via the large sphere, an AFM force–distance plot is recorded to reveal the force-cell deformation behavior, referred to as the cell compression profiles.82 Using chromophores, such as cell tracing dyes, confocal images can be acquired during the compression cycle to reveal the cell deformation, such as nucleus dislocation, blebbing, or bursting of the cellular membrane (Figure 5C). In comparison to the existing cell mechanics measurements, the AFM-based method provides direct and independent force and deformation measurements of the entire range of a cell’s deformation, 0–100%. This method also allows the force (load) to be precisely applied at designated speeds. Attachment of a large sphere enables the whole cell instead of local mechanics to be measured with high consistency. This approach both increases sensitivity to adhesion forces and prevents local membrane rupturing at small deformations (e.g., due to high pressure in AFM local force mapping), thus allowing measurements of membrane elasticity and permeability. This method has high reliability and reproducibility because of its high accuracy of cell positioning and high sensitivity of the AFM probes. More importantly for cell biomechanics, the derived model of cell compression suggests that the cell compression profile provides not only a sensitive measure of mechanical properties of the cytoskeleton but also information on plasma membrane elasticity, permeability, and adhesion.84
Figure 5.
(A) Schematic diagram of the AFM-based single cell compression experiment, which illustrates the concept and (B) confocal side view of T-cell compression. (C) Confocal microscopy snapshots of a single cell under the compression cycle of AFM force curve acquisition. Cell membrane deformation (blue boundaries) followed by the flattening of the nucleus (see pink regions) was captured. At high deformation, the Newton diffraction ring is clearly visible, which demonstrates the microscopic precision in the alignment.
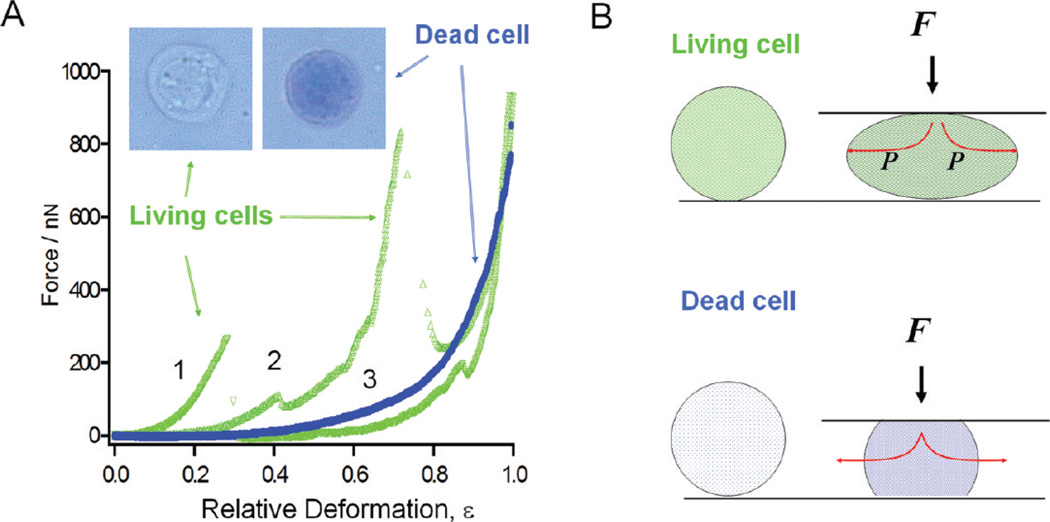
The force sensitivity of AFM-based single cell compression can be seen in Figure 6. The two typical force-deformation curves for living T-cells are shown in Figure 6A. Six separate batches of cultures were measured, and force curves were acquired on at least 20 different cells per experiment. The two curves shown in Figure 6A are representative results (>80%) from single-cell compression measurements. Since the same cantilever is used and the AFM and optical combination enables precise probe–cell alignment, the difference in the force response reflects the individuality of the cells. Despite cell variations, common characteristics were observed from the single-cell compression profiles of living cells. At low applied load and deformation (20–30%), the cell’s “spring” is elastic and fully reversible, which suggests that the cellular membrane remain impermeable. At intermediate deformation, 30–80%, stress peaks appear in the profiles.82 The responses become highly nonlinear with very little reversibility. Simultaneous optical imaging suggests that the cell membrane likely bursts at the first peak, causing leaks of cell fluid and, thus, a sudden drop in spring elastic compliance. In most T-cells, stress peaks appear at 30% deformation, with the observed cell-rupturing force ranging from 0.1 to 0.3 N.
Figure 6.
(A) Typical force versus deformation curves are shown for living (green, curves 1–2) and dead (blue, curve 3) T-cells. Insets show the two types of cells upon the addition of 10 µL of 4% trypan blue solution, where dead cells turn blue under optical microscopy. (B) Mechanical model for living (top) and dead (bottom) T-cell compression.
