SUMMARY
Exercise can improve cognitive function and has been linked to the increased expression of brain-derived neurotrophic factor (BDNF). However, the underlying molecular mechanisms driving the elevation of this neurotrophin remain unknown. Here we show that FNDC5, a previously identified muscle protein that is induced in exercise and is cleaved and secreted as irisin, is also elevated by endurance exercise in the hippocampus of mice. Neuronal Fndc5 gene expression is regulated by PGC-1α and Pgc1a−/− mice show reduced Fndc5 expression in the brain. Forced expression of FNDC5 in primary cortical neurons increases Bdnf expression, whereas RNAi-mediated knockdown of FNDC5 reduces Bdnf. Importantly, peripheral delivery of FNDC5 to the liver via adenoviral vectors, resulting in elevated blood irisin, induces expression of Bdnf and other neuroprotective genes in the hippocampus. Taken together, our findings link endurance exercise and the important metabolic mediators, PGC-1α and FNDC5, with BDNF expression in the brain.
Keywords: Exercise, FNDC5, Pgc1a, Bdnf, transcription, hippocampus
INTRODUCTION
Exercise, especially endurance exercise, is known to have beneficial effects on brain health and cognitive function (Cotman et al., 2007; Mattson, 2012a). This improvement in cognitive function with exercise has been most prominently observed in the aging population (Colcombe and Kramer, 2003). Exercise has also been reported to ameliorate outcomes in neurological diseases like depression, epilepsy, stroke, Alzheimer's and Parkinson's Disease, (Ahlskog, 2011; Arida et al., 2008; Buchman et al., 2012; Russo-Neustadt et al., 1999; Zhang et al., 2012). The effects of exercise on the brain are most apparent in the hippocampus and its dentate gyrus, a part of the brain involved in learning and memory. Specific beneficial effects of exercise in the brain have been reported to include increases in the size of and blood flow to the hippocampus in humans and morphological changes in dendrites and dendritic spines, increased synapse plasticity and, importantly, de novo neurogenesis in the dentate gyrus in various mouse models of exercise (Cotman et al., 2007; Mattson, 2012a). De novo neurogenesis in the adult brain occurs is observed in only two areas; the dentate gyrus of the hippocampus is one of them and exercise is one of the few known stimuli of this de novo neurogenesis (Kobilo et al., 2011).
One important molecular mediator for these beneficial responses in the brain to exercise is the induction of neurotrophins/growth factors, most notably brain-derived neurotrophic factor (BDNF). In animal models, BDNF is induced in various regions of the brain with exercise, most robustly in the hippocampus (Cotman et al., 2007). BDNF promotes many aspects of brain development including neuronal cell survival, differentiation, migration, dendritic arborization, synaptogenesis and plasticity (Greenberg et al., 2009; Park and Poo, 2013). In addition, BDNF is essential for synaptic plasticity, hippocampal function and learning (Kuipers and Bramham, 2006). Highlighting the relevance of BDNF in human, individuals carrying the Val66Met mutation in the BDNF gene, exhibit decreased secretion of BDNF, display a decreased volume of specific brain regions, deficits in episodic memory function as well as increased anxiety and depression (Egan et al., 2003; Hariri et al., 2003). Blocking BDNF signaling with anti-TrkB antibodies attenuates the exercise-induced improvement of in acquisition and retention a spatial learning task, as well as the exercise-induced expression of synaptic proteins (Vaynman et al., 2004; Vaynman et al., 2006). However, the underlying mechanism which induces BDNF in exercise remains to be determined.
PGC-1α is induced in skeletal muscle with exercise, and is a major mediator of the beneficial effects of exercise in this tissue (Finck and Kelly, 2006). PGC-1α was initially discovered as a transcriptional co-activator of mitochondrial biogenesis and oxidative metabolism in brown fat (Puigserver et al., 1998; Spiegelman, 2007). Subsequent work has demonstrated an important role of PGC-1α in the brain. Lack of PGC-1α in the brain is associated with neurodegeneration (Lin et al., 2004; Ma et al., 2010) as well as GABAergic dysfunction and a deficiency in neuronal parvalbumin expression (Lucas et al., 2010). PGC-1α has been shown to be neuroprotective in the MPTP mouse model of Parkinson's disease (St-Pierre et al., 2006). It also negatively regulates extrasynaptic NMDA (N-methyl-D-aspartate) receptor activity and thereby reduces excitotoxicity in rat cortical neurons (Puddifoot et al., 2012). In addition, the involvement of PGC-1 α in the formation and maintenance of neuronal dendritic spines has been reported (Cheng et al., 2012). Interestingly, long-term forced treadmill running over 12 weeks increases Pgc1a expression in various areas of the brain (Steiner et al., 2011).
Recently, our group identified a PGC-1α-dependent myokine, FNDC5, that is cleaved and secreted from muscle during exercise and induces some major metabolic benefits of exercise (Bostrom et al., 2012). FNDC5 is a glycosylated type I membrane protein and is released into the circulation after proteolytic cleavage. The secreted form of FNDC5 contains 112 amino acids and has been named irisin. Irisin acts preferentially on the subcutaneous ‘beige’ fat and causes it to ‘brown’ by increasing the expression of UCP-1 and other thermogenic genes (Bostrom et al., 2012; Wu et al., 2012). Clinical studies in humans have confirmed this positive correlation between increased FNDC5 expression and circulating irisin with the level of exercise performance (Huh et al., 2012; Lecker et al., 2012).
Interestingly, FNDC5 is also expressed in the brain (Dun et al., 2013; Ferrer-Martinez et al., 2002; Teufel et al., 2002) and in rat pheochromocytoma-derived PC12 cells differentiated into neuron-like cells (Ostadsharif et al., 2011). Knockdown of FNDC5 in neuronal precursors impaired the development into mature neurons, suggesting a developmental role of FNDC5 in neurons (Hashemi et al., 2013). This interesting connection of FNDC5 as an important exercise-related factor in the periphery and its expression in the central nervous system, led us to investigate the effects of exercise on FNDC5 expression and function in the brain. Here we show that FNDC5 is elevated by endurance exercise in the hippocampus of mice and that PGC-1α and FNDC5 regulate BDNF expression in the brain.
