Abstract
Nucleosome remodelling enzymes of the ISWI family reposition nucleosomes in eukaryotes. ISWI contains an ATPase and a HAND-SANT-SLIDE (HSS) domain. Conformational changes between these domains have been proposed to be critical for nucleosome repositioning by pulling flanking DNA into the nucleosome. We inserted flexible linkers at strategic sites in ISWI to disrupt this putative power stroke and assess its functional importance by quantitative biochemical assays. Notably, the flexible linkers did not disrupt catalysis. Instead of engaging in a power stroke, the HSS module might therefore assist DNA to ratchet into the nucleosome. Our results clarify the roles had by the domains and suggest that the HSS domain evolved to optimize a rudimentary remodelling engine.
Keywords: ISWI, chromatin remodelling, nucleosome sliding
Introduction
Nucleosomes are the basic packaging units of chromatin in eukaryotes. By binding tightly to ∼146 bp of DNA, they act as physical barriers for the cellular machinery that needs to access the underlying DNA, for example, during transcription, DNA replication and DNA repair. The cell must precisely control the genomic location of nucleosomes to allow for a regulated use of the genetic material in response to different environmental and developmental stimuli.
Mobilizing the nucleosomes is a challenge for the cell as they are inherently stable particles. Dozens of DNA-histone contacts must be broken to rearrange nucleosomes. The cell thus employs dedicated enzymes, so called ATP-dependent nucleosome remodelling factors, to shift the position of nucleosomes along DNA [1]. Remodelling factors of the ISWI and several other families can move nucleosomes along DNA in a process that is termed nucleosome sliding. Elucidating the molecular mechanisms of remodelling enzymes remains a pressing goal.
Early mechanistic clues came from the observation that all remodelling factors contain ATPase engines that are evolutionary related to DNA helicases [2]. Indeed, many remodellers can translocate on DNA much like helicases do [3–5]. However, unlike helicases, they do not separate the DNA strands. Remarkably, the ATPase domains of several remodellers localize to DNA well within the nucleosome, two helical turns away from the nucleosomal dyad, suggesting that helicase-like translocation of DNA takes place inside the nucleosome [5–9].
DNA translocation within the nucleosome begs the question how DNA enters the nucleosome in the first place. For remodelling by ISWI enzymes, it has been proposed that a conformational change mechanically pulls flanking DNA into the nucleosome [10–13]. The energy required for this conformational change would come from hydrolysis of ATP. A step that uses chemical energy to perform mechanical work is often called a power stroke, a terminology that we adopt herein. The carboxy-terminal DNA-binding domain (DBD) of ISWI, which comprises the HAND, SANT and SLIDE (HSS) domains, would be intimately involved in such a power stroke, as it binds to the DNA that flanks the nucleosome [14]. Notably, recent models propose that the power stroke takes place only after the first 7 bp of DNA have been extruded already from the nucleosome’s exit site through the translocase activity of the ATPase domain. The size of the proposed power stroke has been measured to be ≤3 bp [11].
Other data appear to be in conflict but can be reconciled with the power stroke model. We and others have shown that ISWI can remodel nucleosomes even if the HSS module is missing [15, 16]. Similarly, the C-terminal DBD of Chd1, composed of a related SANT-SLIDE module [17], is also not required for remodelling [18, 19]. Nevertheless, the remodelling activity of ISWI decreases an order of magnitude on deletion or mutation of the HSS module [10, 15]. This drop in activity could potentially be attributed to a missing power stroke in the deletion mutants.
Other scenarios, however, can also explain the drop in activity incurred by deletion of the HSS module without invoking a power stroke. As the HSS domain is the nucleosome recognition module [15, 20], the ATPase domain lacks sufficient specificity to dock productively to its binding site on the nucleosome. Lack of specificity can result in lower observed ATPase and remodelling activity. This problem becomes especially apparent with saturating concentrations of ATP [15]. In addition, the removal of the HSS module might allow a polypeptide motif at the C-terminal end of the ATPase domain known as ‘bridge’ or ‘NegC’ to inhibit the enzyme by holding the ATPase domain in a catalytically less active conformation [15, 16].
Here we explore whether a power stroke operating between the ATPase and HSS module constitutes an important part of the catalytic strategy of Drosophila ISWI. As rigidity in the force-transducing regions of the protein is necessary during a power stroke, one can test the functional relevance of the putative power stroke by artificially increasing the flexibility of these enzyme regions [21, 22]. To this end, we inserted glycine-serine rich linkers at several strategic locations in the protein. These linkers act like random coils with a high degree of flexibility [23, 24]. Surprisingly, ISWI enzymes with these artificial, flexible hinges showed no defect in ATPase, restriction enzyme accessibility-based remodelling and nucleosome sliding assays. These results strongly argue against the power stroke model. We instead conclude that the HSS module assumes a more passive role during catalysis in that it mainly increases the time the ATPase engine can productively engage with the proper binding site on the nucleosome. With regards to how DNA enters the nucleosome, we propose that DNA ratchets into the nucleosome once the tension that builds up by extruding base pairs (bp) from the exit site becomes too large.