The complexity in force response by various cellular components is revealed by the appearance of multiple peaks after membrane rupture (see Figure 6A). Beyond 80% deformation, the slope increases rapidly, reaching micro-Newton forces. We infer that this region is a characteristic of further loss of intracellular contents as well as deformation of the intracellular components.
The sensitivity to cell viability is also demonstrated. Prolonged exposure to ambient conditions without cell-growth media led to cell death. The viability of the cells was checked using the trypan blue assay,82 in which dead cells are permeable and turn blue, as visualized under optical microscopy. The dead cells exhibited a distinctively different force response from that of the living cells. A smooth deformation profile of a dead cell is shown in Figure 6A, in contrast to the living cell curves that contain multiple stress peaks. There are also fewer individual variations in the compression profiles of dead cells as compared to those of living cells. For dead cells, the compression process becomes irreversible beyond 10–20% deformation, but for living cells, it is reversible until the first stress peak, deformations up to 30–40% of original height. At high deformations, the curves become very similar among cells, regardless of their initial viability, which is consistent with our explanation of the force curve for living cells.82
In addition to establishing the force–deformation profile of individual cells, quantification of the membrane and cytoskeleton Young’s moduli is attainable by fitting the profiles using a simple analytical formula. The formula was derived from the elastic theory of membranes85 and Hertzian contact mechanics.85,86 This model and derivation are based on the following assumptions: (a) Cells may be treated as a spherical membrane filled with incompressible fluid and cytoskeleton, sandwiched between the parallel plates. (b) At small deformations (i.e., ε <30%) and under our compression speed, the membrane (shell of balloon) is not permeable. (c) Force–deformation arises from membrane stretching and cytoskeletal deformation, with insignificant contributions from other cellular components such as nuclei, during the initial compression process (i.e., ε <30%). These assumptions are based upon an approximation of treating a cell as a nonpermeable balloon filled with fluid. More accurate models and advance calculation methods, such as finite element analysis,87 would be necessary to account for profiles at high deformation as well as detailed heterogeneity of the membrane deformation.
Assumption “a” arises from the fact that the probes are much larger than the equivalent sphere of cells under compression. T-cells, for instance, are relatively spherical with a height below 10 µm (most of them are below 5 µm), whereas the probe’s diameter is 40 µm. When compressing cells along the surface normal, the initial contact may be modeled as a 40 µm sphere compressing a small sphere (D = 5–10 µm; i.e., the cell height).
Assumption “b” is based on a large number of prior experiments.88,89,82 During single-cell compression, two characteristic behaviors, both consistent with membrane nonpermeability, were observed. The force–deformation profiles of T-cells reveal stress peaks, corresponding to bursting of the membrane, as per simultaneous optical microscopy. This global membrane bursting happens because the intracellular pressure builds up during compression due to the impermeability of the membrane. Instead of bursting, other types of cells, such as human breast cancer (MDA-MB-468) and human prostatic epithelial (MLC-SV40) cells, develop 2–8 small blebs as a result of pressure buildup.90 This pressure-induced blebbing is very typical for many types of living cells91 and is attributed to the behavior of nonpermeable plasma membranes under pressure, which causes membrane stretching and detachment from the cytoskeleton.88 It has been shown using M2 melanoma cells that cell volume remains constant during blebbing,88,92 which provides additional proof of membrane nonpermeability during blebbing. Therefore, regardless of global bursting or blebbing behavior, the impermeable balloon model remains valid under the conditions and time scale of our single-cell compression experiments.
A simple analytical expression can be derived, under assumptions a–c to account for the cell deformation profiles,90
| (1) |
where ε is the relative deformation of the cell; R0 and h are the radius of the uncompressed cell and its plasma membrane thickness; Em and Ec represent the Young’s modulus of membrane and cytoskeleton, respectively; and νm and νc represent the Poisson ratio of membrane and cytoskeleton, respectively. The physical meaning of eq 1 is that cellular mechanical resistance arising from cytoskeletal compression follows a F ∝ Ec•ε3/2 relationship and a F ∝ Em•ε3 term from the membrane stretching. The fact that the deformation profiles for the membrane and cytoskeleton follow different power laws (cubic vs 2/3) allows us to extract Em and Ec from one cell compression profile using nonlinear fitting.