RESULTS
Endurance exercise induces hippocampal Fndc5 gene expression
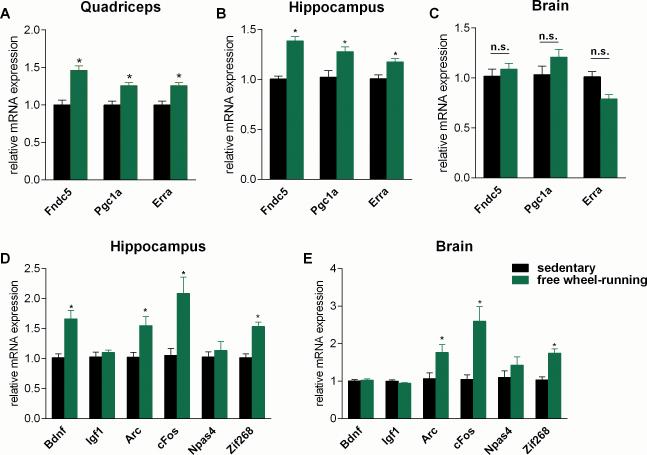
FNDC5 is highly expressed in the brain, as well as in skeletal muscle (Ferrer-Martinez et al., 2002; Teufel et al., 2002). Very little is known about the function of FNDC5 in the brain. We have therefore investigated the effects of exercise on FNDC5 expression and function. We used an established endurance exercise regimen: 30 days of voluntary free running-wheel exercise. This regimen is known to induce BDNF expression, neurogenesis, dendritic spines and improved memory function in mice (Eadie et al., 2005; Kobilo et al., 2011). As has previously been established, this training was sufficient to induce muscle Fndc5 gene expression (Fig. 1A), as well as the transcriptional regulators Pgc1a and Erra, known mediators of the exercise-response in skeletal muscle. In addition, other known genes of the exercise gene program were induced, confirming an adaptive endurance exercise response in the muscle (Fig. S1). Interestingly, the same exercise regime led to a significant elevation of Fndc5 expression in the hippocampus (Fig. 1B) but not in the remainder of the brain (Fig. 1C). The hippocampus is a region of the brain involved in learning and memory and has been identified as a major site where changes induced by exercise occur. Of note, even though genes that are induced by neuronal activity, such as Arc, cFos and Zif268, were upregulated in both the remainder of the brain and the hippocampus, the important exercise-related neurotrophin Bdnf was induced only in the hippocampus (Fig. 1D and E). However, Npas4, an important transcriptional component in hippocampal function and a key regulator of activity-induced Bdnf expression (Lin et al., 2008; Ramamoorthi et al., 2011) was not increased in the exercise regimen used here (Fig. 1D and E).These data suggest that the induction of FNDC5 is part of the transcriptional response to exercise in the hippocampus.
Figure 1. Endurance exercise induces hippocampal Fndc5 gene expression.
(A-E) Male six week old C57/Bl6 wild type mice were individually housed in cages with access to a running-wheel (free wheel-running) or without (sedentary). Mice were exercised for 30 days and sacrificed approximately 10 h after their last bout of exercise. Data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to sedentary control group. See also Figure S1.
Fndc5 gene expression correlates with Pgc1a expression levels in various tissues and developmental stages
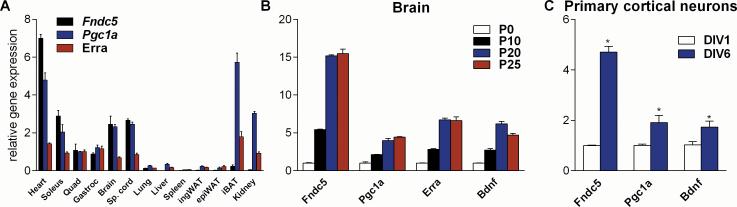
We previously reported that elevations in Fndc5 gene expression in exercised muscle was dependent on PGC-1α (Bostrom et al., 2012). We therefore investigated whether Fndc5 expression in the brain is also regulated by PGC-1α. To first assess if there is a correlation between the gene expression of these two proteins, we isolated 13 different tissues from C57/Bl6 mice, extracted total RNA and measured gene expression for Fndc5 and Pgc1a. Consistent with earlier reports, the highest level of Fndc5 gene expression was detected in heart, skeletal muscle, brain and spinal cord (Ferrer-Martinez et al., 2002; Teufel et al., 2002). When we grouped the different tissues according to their levels of Fndc5 expression, it is clear that most tissues with very high Fndc5 expression also showed relatively high levels of Pgc1a gene expression (Fig. 2A). Notably, Fndc5 and Pgc1a expression levels correlated well, even within very distinct muscle beds. Fndc5 expression was higher in oxidative muscle, such as the soleus muscle, which also contains higher levels of Pgc1a, than in glycolytic or mixed muscles, such as gastrocnemius or quadriceps muscle. Exceptions to this tight correlation of Fndc5 and Pgc1a expression are the interscapular brown adipose tissue and the kidney. Both are tissues with extremely high mitochondrial content, which might explain their requirement for high Pgc1a levels without very high expression of Fndc5.
Figure 2. Fndc5 gene expression correlates with Pgc1a expression levels in various tissues and developmental stages.
(A) The indicated tissues were harvested from male 13 week old C57/Bl6 wild type mice. Quad = quadriceps muscle, Gastroc = gastrocnemius muscle, Sp. Cord = spinal cord, ingWAT = inguinal white adipose tissue, epiWAT = epididymal white adipose tissue, iBAT = interscapular brown adipose tissue.
(B) Brains were harvested from C57/Bl6 wild type mice at the indicated postnatal (P) time points.
(C) Primary cortical neurons were isolated from C57/Bl6 wild type E17 embryos and cultured for the indicated days in vitro. mRNA was prepared and gene expression was assessed by qPCR. All data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to control group.
To examine whether FNDC5 and PGC-1α were developmentally regulated in synchrony during maturation of the brain, we perform a time-course experiment of postnatal development. Brains were harvested from pups at postnatal day 0 (P0), P10, P20, P25, and P30 and gene expression was measured by qPCR. These time-points were chosen because they cover an important time period of postnatal brain developmental, up to the mature state at P30. A two-step pattern of increased Fndc5 gene expression during development was observed, with a first increase between P0 and P10 and second increase between P10 and P20, which then leveled off (Fig. 2B). Pgc1a gene expression followed essentially the same pattern. Of note, we also observed this two-step pattern of increased gene expression during brain development for the key neural regulatory protein, Bdnf. Next, the gene expression patterns for these factors were assessed during the maturation of primary cortical neurons in culture. We observed again this correlation: Fndc5 gene expression increased between in vitro days (DIV) 1 and DIV 6, when the expression levels of Pgc1a and Bdnf were also elevated (Fig. 2C). These data illustrate that, similar to muscle, there is a strong correlation between PGC-1α and FNDC5 gene expression in the brain.
Neuronal Fndc5 gene expression is regulated by PGC-1α
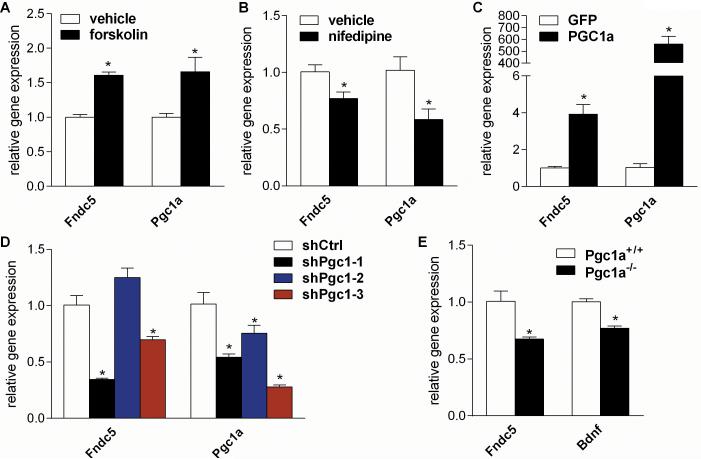
To investigate whether PGC-1α is a transcriptional regulator of Fndc5 gene expression in the brain, we turned to dissociated primary cortical neurons in culture. Although more heterogeneous than neurons from the dentate gyrus of the hippocampus, these cultures can be isolated in sufficient quantities for molecular studies and can be readily manipulated. Primary cortical neurons were stimulated with forskolin (10μM), a strong inducer of intracellular cAMP, which is known to increase Pgc1a gene expression in cell types as diverse as brown adipocytes, hepatocytes and Schwann cells (Cowell et al., 2008; Herzig et al., 2001; Yoon et al., 2001). This increase in Pgc1a gene expression was accompanied by a significant increase in Fndc5 gene expression (Fig. 3A). On the other hand, treatment of cortical neurons with nifedipine (5μM), a selective L-type calcium channel blocker, which leads to decreased intracellular calcium levels and decreased Pgc1a gene expression, was accompanied by decreased Fndc5 gene expression (Fig. 3B).