Results
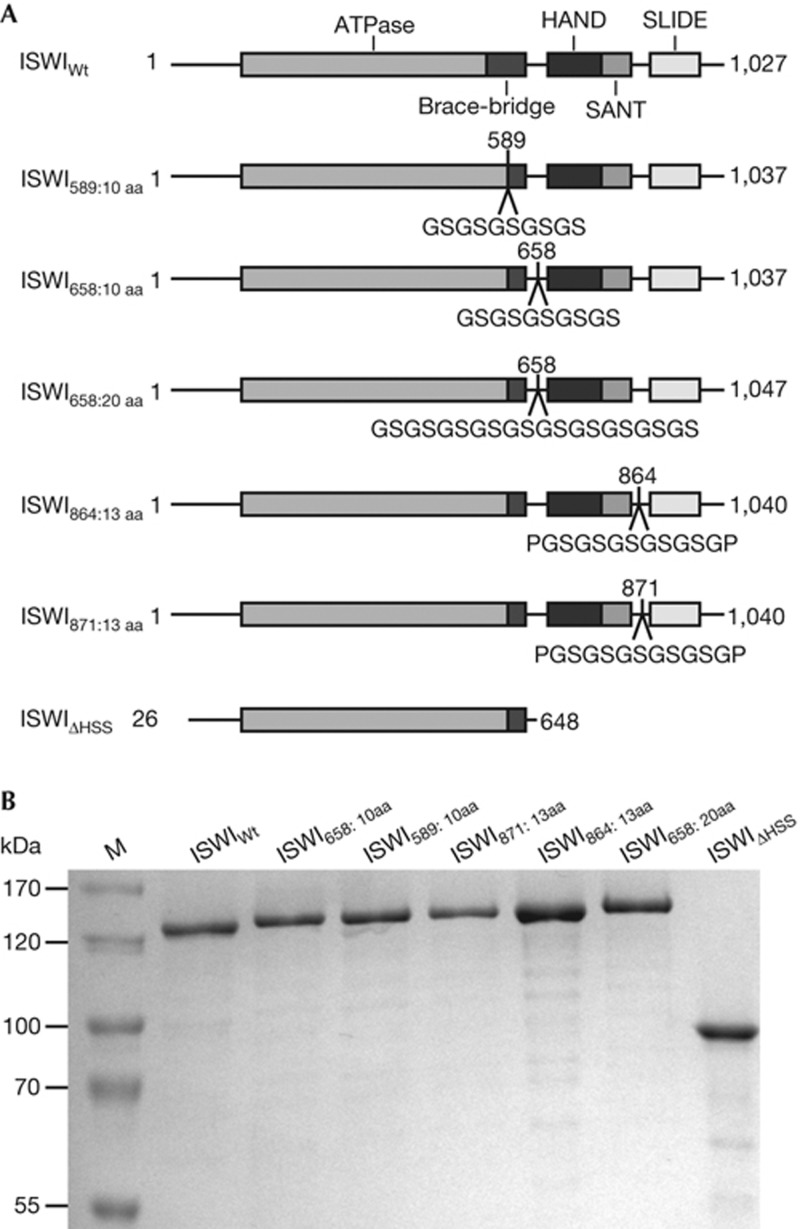
To probe for the importance of the putative ATP-dependent power stroke, we inserted glycine- and serine-rich flexible linkers [23, 24] into regions of ISWI that could conceivably transmit the force. The ‘brace’ and ‘bridge’ at the C-terminal end of the ATPase domain could be such elements, because they intimately contact both ATPase lobes and thus could directly react to the ATPase cycle [2, 15, 16, 19, 25]. The connection between the ATPase and HSS modules is another prime candidate, as the force generated by the ATPase domain must reach the HSS module. Force transmitted from the HAND-SANT to the SLIDE domain would have to go through the connecting spacer helix, as no tertiary contacts between SANT and SLIDE exist [20]. We chose altogether four insertion points (Fig 1A). Linker lengths varied between 10 and 20 amino acids. When fully extended, these linkers can reach ∼4–8 nm, a significant range considering the size of the proposed power stroke (≤3 bp, equivalent to ≤1 nm; [11]). All ISWI preparations (Fig 1B) were monodisperse as judged by size exclusion chromatography (supplementary Fig S1 online). The monodispersity attests to the overall structural integrity of the enzymes.
Figure 1.