For T-cells at low deformation, the cytoskeleton contribution is insignificant. The observed force profile of living cells follows a cubic relationship with respect to deformation (ε3) at small loads. The nonlinear fitting to the force curves of living cells allows the single cell elasticity to be extrapolated. The fitting is excellent for deformations less than 30%, with a typical standard deviation of ±0.4 MPa. Assuming the Poisson ratio ν = 1/2 for the cell membrane, that is, it is incompressible, then the Young’s modulus of the membrane ranges from 10 to 35 MPa for the typical living T-cell compression profiles. This value is comparable to the Young’s modulus of highly elastic polymers (typically 1–100 MPa).93 Young’s modulus of the protein-coated lipid vesicles in the micropipet studies were found to be 10–20 MPa.94
The mechanical model for dead and fixed cells is a balloon filled with fluid; however, that is different for cells with a permeable skin, as illustrated by the experiments shown in Figure 6. During single cell compression, fluid may be squeezed out, thus making the membrane stretching term become insignificant. Under our compression conditions, hydrodynamic contribution of the liquid squeezed from the cell may be neglected because little force-speed dependence is observed. Thus, the deformation of cells, according to the Hertzian theory,85 can be calculated using
| (2) |
where R0 represents the cell radius before deformation and Ec and νc are the Young’s modulus and Poisson ratio of the whole cell’s cytoskeleton, respectively. At 30% deformation or less, fitting the force curves of dead cells using eq 2 allows the extraction of the cell elasticity, giving the Young’s modulus of the T-cell mainly attributed to the cytoskeleton as 3–6 kPa (for ν = 0; i.e., fully compressible material). The standard deviation of these fitting is ±0.16 kPa. The resulting elasticity is in good agreement with the cytoskeleton or cell nuclei (1–10 kPa), as estimated from previous cell mechanics studies.95
It is known that during the fixation process, aldehyde groups from the fixative react with primary amines in the membrane and cytoskeletal proteins, forming covalent imine bonds.96 The cross-linking of proteins dramatically changes the elasticity and the mechanical behavior of cells. The most evident observation is the dramatic increase in cell elasticity; that is, the fixed cells are almost 20–50 times more rigid than living cells. Additionally, the compression can no longer rupture the cells to cause irreversible responses during loading and unloading cycles; that is, deformation was completely reversible for fixed cells. The bending force contribution of the cross-linked cell membrane is less than 10 nN, even assuming the Young’s modulus of membrane of 1 GPa, and thus, the bending contribution may be ignored.90 The single cell mechanics is therefore similar to that of dead cells without fixation in terms of the fact that the cell mechanics is dictated by cytoskeleton compression instead of membrane stretching. The least-squares fitting of force curves using eq 2 (derived from the Hertzian model) yields the Young’s modulus of these cells in the range of 120–180 kPa.
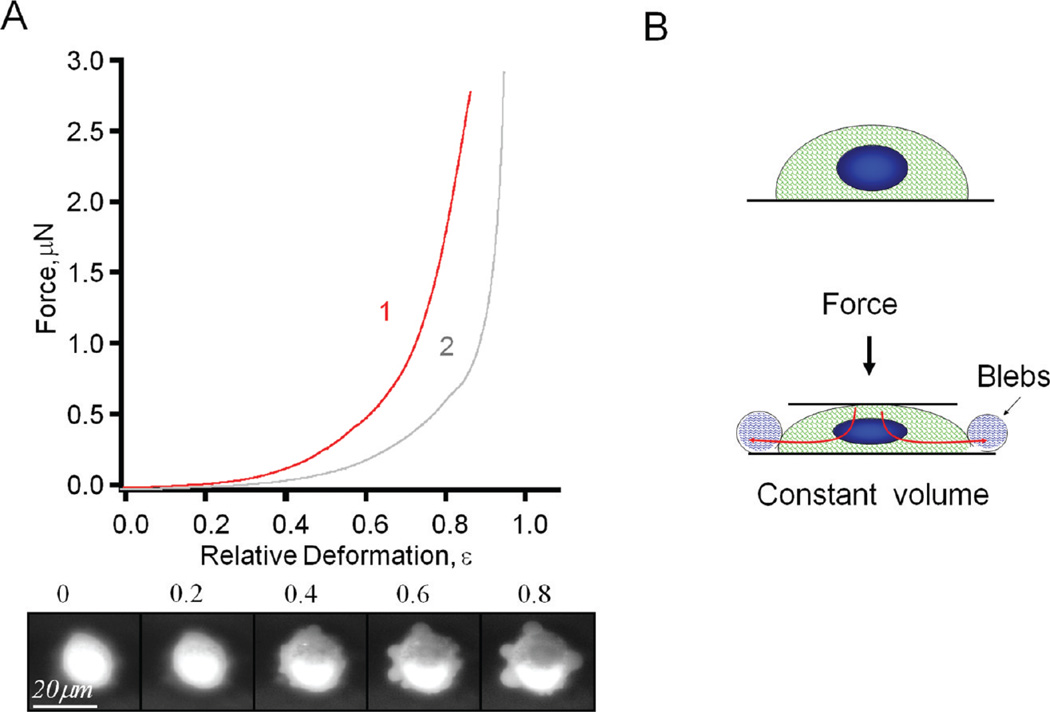
Various types of cells derived from different organs were tested using AFM-based single cell compression. Figure 7 shows typical force deformation profile for NIH 3T3 (mouse embryonic fibroblast cells) and immortalized human prostatic epithelial cell line MLC-SV40. In contrast to living T-cells, the overall profile for fibroblast and prostate epithelia cells is smooth without obvious stress peaks. When deformation exceeds approximately 50%, cells die (similar to T-cells); however, even at high deformations, cells retain significant mechanical resistance. This suggests a higher strength of their cytoskeleton.