Figure 3. Neuronal Fndc5 gene expression is regulated by a PGC-1α.
(A) Primary cortical neurons at DIV 7 were treated with either forskolin (10 μM), a stimulator intracellular cAMP levels, or vehicle for overnight.
(B) Primary cortical neurons at DIV 7 were treated with nifedipine (5 μM), a L-type calcium channel blocker, or vehicle for overnight.
(C) Primary cortical neurons at DIV 7 were transduced with either PGC-1α or GFP adenovirus and harvested 48 hrs later.
(D) Primary cortical neurons at DIV 5 were transduced with lentivirus carrying the specified shRNA hairpins against Pgc1a or luciferase (Luc) as control and were harvested four days later.
(E) Cortices were harvested from either male five months old Pgc1a KO (Pgc1a−/−) or wild type mice (Pgc1a+/+). mRNA was prepared and gene expression was assessed by qPCR. All data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to corresponding control group. See also Figure S2.
Next, genetic gain- and loss-of-function approaches were used to test causality. Forced expression of PGC-1α by adenoviral delivery in primary cortical neurons resulted in a 4-fold increased Fndc5 gene expression (Fig. 3C). Immunoblotting confirmed that the increase in Fndc5 mRNA translated into elevated FNDC5 protein levels (Fig. S2). Conversely, reducing Pgc1a gene expression with lentiviral-mediated shRNA knockdown by more than 40 % significantly decreased Fndc5 gene expression by 66 % and 31%, respectively (Fig. 3D). As an additional loss-of-function model, the brains of global Pgc1a knockout mice (Pgc1a−/−) were used. We observed the same requirement of PGC-1α for Fndc5 gene expression in brains of these mice, which display a reduction in Fndc5 gene expression by 32 % (Fig. 3E). Taking together, these results demonstrate that PGC-1α is a regulator of neuronal Fndc5 gene expression in neural cultures and in the brain.
ERRα is a key interacting transcription factor with PGC-1α for regulating Fndc5 gene expression in neurons
PGC-1α is a transcriptional co-activator, meaning it does not bind to the DNA itself but interacts with transcription factors to execute its effects on gene expression (Spiegelman, 2007). The orphan nuclear receptor estrogen-related receptor alpha (ERRα; also known as NR3B1) is a central metabolic regulator (Giguere et al., 1988; Luo et al., 2003) and a very important interactor with PGC-1α (Laganiere et al., 2004; Mootha et al., 2004; Schreiber et al., 2004). The interaction of Errα with PGC-1α has been best studied in skeletal muscle, where it is required for mitochondrial biogenesis, induction of angiogenesis, oxidative metabolism, and oxidative muscle fibers (Arany et al., 2008; Mootha et al., 2004; Schreiber et al., 2004).
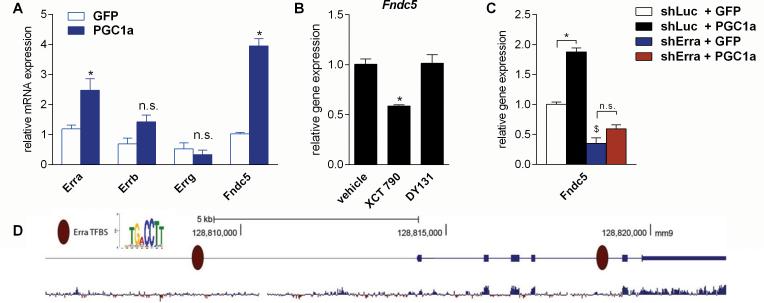
Interestingly, Erra follows the exercise-induced gene expression pattern of Fndc5 in the brain. Erra is up-regulated in the hippocampus upon exercise but not in the rest of the brain (Fig. 1B and C). In addition, there was a correlation between Fndc5 and Erra gene expression in our tissue-panel (Fig. 2A) as well as our developmental time-course (Fig.2B). PGC-1α is well-known to often increase the expression of transcription factors that it interacts with, thereby positively regulating its own regulators (Handschin et al., 2003; Mootha et al., 2004). We therefore asked if forced expression of PGC-1α in primary cortical neurons results in an increase Erra mRNA. Indeed, adenoviral expression of PGC-1α significantly increased Erra gene expression, but not Errb or Errg gene expression (Fig. 4A). However, mRNA for other common binding partners of PGC-1α, such as Mef2, Ppara, Nrf1 or Gabpa/b was not induced in these experiments (Fig. S3A).
Figure 4. ERRα is a key interacting transcription factor with PGC-1α for regulating Fndc5 gene expression in neurons.
(A) Primary cortical neurons at DIV 7 were transduced with either PGC-1α or GFP adenovirus and harvested 48 hrs later. *P < 0.05 compared to control group.
(B) Primary cortical neurons at DIV 7 were treated with either XCT 790 (1 μM), a selective inverse ERRα agonist, DY131 (1 μM), a selective ERRβ and ERRγ agonist, or vehicle for overnight. *P < 0.05 compared to vehicle only group.
(C) Primary cortical neurons at DIV 4 were transduced with lentivirus carrying shRNA hairpins against either Erra or luciferase (Luc) as control. Three days later were cells were transduced with either PGC-1α or GFP adenovirus and harvested 48 hrs later. mRNA was prepared and gene expression was assessed by qPCR. Data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to corresponding shLuc expressing control group. $P < 0.05 compared to corresponding GFP expressing control group. Data (A-C) are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM.
(D) Analysis of the murine Fndc5 promoter for putative ERREs. The murine Fndc5 gene and 6 kb of its upstream promoter were searched for the canonical ERRE: TGA CCTT. Genomic coordinates are given according to the assembly mm9 from the UCSC Genome Browser. The bottom diagram indicates the degree of mammalian conservation across the genomic locus. The presented motif was modified from www.factorbook.org (Wang et al., 2012). See also Figure S3.