ISWI derivatives used in this study. (A) Schematic representation of ISWI derivatives. Glycine-rich inserts were introduced behind the indicated amino-acid positions. Numbers in subscript refer to the insertion position and the size of the insert. ISWIΔHSS spans the amino acids 26–648 and lacks the C-terminal HSS domains. (B) Coomassie-stained SDS–PAGE gel showing the purified ISWI derivatives from A. aa, amino acid; HSS, HAND, SANT and SLIDE; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; WT, wild type.
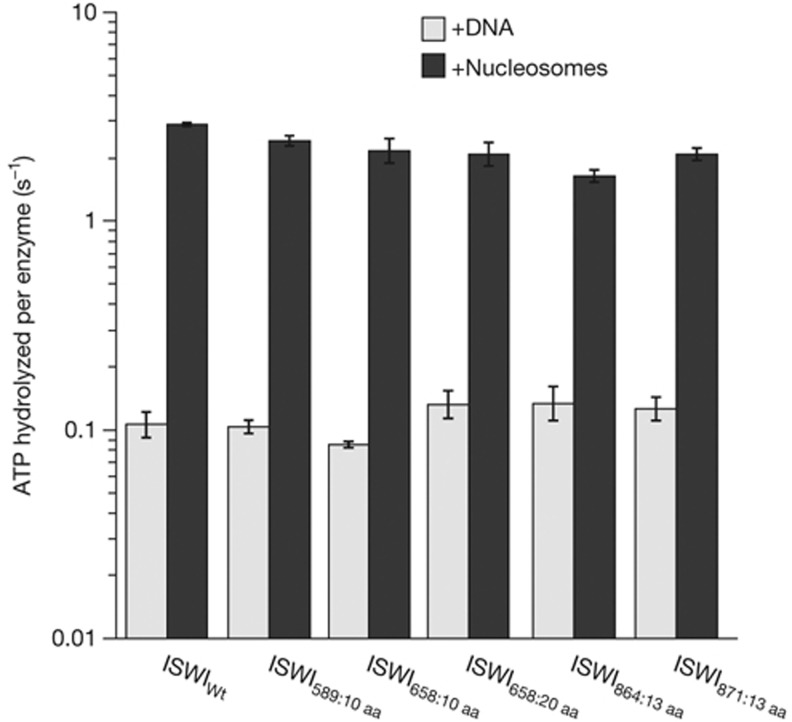
The HSS domain has been proposed to communicate to the ATPase domain and modulate its ATP hydrolysis. Mutations in the SLIDE domain, for instance, can allosterically affect ATP hydrolysis [10]. Moreover, nucleosomes no longer stimulate ATP hydrolysis better than naked DNA when the HSS domain is removed with saturating, although not with sub-saturating, concentrations of ATP [15]. We therefore tested if the ISWI derivatives that have a more flexible link between the ATPase and C-terminal domains could efficiently hydrolyse ATP. We used saturating ATP concentrations to measure ATP turnover, and in fact throughout this study, as defects in the function of the HSS domain become maximally apparent under these conditions [15].
DNA-stimulated ATPase rates of all mutants were indistinguishable from wild-type ISWI (ISWIWt), deviating no more than 1.3-fold (Fig 2). All mutants, just as the wild type, hydrolysed ATP an order of magnitude faster when bound to nucleosomes than to DNA. Importantly, absolute rates for the nucleosome stimulated reaction varied by no more than 1.8-fold between ISWIWt and all its derivatives. As ATPase rates were largely unaffected, we conclude that the artificial flexible joints did not disrupt the putative communication between the domains and suggest that force transduction is not necessary for efficient ATP hydrolysis. In addition, we conclude that all mutants were properly folded and recognized DNA and nucleosomes like their wild-type counterpart. Indeed, similar concentrations of DNA and nucleosomes saturated the wild type and insertion mutants. For comparison, an order of magnitude higher concentrations had to be used to saturate ISWI that completely lacked the HSS domain (ISWIΔHSS; [15] and data not shown).
Figure 2.
Insertion of flexible polypeptide linkers does not disrupt DNA- and nucleosome-stimulated ATP hydrolysis. ATPase rates were measured in the presence of saturating concentrations of ATP, DNA or nucleosomes. Errors are s.d. for ISWIWt (n=3) and minimal and maximal values of two independent measurements for all other enzyme derivatives. ATPase rates in the absence of DNA were <0.02 s−1 under otherwise identical conditions (data not shown). ISWIWt, wild-type ISWI.
The ATPase results do not favour but also do not rule out the power stroke hypothesis. For example, even though the insertion mutants efficiently hydrolysed ATP, a power stroke might be necessary to couple hydrolysis to remodelling. To test this scenario, we performed remodelling assays.