Figure 7.
(A) Typical profiles, that is, force versus relative deformation, for fibroblast NIH-3T3 (curve 2, gray) and prostate MLC-SV40 (curve 1, red) cells. Row below shows bottom view of the prostate cell deformation process. Pressure-induced bubbles (blebs) are clearly visible. (B) Simplified model of the formation of blebs.
As shown in the snapshots at the bottom of Figure 7A, when deformation exceeds 50%, cells typically develop 4–10 membrane extrusions, or blebs, ranging 4–12 µm in diameter. These are similar to the blebs occurring during apoptosis or cytokinesis.88,92 These blebs represent detachment of the plasma membrane from the cytoskeleton due to an increase in intracellular pressure driven by the external loading force. In contrast to a global expansion of membrane (as in the case of T-cells), local blebbing is an indication of membrane heterogeneity, a result of local compositional and structural variations, such as membrane protein, cholesterol, or lipid raft distribution.
Using eq 1 to fit deformation profiles in the range 0–30%, we found that Young’s modulus for typical MLC-SV40 cells was in the range of 1.7–4.9 MPA for the membrane and 13–26 kPa for the cytoskeleton. NIH 3T3 cells have a 1.5–5.9 MPa range for Em and 6–14.5 kPa for Ec; therefore, the difference in their force profiles can be attributed mostly to the strength of their cytoskeleton. The results discussed above revealed the high sensitivity of the single cell compression method to the molecular to microscopic level structural changes of cells, such as the cross-linking of proteins within the cytoskeleton, membrane- to-cytoskeleton attachment, membrane elasticity and permeability, and adhesion. The reproducibility in cell compression experiments demonstrates that the AFM-based single cell compression technique with independent high-resolution optical monitoring allows unprecedented accuracy in cell positioning, mechanical perturbation, and force measurements. A simple model which assumes that a cell’s mechanical resistance arises from cytoskeleton compression and global plasma membrane stretching may be utilized to rationalize force–deformation measurements for small deformations, resulting in Young’s modulus of the cell membrane and cytoskeleton. We envision that such a simple and quantitative approach can be extended to investigate cell mechanics under a broad range of environments and conditions. Work is in progress to develop single cell compression into a general platform for cell mechanic and adhesion studies and accurate cell manipulations in cancer research and in various steps of tissue engineering.
V. Concluding Remarks
This article reveals three enabling aspects of AFM in cellular biophysical chemistry: (1) visualization of structural features with nanometer resolution on cellular membrane, such as microvilli, ridges, porosomes, lamellapodia, and filopodia; (2) revealing cellular singling pathways by time-dependent and high-resolution imaging of cellular membrane in correlation with intracellular components; and (3) qualitative and quantitative measurements of single cell mechanics by acquisition of force–deformation profiles and extraction of Young’s moduli for the membrane as well as the cytoskeleton. We find these three enabling aspects take best advantage of the current state-of-the-art AFM technology, which is well-positioned to truly reveal new insights for cellular biology research. In these regards, we hope to see more cell types and signaling pathways investigated using AFM. To fully realize further enabling aspects of AFM in cellular biology research, we envision further development and research in the following fields: (a) Advanced imaging and data analysis for high-resolution AFM imaging in vitro and under cellular culture conditions, 37 °C and 5% CO2. (b) High-speed imaging for time-dependent investigation of dynamic processes. Although high-speed AFM imaging instrumentation is emerging and advancing, the biocompatibility and spatial resolution for imaging biosystems is not up to the requirements for biostructural characterization. We anticipate active areas of research and development in this arena. (c) Multifunctional and multidimensional imaging of cellular structure and mechanics in conjunction with theory, such as finite element analysis.97 With the advances of AFM probes and imaging modes developed for biosystems, this objective can be attained in the imminent future.