The murine Fndc5 gene and 6 kb of its upstream promoter were searched for putative ERRα transcription factor binding sites, (ERRE), with the canonical ‘TGACCTT’ sequence (Charest-Marcotte et al., 2010; Mootha et al., 2004; Wang et al., 2012). We identified two putative ERRE's: one around 5.3 kb upstream of the transcriptional start site and one in the fourth intron of the Fndc5 gene (Fig. 4B). ERRα had been previously reported to also bind to intronic sequences to exert its biological function (Arany et al., 2008). This further suggests that ERRα could be important in FNDC5 gene regulation. Treatment of primary cortical neurons with XCT 790 (1 μM), a selective ERRα inhibitor (inverse agonist), which disrupts the ERRα/PGC-1α transcriptional complex (Mootha et al., 2004), significantly reduced Fndc5 gene expression compared to vehicle treated cells (Fig. 4C). However, stimulation with DY131 (1 μM), a selective ERRβ and ERRγ agonist, had no effect on Fndc5 gene expression. This suggests certain specificity for the involvement of ERRα compared to other ERR subfamily members. Since the nuclear receptor PPARα, another common binding partner of PGC-1α, was slightly induced by forced expression of PGC-1α, we tested the effect of GW7647, a potent and highly selective PPARα agonist, and GW0742, a potent and highly selective PPARδ agonist on Fndc5 gene expression. However, under the conditions tested, no effect on Fndc5 gene expression in primary cortical neurons by these compounds was observed (Fig. S3B).
The results from the treatment with ERRα antagonist suggest that interaction of the PGC-1α with ERRα is required for the PGC-1α-dependent induction of Fndc5 gene expression. To test this, we first knocked-down ERRα in primary cortical neurons using lentivirally expressed shRNA hairpins and then three days later transduced with the cells with either the PGC-1α adenovirus or GFP expressing adenovirus. Erra mRNA was efficiently knocked-down by this hairpin (70%) and forced expression of PGC-1 α did not affect the efficiency of the knock-down (Fig. S2C). Knockdown of ERRα significantly reduced Fndc5 gene expression at base line (Fig. 4D). Furthermore, forced expression of PGC-1α by adenovirus in the cells with reduced ERRα failed to significantly increase Fndc5 gene expression (Fig. 4D). However, this failure to increase Fndc5 gene expression was not due to a lack over expression of PGC-1α in the shErra treated neurons (Fig. S3C). Together these data suggest an involvement of ERRα in the induction of FNDC5 by PGC-1α. The precise role of the individual ERRα binding sites in the Fndc5 gene remains to be determined.
FNDC5 regulates Bdnf gene expression in a cell-autonomous manner and recombinant BDNF decreases Fndc5 gene expression as part of potential feedback loop
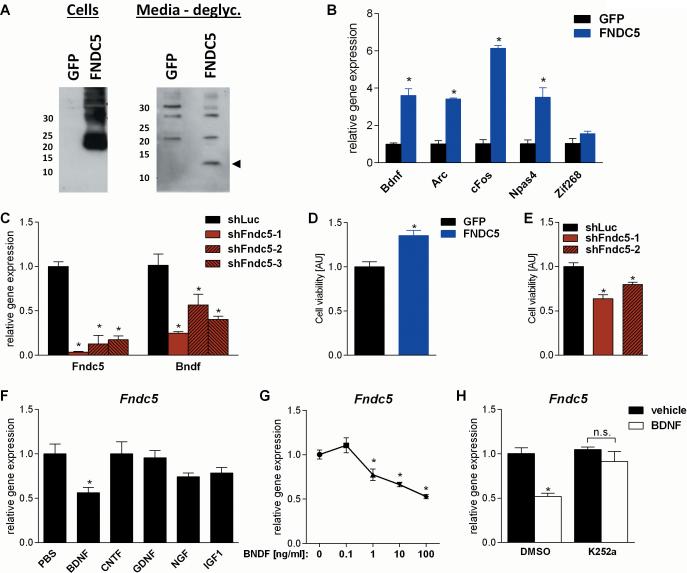
As mentioned earlier, BDNF is a major mediator of certain beneficial effects on the brain. In addition, an increase in the Bdnf gene expression in the hippocampus was observed, where Fndc5 gene expression was also induced (Fig. 1B and D), but not in the rest of the brain, where Fndc5 was not induced (Fig. 1C and E). We therefore tested whether FNDC5 could be a regulator of Bdnf gene expression in a cell culture model. Primary cortical neurons were transduced with either FNDC5 adenovirus or a GFP adenovirus as control. Forced expression of FNDC5 resulted in a clear increase in FNDC5 protein in the whole cell lysate, as well as an increase in the secreted form of FNDC5 (irisin) in the cell culture supernatant (Fig. 5A). After deglycosylation, this protein has the same apparent molecular mass (12kDa) as predicted for irisin (Fig. 5A). In addition, forced expression of FNDC5 significantly upregulated Bdnf gene expression by four fold (Fig.5B). Importantly, FNDC5 expression also induced other important activity-induced genes involved in hippocampal function including Npas4, cFos, andArc; Zif268 however, was only slightly elevated.
Figure 5. FNDC5 regulates Bdnf gene expression in a cell-autonomous manner and recombinant BDNF decreases Fndc5 gene expression as part of negative feedback loop.
(A) Primary cortical neurons at DIV 6 were transduced with either FNDC5 or GFP adenovirus. Whole cell lysates and conditioned media were harvested and analyzed by immunoblotting. Intensity of unspecific bands and Ponceau staining were used to assess equal loading. deglyc. = deglycosylation.
(B) Primary cortical neurons at DIV 7 were transduced with either FNDC5 or GFP adenovirus. Forty-eight hours later mRNA was prepared and gene expression was assessed by qPCR.
(C) Primary cortical neurons at DIV 5 were transduced with lentivirus carrying the specified shRNA hairpins against Fndc5 or luciferase (Luc) as control. Four days later mRNA was prepared and gene expression was assessed by qPCR.
(D) Primary cortical neurons at DIV 7 were transduced with either FNDC5 or GFP adenovirus. Cell viability was assessed three days later using the CellTiter-Glo® Luminescent Cell Viability Assay. AU = arbitrary unit.
(E) Primary cortical neurons at DIV 5 were transduced with lentivirus carrying the specified shRNA hairpins against Fndc5 or luciferase (Luc) as control. Cell viability was assessed three days later using the CellTiter-Glo® Luminescent Cell Viability Assay. AU = arbitrary unit.
(F) Primary cortical neurons at DIV 7 were stimulated with the indicated recombinant neurotrophins and growth factors (100ng/ml) for overnight. mRNA was prepared and gene expression was assessed by qPCR.
(G) Primary cortical neurons at DIV 7 were stimulated with human recombinant BDNF at the indicated concentrations or vehicle for overnight. mRNA was prepared and gene expression was assessed by qPCR.
(H) Primary cortical neurons at DIV 6 were treated either with the TrkB inhibitor K252a (50nM) or vehicle. Twenty-four hours later human recombinant BDNF (100ng/ml) or vehicle was added for overnight stimulation. mRNA was prepared and gene expression was assessed by qPCR. Data (B, C and F- H) are shown as mRNA levels relative to Rsp18 expression. All data are expressed as mean ± SEM. *P < 0.05 compared to corresponding control group.