Acknowledgment
We thank Mr. Alan Hicklin at U.C. Davis, Anthony Winson at Olympus America, and Dr. Deron Walters at Asylum Research for technical assistance in combined AFM and confocal microscopy. We are also grateful to Dr. Huan- Yuan Chen, Ms. Tiffany Zink, and Ms. I-chun Weng for collaborations on the preparation of mast and T cells in investigations published previously. This work is supported by NIH (R21 GM077850-01), NSF(CHE-0809977, DMR-0723118), and the University of California, Davis.
Biographies

Zhao Deng received his B.S. and M.S. degrees in Chemical Engineering from Shanghai Jiao Tong University, Shanghai, China, in 1999 and in 2002. His master’s thesis was about tribological studies of microelectromechanical system devices. In 2004, he began his Ph.D. study in analytical chemistry at the University of California, Davis and joined Professor Gang-yu Liu’s research group, where he worked extensively on bioimaging and nanolithography using atomic force microscopy. His Ph.D. dissertation focused on engineering antigen nanostructures to regulate the activation of mast cells, which involves the AFM-based nanografting technique and combined atomic force and confocal microscope developed in the Liu lab.

Valentin Lulevich received his B.S. and M.S. degrees from Moscow State University in 1998 and 2001, respectively, specializing in polymer physics and, in particular, in the mechanical properties of polymer films. His Ph.D. work was performed at Max Plank Institute for Polymer Research in Germany and focused on using precise force measurements to characterize electrostatic self-assembly (polyelectrolyte multilayer) capsules. During 2005–2008 he was a postdoctoral research fellow at the University of California, Davis, where he worked on establishing a new AFM approach to measure mechanical properties of living cells. He is currently a research associate at the University of California, Berkeley working on carbon nanotube-based membrane characterization.

Fu-Tong Liu received a B.S. degree in Chemistry from National Taiwan University and his Ph.D. degree in Chemistry from the University of Chicago. He performed postdoctoral work first in organic chemistry and then in immunology. He became a faculty member at the Scripps Research Institute and established an NIH-funded research laboratory. He obtained his M.D. degree from the two-year Ph.D. to M.D. program at the University of Miami. He was previously Head of the Allergy Research Section at the Scripps Research Institute, and Head of the Division of Allergy at La Jolla Institute for Allergy and Immunology. He has been Professor and Chair of the Department of Dermatology at the University of California, Davis since 2001. Dr. F. T. Liu has been instrumental in the discovery of the family of animal lectin galectins and is a leading investigator in the study of these family members. His current research is focused on the role of galectins in allergic inflammation, innate immunity, cancer progression, and wound healing. His research group also studies molecular and cellular mechanisms of allergic skin inflammation.

Gang-yu Liu received her B.S. from Peking University in China in 1985. She received her Ph.D. at Princeton University in 1992 studying helium scattering from thin film materials. Following a 2-year postdoctoral research appointment at Lawrence Berkeley National Laboratory under a Miller Research Fellowship, she joined Wayne State University in 1994 to investigate size-dependent chemical reactions using scanning probe nanolithography. She is currently Professor of Chemistry at the University of California, Davis. Her research interests include using micro- and nanostructures of designed functionalities to mimic extracellular matrixes and regulate cellular signaling processes. Advanced micro- and nanolithography methods have been developed for design and characterization of nanostructures. High resolution bioimaging is frequently used for monitoring surface and cellular structures and signaling processes.
References and Notes
- 1.Florian J. Nature. 1932;130:634. [Google Scholar]
- 2.Ruska E. Rev. Mod. Phys. 1987;59:627. [Google Scholar]
- 3.Anonymous. Lancet. 1945;248:792. [Google Scholar]
- 4.McIntosh JR. J. Cell Biol. 2001;153:F25. doi: 10.1083/jcb.153.6.f25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palade GE. J. Histochem. Cytochem. 1953;1:188. doi: 10.1177/1.4.188. [DOI] [PubMed] [Google Scholar]
- 6.Sabatini DD. Trends Cell Biol. 1999;9:413. doi: 10.1016/s0962-8924(99)01633-5. [DOI] [PubMed] [Google Scholar]
- 7.Palade GE. J. Exp. Med. 1952;95:285. doi: 10.1084/jem.95.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter KR. Anat. Rec. 1950;106:311. [Google Scholar]
- 9.Taylor KA. J. Microsc. (Oxford, U.K.) 1978;112:115. doi: 10.1111/j.1365-2818.1978.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 10.Binnig G, Quate CF, Gerber C. Phys. Rev. Lett. 1986;56:930. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 11.Drake B, Prater CB, Weisenhorn AL, Gould SAC, Albrecht TR, Quate CF, Cannell DS, Hansma HG, Hansma PK. Science. 1989;243:1586. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- 12.Ohnesorge F, Binnig G. Science. 1993;260:1451. doi: 10.1126/science.260.5113.1451. [DOI] [PubMed] [Google Scholar]
- 13.Muller DJ, Buldt G, Engel A. J. Mol. Biol. 1995;249:239. doi: 10.1006/jmbi.1995.0292. [DOI] [PubMed] [Google Scholar]
- 14.Butt HJ, Downing KH, Hansma PK. Biophys. J. 1990;58:1473. doi: 10.1016/S0006-3495(90)82492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radmacher M, Tillamnn RW, Fritz M, Gaub HE. Science. 1992;257:1900. doi: 10.1126/science.1411505. [DOI] [PubMed] [Google Scholar]
- 16.Haberle W, Horber JKH, Binnig G. Force Microscopy on Living Cells. 1991 [Google Scholar]
- 17.Hofmann UG, Rotsch C, Parak WJ, Radmacher M. J. Struct. Biol. 1997;119:84. doi: 10.1006/jsbi.1997.3868. [DOI] [PubMed] [Google Scholar]
- 18.Schneider SW, Sritharan KC, Geibel JP, Oberleithner H, Jena BP. Proc. Natl. Acad. Sci. U.S.A. 1997;94:316. doi: 10.1073/pnas.94.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jena BP. J. Cell. Mol. Med. 2004;8:1. doi: 10.1111/j.1582-4934.2004.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jena BP. Mol. Cells. 2008;26:517. [PubMed] [Google Scholar]
- 21.Fritz M, Radmacher M, Petersen N, Gaub HE. J. Vac. Sci. Technol. B. 1994;12:1526. [Google Scholar]
- 22.Schoenenberger CA, Hoh JH. Biophys. J. 1994;67:929. doi: 10.1016/S0006-3495(94)80556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotsch C, Braet F, Wisse E, Radmacher M. Cell Biol. Int. 1997;21:685. doi: 10.1006/cbir.1997.0213. [DOI] [PubMed] [Google Scholar]
- 24.Ebner A, Madl J, Kienberger F, Chtcheglova LA, Puntheeranurak T, Zhu R, Tang JL, Gruber HJ, Schutz GJ, Hinterdorfer P. Curr. Nanosci. 2007;3:49. [Google Scholar]
- 25.Dufrene YF, Hinterdorfer P. Pflugers Arch. 2008;456:237. doi: 10.1007/s00424-007-0413-1. [DOI] [PubMed] [Google Scholar]
- 26.Pfister G, Stroh CM, Perschinka H, Kind M, Knoflach M, Hinterdorfer P, Wick G. J. Cell Sci. 2005;118:1587. doi: 10.1242/jcs.02292. [DOI] [PubMed] [Google Scholar]
- 27.Burdon RH. Biochem. J. 1986;240:313. doi: 10.1042/bj2400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kailas L, Ratcliffe EC, Hayhurst EJ, Walker MG, Foster SJ, Hobbs JK. Ultramicroscopy. 2009;109:775. doi: 10.1016/j.ultramic.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Dufrene YF. Nat. Protoc. 2008;3:1132. doi: 10.1038/nprot.2008.101. [DOI] [PubMed] [Google Scholar]
- 30.Wadu-Mesthrige K, Amro NA, Garno JC, Cruchon-Dupeyrat S, Liu GY. Appl. Surf. Sci. 2001;175:391. [Google Scholar]
- 31.Moreno-Herrero F, Colchero J, Gomez-Herrero J, Baro AM. Phys. Rev. E. 2004;69 doi: 10.1103/PhysRevE.69.031915. [DOI] [PubMed] [Google Scholar]
- 32.Sun YX, Lin DY, Rui YF, Han D, Ma WY. J. Electron Microsc. 2006;55:165. [Google Scholar]
- 33.Adamsmair S, Ebner A, Hinterdorfer P, Zagar B, Kepler J. Tech. Mess. 2007;74:196. [Google Scholar]
- 34.Dupres V, Menozzi FD, Locht C, Clare BH, Abbott NL, Cuenot S, Bompard C, Raze D, Dufrene YF. Nat. Methods. 2005;2:515. doi: 10.1038/nmeth769. [DOI] [PubMed] [Google Scholar]