To investigate if FNDC5 is required for Bdnf gene expression, lentivirally delivered shRNA was used to knockdown FNDC5 in primary cortical neurons. To address possible off-targets of a single hairpin, we tested a total of five hairpins of which three significantly knocked down Fndc5 mRNA (Fig. 5C). The same three hairpins also significantly reduced Bdnf gene expression. We also assessed the role of PGC-1α in controlling Bdnf gene expression in vivo. To do this, we used the brains of global Pgc1a knockout mice (Pgc1a−/−). As shown in Fig 3E, Bdnf gene expression was significantly reduced in the brains of Pgc1a−/− mice (Fig. 3E).
BDNF is well-known for its ability to improve survival of neurons in culture. We therefore assessed the effects of gain- and loss-of-function of FNDC5 on cell viability of cultured neurons using a luminescence/ATP-based assay. Gain-of-function of FNDC5 significantly improved neuron survival in culture (Fig. 5D) while loss-of-function of FNDC5 using shRNA mediated knockdown of FNDC5 with two different hairpins significantly impaired the survival of neurons in culture (Fig. 5E).
To examine how BDNF might, in turn, alter FNDC5 gene expression, primary cortical neurons were stimulated with recombinant BDNF overnight at various concentrations at physiological and pharmacological dosages (0.1-100 ng/ml). BDNF concentrations as low as 1 ng/ml significantly reduced Fndc5 gene expression (Fig. 5F) and a dose-response was observed. To ask whether the reduction in Fndc5 gene expression was specific to BDNF, we treated primary cortical neurons with a variety of central and peripheral neurotrophic factors in addition to BDNF, such as CNTF (ciliary neurotrophic factor), GDNF (glial cell-derived neurotrophic factor), NGF (nerve growth factor), and IGF-1 (insulin-like growth factor 1) at 100ng/ml for overnight. However, only BDNF stimulation significantly reduced Fndc5 mRNA expression Fig. 5G). This effect was abolished by pre-incubating the cortical neurons with a low dose (50nM) of K252a, well-characterized inhibitor of TrkB, the receptor of BDNF signaling (Gimenez-Cassina et al., 2012; Tapley et al., 1992) (Fig. 5H). In summary, these data suggest a homeostatic FNDC5/BDNF feed-back loop.
Peripheral delivery of FNDC5 by adenoviral vectors increases Bdnf expression in the hippocampus
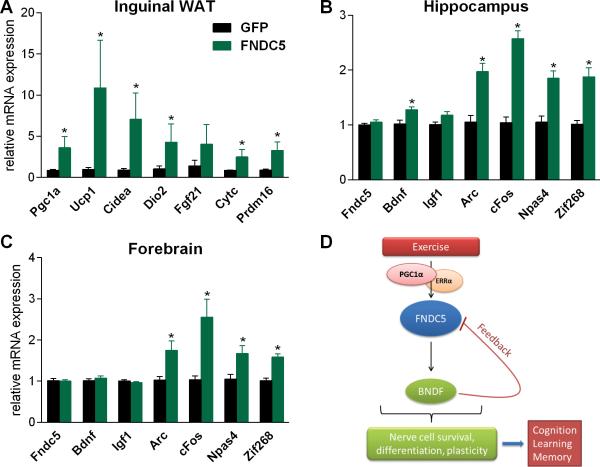
We had previously shown that adenoviral overexpression of FNDC5 in the liver, a major secretory organ, increases circulating levels of irisin, the secreted form of FNDC5 (Bostrom et al., 2012). This resulted in the activation of a thermogenic gene program in certain fat tissues. To determine if peripheral delivery of FNDC5/irisin could elevate central BDNF levels, we repeated this experiment and measured Bdnf gene expression in the hippocampus seven days later. As previously shown forced expression of FNDC5 in the liver in the induced ‘browning’ of the inguinal fat depot (Fig. 6A), including increased expression of mRNA for a group of key thermogenic genes, such as Pgc1a, Ucp1 and Cidea. In addition, plasma levels of irisin were elevated in mice overexpressing FNDC5 as compared to GFP-overexpressing control mice (Fig. S4A). Interestingly, Bdnf expression in the hippocampus was significantly increased, as was expression of Npas4, cFos, Arc, and Zif268, all part of the activity-induced immediate early gene (IEG) program as mentioned before. Importantly, this was not caused by any viral mediated expression of Fndc5 in the brain or hippocampus. (Fig. 6B), strongly suggesting that the secreted form of the peripherally expressed FNDC5 was responsible for this effect. This effect of increased Bdnf expression was specific to the hippocampus it was not observed in the forebrain (Fig. 6C) whereas the IEG response was observed in both; this is consistent with our earlier findings of the exercise effects (Fig. 1D and E).
Figure 6. Peripheral delivery of FNDC5 by adenoviral vectors increases Bdnf expression in the hippocampus.
(A-C) Five week old male wild-type BALB/c mice were injected with GFP- or FNDC5-expressing adenoviral particles intravenously. Animals were sacrificed seven days later and (A) inguinal/subcutaneous fat pads (WAT=white adipose tissues), (B) hippocampus, and forebrain (C) were collected and mRNA was prepared and gene expression was assessed by qPCR. Data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to wild type control group. (D) Model of the hippocampal PGC-1α/FNDC5/BDNF pathway in exercise. Endurance exercise stimulates increases hippocampal Fndc5 gene expression through a PGC-1α/Errα transcriptional complex. This elevated Fndc5 gene expression stimulates in turn Bdnf gene expression. BDNF is the master regulator of nerve cell survival, differentiation and plasticity in the brain. This will lead to improved cognitive function, learning and memory, which are known beneficial effects of exercise on the brain. See also Figure S4.
PGC-1α/FNDC5/BDNF pathway in primary hippocampal neurons
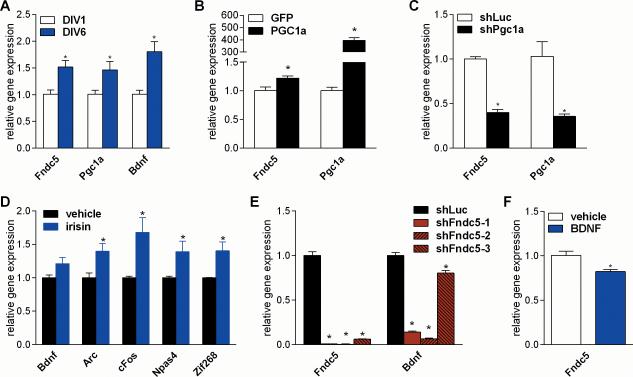
We used cortical neurons in the experiments above because this is the most widely used system of primary CNS cultures and because reasonable numbers of cells can be obtained. However, since some of our observations in vivo were made in the hippocampus, we sought to validate our findings in primary hippocampal neurons. Therefore, a key set of experiments were repeated in primary hippocampal neuron cultures. We confirmed that Fndc5 gene expression is significantly increased in primary hippocampal neurons cultured in vitro from DIV 1 to DIV6; the expression of Pgc1a and Bdnf mRNA is similarly increased (Fig. 7A). To test whether PGC-1α regulates Fndc5 gene expression in hippocampal neurons, gain- and loss of function studies were performed. Forced expression of PGC-1α significantly induced Fndc5 gene expression (Fig. 7B). Interestingly, stimulation with forskolin (10μM) failed to induce Pgc1a gene expression, but decreased the expression of Erra and Fndc5 (Fig. S5). Efficient knockdown of Pgc1a by lentivirally delivered shRNA significantly reduced Fndc5 gene expression (Fig. 7C). Stimulation of primary hippocampal neurons with commercially available recombinant irisin induced a similar gene program (Arc, cFos, Npas4, and Zif268) as was found in the in vivo adenoviral experiments (Fig. 7D). However, the increase in Bdnf gene expression did not reach statistical significance. Loss-of-function of FNDC5 by shRNA mediated knockdown with three different hairpins against Fndc5 significantly reduced Bdnf gene expression in hippocampal neurons (Fig. 7E). In addition, treatment of hippocampal neurons with recombinant BDNF reduced Fndc5 gene expression (Fig. 7F). Together, these data demonstrate that the basic observations made in the primary cortical neurons also apply to primary hippocampal neuron cultures.