- 35.Puntheeranurak T, Wildling L, Gruber HJ, Kinne RKH, Hinterdorfer P. J. Cell Sci. 2006;119:2960. doi: 10.1242/jcs.03035. [DOI] [PubMed] [Google Scholar]
- 36.Muller DJ, Dufrene YF. Nat. Nanotechnol. 2008;3:261. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 37.Kada G, Kienberger F, Hinterdorfer P. Nano Today. 2008;3:12. [Google Scholar]
- 38.Dufrene YF. Nat. Rev. Microbiol. 2008;6:674. doi: 10.1038/nrmicro1948. [DOI] [PubMed] [Google Scholar]
- 39.Albrecht TR, Quate CF. J. Appl. Phys. 1987;62:2599. [Google Scholar]
- 40.Xu S, Liu GY. Langmuir. 1997;13:127. [Google Scholar]
- 41.Muller DJ, Schabert FA, Buldt G, Engel A. Biophys. J. 1995;68:1681. doi: 10.1016/S0006-3495(95)80345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansma PK, Cleveland JP, Radmacher M, Walters DA, Hillner PE, Bezanilla M, Fritz M, Vie D, Hansma HG, Prater CB, Massie J, Fukunaga L, Gurley J, Elings V. Appl. Phys. Lett. 1994;64:1738. [Google Scholar]
- 43.Zink T, Deng Z, Chen H, Yu L, Liu FT, Liu GY. Ultramicroscopy. 2008;109:22. doi: 10.1016/j.ultramic.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kassies R, Van der Werf KO, Lenferink A, Hunter CN, Olsen JD, Subramaniam V, Otto C. J. Microsc. (Oxford, U.K.) 2005;217:109. doi: 10.1111/j.0022-2720.2005.01428.x. [DOI] [PubMed] [Google Scholar]
- 45.Madl J, Rhode S, Stangl H, Stockinger H, Hinterdorfer P, Schutz GJ, Kada G. Ultramicroscopy. 2006;106:645. doi: 10.1016/j.ultramic.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Deng Z, Zink T, Chen HY, Walters D, Liu FT, Liu GY. Biophys. J. 2009;96:1629. doi: 10.1016/j.bpj.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horber JKH, Miles MJ. Science. 2003;302:1002. doi: 10.1126/science.1067410. [DOI] [PubMed] [Google Scholar]
- 48.Schneider SW. J. Membr. Biol. 2001;181:67. [PubMed] [Google Scholar]
- 49.Frankel DJ, Pfeiffer JR, Surviladze Z, Johnson AE, Oliver JM, Wilson BS, Burns AR. Biophys. J. 2006;90:2404. doi: 10.1529/biophysj.105.073692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeiffer JR, Seagrave JC, Davis BH, Deanin GG, Oliver JM. J. Cell Biol. 1985;101:2145. doi: 10.1083/jcb.101.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou X, Block M, Lofman C, Nilsson G. J. Allergy Clin. Immunol. 2001;108:116. doi: 10.1067/mai.2001.116124. [DOI] [PubMed] [Google Scholar]
- 52.Stossel TP. Science. 1993;260:1086. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 53.Fackler OT, Grosse R. J. Cell Biol. 2008;181:879. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jena BP. Curr. Opin. Struct. Biol. 2007;17:437. doi: 10.1016/j.sbi.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeremic A, Kelly M, Cho SJ, Stromer MH, Jena BP. Biophys. J. 2003;85:2035. doi: 10.1016/S0006-3495(03)74631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jena BP. Ultramicroscopy. 2006;106:663. doi: 10.1016/j.ultramic.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Brown DA. Physiology. 2006;21:430. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- 58.Magee AI, Parmryd I. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai EC. J. Cell Biol. 2003;162:365. doi: 10.1083/jcb.200307087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munro S. Cell. 2003;115:377. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 61.Giocondi MC, Vie V, Lesniewska E, Goudonnet JP, Le Grimellec C. J. Struct. Biol. 2000;131:38. doi: 10.1006/jsbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 62.Shevchuik AI, Frolenkov GI, Sanchez D, James PS, Freedman N, Lab MJ, Jones R, Klenerman D, Korchev YE. Angew. Chem. Int. Ed. 2006;45:2212. doi: 10.1002/anie.200503915. [DOI] [PubMed] [Google Scholar]
- 63.Proksch R, Lal R, Hansma PK, Morse D, Stucky G. Biophys. J. 1996;71:2155. doi: 10.1016/S0006-3495(96)79416-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansma PK, Drake B, Marti O, Gould SAC, Prater CB. Science. 1989;243:641. doi: 10.1126/science.2464851. [DOI] [PubMed] [Google Scholar]
- 65.Gilfillan AM, Tkaczyk C. Nat. Rev. Immunol. 2006;6:218. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- 66.Rivera J, Gilfillan AM. J. Allergy Clin. Immunol. 2006;117:1214. doi: 10.1016/j.jaci.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 67.Mousli M, Hugli TE, Landry Y, Bronner C. Immunopharmacology. 1994;27:1. doi: 10.1016/0162-3109(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 68.Berlin G. Agents Actions. 1984;15:482. doi: 10.1007/BF01966760. [DOI] [PubMed] [Google Scholar]
- 69.Lagunoff D, Martin TW, Read G. Annu. Rev. Pharmacol. Toxicol. 1983;23:331. doi: 10.1146/annurev.pa.23.040183.001555. [DOI] [PubMed] [Google Scholar]
- 70.Ortner MJ, Chignell CF. Biochem. Pharmacol. 1981;30:1587. doi: 10.1016/0006-2952(81)90385-3. [DOI] [PubMed] [Google Scholar]
- 71.Bueb JL, Dasilva A, Mousli M, Landry Y. Biochem. J. 1992;282:545. doi: 10.1042/bj2820545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishida K, Yamasaki S, Ito Y, Kabu K, Hattori K, Tezuka T, Nishizumi H, Kitamura D, Goitsuka R, Geha RS, Yamamoto T, Yagi T, Hirano T. J. Cell Biol. 2005;171:177. doi: 10.1083/jcb.200501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Proksch R. Appl. Phys. Lett. 2006;89 [Google Scholar]
- 74.Parish CR. Immunol. Cell Biol. 1999;77:499. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- 75.Albillos A, Dernick G, Horstmann H, Almers W, de Toledo GA, Lindau M. Nature. 1997;389:509. doi: 10.1038/39081. [DOI] [PubMed] [Google Scholar]
- 76.Williams RM, Webb WW. J. Cell Sci. 2000;113:3839. doi: 10.1242/jcs.113.21.3839. [DOI] [PubMed] [Google Scholar]
- 77.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Cell. 2004;118:363. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 78.Tobacman LS, Korn ED. J. Biol. Chem. 1983;258:3207. [PubMed] [Google Scholar]
- 79.Dvorak AM. J. Allergy Clin. Immunol. 1994;94:1103. doi: 10.1016/0091-6749(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 80.Sackmann E, Duwe HP, Engelhardt H. Faraday Discuss. 1986;81:281. doi: 10.1039/dc9868100281. [DOI] [PubMed] [Google Scholar]
- 81.Friedrichs J, Taubenberger A, Franz CM, Muller DJ. J. Mol. Biol. 2007;372:594. doi: 10.1016/j.jmb.2007.06.078. [DOI] [PubMed] [Google Scholar]
- 82.Lulevich V, Zink T, Chen HY, Liu FT, Liu GY. Langmuir. 2006;22:8151. doi: 10.1021/la060561p. [DOI] [PubMed] [Google Scholar]
- 83.Bowen WR, Hilal N, Lovitt RW, Wright CJ. Colloids Surf. A. 1999;157:117. [Google Scholar]
- 84.Lulevich V, Yang HY, Isseroff RR, Liu GY. Ultramicroscopy. 2009 submitted. [Google Scholar]
- 85.Landau LD, Lifshits EM. Theory of Elasticity. 3rd ed. New York: Oxford, Pergamon Press; 1965. [Google Scholar]
- 86.Mofrad MR, Kamm RD, editors. Cytoskeletal Mechanics. Cambridge University Press; 2006. [Google Scholar]
- 87.Caille N, Thoumine O, Tardy Y, Meister JJ. J. Biomech. 2002;35:177. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 88.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Nature. 2005;435:365. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karp G. Cell and Molecular Boilogy. New York: John Willey & Sons Inc; 2003. [Google Scholar]
- 90.Lulevich V, Shih YP, Lo SH, Liu GY. J. Phys. Chem. B. 2009;113:6511. doi: 10.1021/jp8103358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peeters EAG, Oomens CWJ, Bouten CVC, Bader DL, Baaijens FPT. J. Biomech. 2005;38:1685. doi: 10.1016/j.jbiomech.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 92.Charras GT, Hu CK, Coughlin M, Mitchison TJ. J. Cell Biol. 2006;175:477. doi: 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lulevich V, Andrienko D, Vinogradova OI. J. Chem. Phys. 2004;120:3822. doi: 10.1063/1.1644104. [DOI] [PubMed] [Google Scholar]
- 94.Ratanabanangkoon P, Gropper M, Merkel R, Sackmann E, Gast AP. Langmuir. 2003;19:1054. [Google Scholar]
- 95.Caille N, Thoumine O, Tardy Y, Meister JJ. J. Biomech. 2002;35:177. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 96.Baker A, Zidek L, Wiesler D, Chmelik J, Pagel M, Novotny MV. Chem. Res. Toxicol. 1998;11:730. doi: 10.1021/tx970167e. [DOI] [PubMed] [Google Scholar]
- 97.Charras GT, Lehenkari PP, Horton MA. “Atomic force microscopy can be used to mechanically stimulate osteoblasts and evaluate cellular strain distributions”. doi: 10.1016/s0304-3991(00)00076-0. [DOI] [PubMed] [Google Scholar]