Figure 7. PGC-1α/FNDC5/BDNF pathway in primary hippocampal neurons.
(A) Primary hippocampal neurons were isolated from C57/Bl6 wild type E17 embryos and cultured for the indicated days in vitro.
(B) Primary hippocampal neurons at DIV 7 were transduced with either PGC-1α or GFP adenovirus and harvested 48 hrs later.
(C) Primary hippocampal neurons at DIV 5 were transduced with lentivirus carrying the specified shRNA hairpin against Pgc1a or luciferase (Luc) as control and harvested four days later.
(D) Primary hippocampal neurons were stimulated with recombinant irisin (1ug/ml) at DIV 5 and 6 and harvested 24 hrs later
(E) Primary hippocampal neurons at DIV 5 were transduced with lentivirus carrying the specified shRNA hairpins against Fndc5 or luciferase (Luc) as control and harvested four days later.
(F) Primary hippocampal neurons at DIV 7 were stimulated with recombinant BDNF (100ng/ml) for overnight. mRNA was prepared and gene expression was assessed by qPCR. Data are shown as mRNA levels relative to Rsp18 expression, expressed as mean ± SEM. *P < 0.05 compared to corresponding control group. See also Figure S5.
DISCUSSION
A recent study has reported a positive correlation between human brain size and endurance exercise capacity suggesting a co evolution between human cognition and locomotion (Raichlen and Gordon, 2011). More complex tasks require a more complex brain and foraging in wide and open spaces in the savannas put high demands on spatial orientation, as well as the ability to acquire and retain new information. Therefore individuals with a more complex brain who performed better at these tasked might have had an evolutionary advantage. On the other hand, since endurance exercise clearly increases expression of BDNF in the brain, improvements in the exercise capacity might have positively enforced brain growth (Mattson, 2012b), especially in the hippocampus.
In this study we report a PGC-1α/FNDC5/BDNF pathway that is activated in the hippocampus by endurance exercise (Fig. 6). In our current model, exercise leads to increased transcription of Pgc1a and Erra. It has been observed previously that PGC-1α often induces the expression of transcription factors to which it binds and co-activates (Handschin et al., 2003; Mootha et al., 2004). Indeed, the ability of PGC-1α to induce FNDC5 gene expression depends on ERRα availability (Fig. 4D). This PGC-1 α / Errα complex, in turn, may bind to one or more of the canonical ERRE's found in or near the Fndc5 gene, thus activating Fndc5 gene expression. As shown in a cell culture model in Fig. 5A, FNDC5 is a positive regulator of BDNF expression. Based on this, it seems likely that the increased Fndc5 gene expression in exercise will lead to increased BDNF levels. Interestingly, BDNF also can signal to reduce the expression of FNDC5 as part of an apparent homeostatic loop. Both FNDC5-dependent and FNDC5-independent pathways by which exercise induces BDNF expression seem plausible. For example, CREB and NF-kB are two other transcription factors known to induce BDNF expression in exercise (Mattson, 2012b). These may act upstream or downstream of FNDC5, or in an independent pathway.
The induction of FNDC5 by exercise in the hippocampus is quantitatively comparable to the induction observed in skeletal muscle. Interestingly, it is also in the same quantitative range as the induction of BDNF, a neurotrophic mediator of exercise in the brain, as well as cFos, Arc, and Zif268, important indicators for the activity state of neurons (Hunt et al., 1987; Lyford et al., 1995; Rusak et al., 1990; Saffen et al., 1988). This places FNDC5 induction in a similar range to other known important regulators in the brain.
In our study of 30 days of free-wheel running exercise, Fndc5 and Pgc1a was induced in the hippocampus but not in the rest of the brain (Fig 1B) when taken as one unit. Therefore it is possible that Fndc5 and Pgc1a were induced in relatively small numbers of neurons elsewhere, but that that change was not detectable because it is occurring in the background of little or no change in larger brain structures. Indeed, using a longer and more intense exercise regimen exercise protocol and more detailed dissections, Steiner et al. reported an upregulation of Pgc1a expression in various other parts of the brain, in addition to the hippocampus (Steiner et al., 2011).
A central question arising from our study is how does the PGC-1α/FNDC5/BDNF pathway get initiated in exercise? This question is closely linked to the more central and open question in the field: how is exercise sensed by the brain? One obvious initiator could be increased neuronal activity in areas of the brain that are involved in spatial orientation, learning and memory, since BDNF gene expression is well known to be stimulated by neural activity (West and Greenberg, 2011). Increased sympathetic tone, namely higher norepinephrine levels (Garcia et al., 2003) and increased IGF-1 levels from periphery crossing the blood-brain-barrier have also been discussed as exercise-related inducers of BDNF (Ding et al., 2006). However, because exercise is known to change the metabolic state of the whole body, another important factor could be changes in the energy state or oxygen levels within the brain, both signals to which PGC-1α gene expression is known to respond in other tissues (Arany et al., 2008; St-Pierre et al., 2006). In our study we linked the activation of a metabolic regulator, PGC-1α, via FNDC5 to increased BDNF levels in the neurons in response to exercise (Fig. 6). Of note, there are other important metabolic regulators, such as AMPK or PPARgamma, which have not been part of this study.
FNDC5 in the periphery is cleaved and secreted as irisin and secreted irisin can cause the ‘browning’ of adipose tissues (Bostrom et al., 2012; Shan et al., 2013; Wu et al., 2012). Therefore several important questions arise from our studies. First, is FNDC5 functioning mainly as a membrane-bound molecule in the brain or is it secreted by neurons? Secondly, if FNDC5 is secreted, is it secreted as irisin (amino acids 29-140) or as a different peptide species? Perhaps the most exciting result overall is that peripheral delivery of FNDC5 with adenoviral vectors is sufficient to induce central expression of Bdnf and others genes with potential neuroprotective functions or those involved in learning and memory. This suggests that a secreted, circulating form of FNDC5 has these effects on these neurons and that it crosses the blood brain barrier. Whether this is the full-length irisin protein or a further modified form remains to be determined. The therapeutic implications of this are obvious since it suggests that a polypeptide might be developed as a drug capable of giving neuroprotection in disease states or improved cognition in aging populations.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant human BDNF was purchased from PeproTech, recombinant human GDNF and CNTF and forskolin from Sigma, and recombinant mouse IGF-1 was obtained from R&D Systems. Recombinant mouse NGF and K252a were obtained from EMD Millipore. Nifedipine, XCT 790, DY131, GW7647, and GW0742 were purchased from Tocris. Recombinant irisin (human, rat, mouse, canine) was obtained from Phoenix Pharmaceuticals (Burlingame, CA).
Primers used for qPCR
All primers used are listed with their sequences in Supplemental Table 1.
Animal studies
All animal experiments were performed according to procedures approved by the IACUC of Dana-Farber Cancer Institute and the BIDMC. Pgc1a−/− mice have been described previously (Lin et al., 2004). Mice were housed and exercised as previously described (Bostrom et al., 2012).
Cell culture
Primary cortical and hippocampal neurons were isolated as described previously (Bartlett and Banker, 1984).
RNA and protein preparation and analysis
RNA and protein analyses were performed as described previously (Bostrom et al., 2012).
Forced expression and knockdown of target genes
Generation and delivery of the PGC-1α, GFP, and FNDC5 adenovirus has been describes before (Bostrom et al., 2012; Lustig et al., 2011). For knockdown studies, primary cortical neurons were transduced with lentiviral viral supernatants
Cell viability assay
Cell viability of cultured neurons was assessed using CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI) according to the manufacturer's instructions
Analysis of the murine Fndc5 promoter for Erra transcription factor binding sites
The genomic sequence of the murine Fndc5 gene and 6kb of its upstream promoter was retrieved from the USCS Genome browser (www.genome.ucsc.edu; assembly mm9). This genomic sequence was searched for the canonical Erra transcription factor binding motif: TGACCTT. This motif had been identified and established in previous studies (Charest-Marcotte et al., 2010; Mootha et al., 2004; Wang et al., 2012).
Peripheral delivery of FNDC5 by adenoviral vectors
High titer GFP- or FNDC5-expressing adenoviral particles were obtained by ViraQuest Inc. (North Liberty, IA). Five week old male wild-type BALB/c mice were injected with GFP- or FNDC5-expressing adenoviral particles (1011/animal) intravenously. Animals were sacrificed seven days later and the indicated tissues were harvested for gene expression analyses using qPCR.
Supplementary Material
HIGHLIGHTS.
Exercise induces FNDC5 in the hippocampus
PGC-1α regulates neuronal Fndc5 gene expression in vitro and in vivo
FNDC5 positively regulates the expression of the important neurotrophin BDNF
Peripheral delivery of FNDC5 via adenoviral vectors induces Bdnf in the hippocampus
ACKNOWLEGMENTS
We are grateful to Alfredo Cassina-Gimenez (Dana-Farber Cancer Institute) and Alan R. Mardinly (Harvard Medical School) for helpful experimental advice and useful discussions. We thank Pere Puigserver for the plasmids for the lentiviral hairpins against Ppargc1a. C.D.W. was supported by a fellowship from the German Research Foundation (DFG, WR 157/3-1). This work is funded by the JPB Foundation and NIH grants (DK31405 and DK90861) to B.M.S. B.M.S. is a shareholder and consultant to Ember Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahlskog JE. Does vigorous exercise have a neuroprotective effect in Parkinson disease? Neurology. 2011;77:288–294. doi: 10.1212/WNL.0b013e318225ab66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- Arida RM, Cavalheiro EA, da Silva AC, Scorza FA. Physical activity and epilepsy: proven and predicted benefits. Sports medicine (Auckland, NZ) 2008;38:607–615. doi: 10.2165/00007256-200838070-00006. [DOI] [PubMed] [Google Scholar]
- Bartlett WP, Banker GA. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. I. Cells which develop without intercellular contacts. J Neurosci. 1984;4:1944–1953. doi: 10.1523/JNEUROSCI.04-08-01944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012 doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AS, Boyle PA, Yu L, Shah RC, Wilson RS, Bennett DA. Total daily physical activity and the risk of AD and cognitive decline in older adults. Neurology. 2012;78:1323–1329. doi: 10.1212/WNL.0b013e3182535d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, Mootha VK, Giguere V. The homeobox protein Prox1 is a negative modulator of ERR{alpha}/PGC-1{alpha} bioenergetic functions. Genes Dev. 2010;24:537–542. doi: 10.1101/gad.1871610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Wan R, Yang JL, Kamimura N, Son TG, Ouyang X, Luo Y, Okun E, Mattson MP. Involvement of PGC-1alpha in the formation and maintenance of neuronal dendritic spines. Nature communications. 2012;3:1250. doi: 10.1038/ncomms2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombe S, Kramer AF. Fitness effects on the cognitive function of older adults: a meta-analytic study. Psychological science. 2003;14:125–130. doi: 10.1111/1467-9280.t01-1-01430. [DOI] [PubMed] [Google Scholar]
- Cotman CW, Berchtold NC, Christie LA. Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends in neurosciences. 2007;30:464–472. doi: 10.1016/j.tins.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Cowell RM, Blake KR, Inoue T, Russell JW. Regulation of PGC-1alpha and PGC-1alpha-responsive genes with forskolin-induced Schwann cell differentiation. Neurosci Lett. 2008;439:269–274. doi: 10.1016/j.neulet.2008.04.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Vaynman S, Akhavan M, Ying Z, Gomez-Pinilla F. Insulin-like growth factor I interfaces with brain-derived neurotrophic factor-mediated synaptic plasticity to modulate aspects of exercise-induced cognitive function. Neuroscience. 2006;140:823–833. doi: 10.1016/j.neuroscience.2006.02.084. [DOI] [PubMed] [Google Scholar]
- Dun SL, Lyu RM, Chen YH, Chang JK, Luo JJ, Dun NJ. Irisin immunoreactivity in neural and non-neural cells of the rodent. Neuroscience. 2013 doi: 10.1016/j.neuroscience.2013.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie BD, Redila VA, Christie BR. Voluntary exercise alters the cytoarchitecture of the adult dentate gyrus by increasing cellular proliferation, dendritic complexity, and spine density. The Journal of comparative neurology. 2005;486:39–47. doi: 10.1002/cne.20493. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Ferrer-Martinez A, Ruiz-Lozano P, Chien KR. Mouse PeP: a novel peroxisomal protein linked to myoblast differentiation and development. Developmental dynamics : an official publication of the American Association of Anatomists. 2002;224:154–167. doi: 10.1002/dvdy.10099. [DOI] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C, Chen MJ, Garza AA, Cotman CW, Russo-Neustadt A. The influence of specific noradrenergic and serotonergic lesions on the expression of hippocampal brain-derived neurotrophic factor transcripts following voluntary physical activity. Neuroscience. 2003;119:721–732. doi: 10.1016/s0306-4522(03)00192-1. [DOI] [PubMed] [Google Scholar]
- Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- Gimenez-Cassina A, Lim F, Diaz-Nido J. Chronic inhibition of glycogen synthase kinase-3 protects against rotenone-induced cell death in human neuron-like cells by increasing BDNF secretion. Neurosci Lett. 2012;531:182–187. doi: 10.1016/j.neulet.2012.10.046. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Xu B, Lu B, Hempstead BL. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29:12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi MS, Ghaedi K, Salamian A, Karbalaie K, Emadi-Baygi M, Tanhaei S, Nasr-Esfahani MH, Baharvand H. Fndc5 knockdown significantly decreased neural differentiation rate of mouse embryonic stem cells. Neuroscience. 2013;231:296–304. doi: 10.1016/j.neuroscience.2012.11.041. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Huh JY, Panagiotou G, Mougios V, Brinkoetter M, Vamvini MT, Schneider BE, Mantzoros CS. FNDC5 and irisin in humans: I. Predictors of circulating concentrations in serum and plasma and II. mRNA expression and circulating concentrations in response to weight loss and exercise. Metabolism. 2012;61:1725–1738. doi: 10.1016/j.metabol.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328:632–634. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- Kobilo T, Liu QR, Gandhi K, Mughal M, Shaham Y, van Praag H. Running is the neurogenic and neurotrophic stimulus in environmental enrichment. Learning & memory (Cold Spring Harbor, NY) 2011;18:605–609. doi: 10.1101/lm.2283011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers SD, Bramham CR. Brain-derived neurotrophic factor mechanisms and function in adult synaptic plasticity: new insights and implications for therapy. Current opinion in drug discovery & development. 2006;9:580–586. [PubMed] [Google Scholar]
- Laganiere J, Tremblay GB, Dufour CR, Giroux S, Rousseau F, Giguere V. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor alpha (ERRalpha) promoter dictates peroxisome proliferator-activated receptor gamma coactivator-1alpha control of ERRalpha expression. J Biol Chem. 2004;279:18504–18510. doi: 10.1074/jbc.M313543200. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Zavin A, Cao P, Arena R, Allsup K, Daniels KM, Joseph J, Schulze PC, Forman DE. Expression of the irisin precursor FNDC5 in skeletal muscle correlates with aerobic exercise performance in patients with heart failure. Circulation Heart failure. 2012;5:812–818. doi: 10.1161/CIRCHEARTFAILURE.112.969543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas EK, Markwardt SJ, Gupta S, Meador-Woodruff JH, Lin JD, Overstreet-Wadiche L, Cowell RM. Parvalbumin deficiency and GABAergic dysfunction in mice lacking PGC-1alpha. J Neurosci. 2010;30:7227–7235. doi: 10.1523/JNEUROSCI.0698-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol Cell Biol. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig Y, Ruas JL, Estall JL, Lo JC, Devarakonda S, Laznik D, Choi JH, Ono H, Olsen JV, Spiegelman BM. Separation of the gluconeogenic and mitochondrial functions of PGC-1{alpha} through S6 kinase. Genes Dev. 2011;25:1232–1244. doi: 10.1101/gad.2054711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Ma D, Li S, Lucas EK, Cowell RM, Lin JD. Neuronal inactivation of peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) protects mice from diet-induced obesity and leads to degenerative lesions. J Biol Chem. 2010;285:39087–39095. doi: 10.1074/jbc.M110.151688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Energy intake and exercise as determinants of brain health and vulnerability to injury and disease. Cell Metab. 2012a;16:706–722. doi: 10.1016/j.cmet.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Evolutionary aspects of human exercise--born to run purposefully. Ageing research reviews. 2012b;11:347–352. doi: 10.1016/j.arr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostadsharif M, Ghaedi K, Hossein Nasr-Esfahani M, Mojbafan M, Tanhaie S, Karbalaie K, Baharvand H. The expression of peroxisomal protein transcripts increased by retinoic acid during neural differentiation. Differentiation; research in biological diversity. 2011;81:127–132. doi: 10.1016/j.diff.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nature reviews Neuroscience. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- Puddifoot C, Martel MA, Soriano FX, Camacho A, Vidal-Puig A, Wyllie DJ, Hardingham GE. PGC-1alpha negatively regulates extrasynaptic NMDAR activity and excitotoxicity. J Neurosci. 2012;32:6995–7000. doi: 10.1523/JNEUROSCI.6407-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Raichlen DA, Gordon AD. Relationship between exercise capacity and brain size in mammals. PLoS ONE. 2011;6:e20601. doi: 10.1371/journal.pone.0020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, Lin Y. Npas4 Regulates a Transcriptional Program in CA3 Required for Contextual Memory Formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusak B, Robertson HA, Wisden W, Hunt SP. Light pulses that shift rhythms induce gene expression in the suprachiasmatic nucleus. Science. 1990;248:1237–1240. doi: 10.1126/science.2112267. [DOI] [PubMed] [Google Scholar]
- Russo-Neustadt A, Beard RC, Cotman CW. Exercise, antidepressant medications, and enhanced brain derived neurotrophic factor expression. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 1999;21:679–682. doi: 10.1016/S0893-133X(99)00059-7. [DOI] [PubMed] [Google Scholar]
- Saffen DW, Cole AJ, Worley PF, Christy BA, Ryder K, Baraban JM. Convulsant-induced increase in transcription factor messenger RNAs in rat brain. Proceedings of the National Academy of Sciences. 1988;85:7795–7799. doi: 10.1073/pnas.85.20.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T, Liang X, Bi P, Kuang S. Myostatin knockout drives browning of white adipose tissue through activating the AMPK-PGC1alpha-Fndc5 pathway in muscle. Faseb J. 2013 doi: 10.1096/fj.12-225755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Foundation symposium. 2007;287:60–63. discussion 63-69. [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Steiner JL, Murphy EA, McClellan JL, Carmichael MD, Davis JM. Exercise training increases mitochondrial biogenesis in the brain. J Appl Physiol. 2011;111:1066–1071. doi: 10.1152/japplphysiol.00343.2011. [DOI] [PubMed] [Google Scholar]
- Tapley P, Lamballe F, Barbacid M. K252a is a selective inhibitor of the tyrosine protein kinase activity of the trk family of oncogenes and neurotrophin receptors. Oncogene. 1992;7:371–381. [PubMed] [Google Scholar]
- Teufel A, Malik N, Mukhopadhyay M, Westphal H. Frcp1 and Frcp2, two novel fibronectin type III repeat containing genes. Gene. 2002;297:79–83. doi: 10.1016/s0378-1119(02)00828-4. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. The European journal of neuroscience. 2004;20:2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- Vaynman SS, Ying Z, Yin D, Gomez-Pinilla F. Exercise differentially regulates synaptic proteins associated to the function of BDNF. Brain Res. 2006;1070:124–130. doi: 10.1016/j.brainres.2005.11.062. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wu Y, Zhang P, Sha H, Jia J, Hu Y, Zhu J. Exercise induces mitochondrial biogenesis after brain ischemia in rats. Neuroscience. 2012;205:10–17. doi: 10.1016/j.neuroscience.2011.12.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.