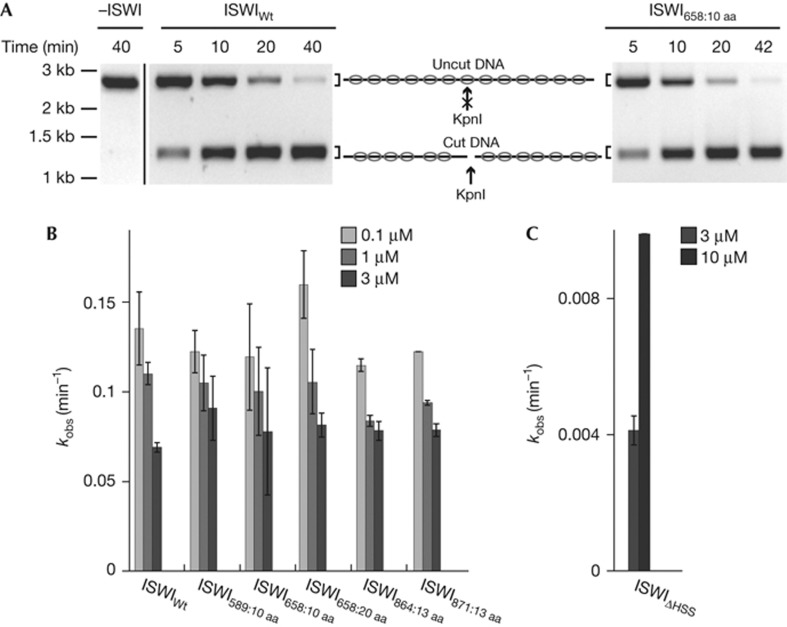
Remodelling leads to exposure of nucleosomal DNA to solvent and can be detected with restriction endonucleases that cut the exposed DNA. We used a quantitative assay that monitors exposure of a unique KpnI site that is occluded by the central nucleosome in a 13-mer nucleosomal array [15]. Rate constants for remodelling (kobs) were determined by measuring exposure of the KpnI site over time and fitting the data to single exponential functions (Fig 3A). Several remodeller concentrations were used to control for possible differences in binding affinities between the ISWI derivatives and the known property of full-length ISWI to inhibit its own catalysis at higher concentrations (Fig 3B) [15].
Figure 3.
Insertion of flexible polypeptide linkers does not disrupt nucleosome remodelling. (A) Remodelling activity was probed by following the accessibility of a unique, central KpnI restriction site in a 13-mer nucleosomal array (see schematic). Exemplary time courses for remodelling by ISWIWt and ISWI658:10 aa (both 3 μM). In mock-treated samples (−ISWI), the KpnI site was not accessible. The original unspliced gel picture containing the mock-treated sample and the ISWIWt time-course is provided as supplementary Fig S2 online. (B,C) The observed rate constants for remodelling, kobs, were determined for ISWI derivatives at the indicated concentrations by fitting time courses as in A to single exponential functions. Errors are s.d. for ISWI589:10 aa and ISWI658:10 aa (n=4 for 0.1 μM and 1 μM; n=3 for 3 μM) and 1 μM ISWIWt (n=3). In all other instances, minimal and maximal values of two independent measurements are shown. aa, amino acid; ISWIWt, wild-type ISWI; kb, kilobases.
Unexpectedly, none of the ISWI derivatives containing flexible linkers showed remodelling defects. Used at the same concentration, they all exposed the KpnI site as efficiently as ISWIWt, with kobs differing by no more than a factor of 1.3. For comparison, ISWIΔHSS exposed nucleosomal DNA an order of magnitude more slowly (Fig 3C), confirming previous results [15].
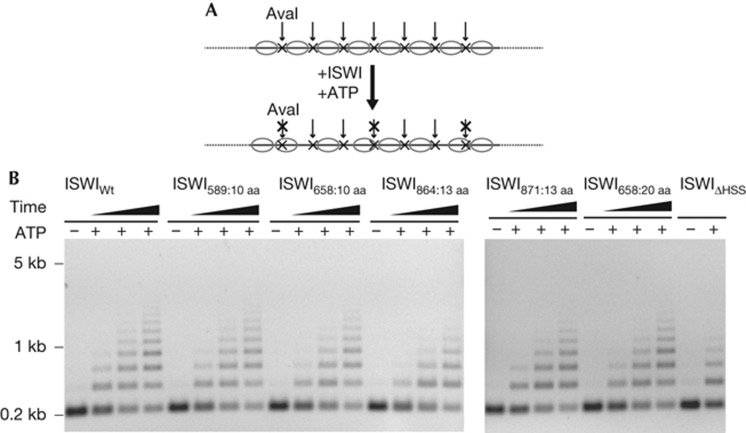
As exposed nucleosomal DNA might be an early intermediate during nucleosome sliding, it was important to test if formation of these intermediates was successfully coupled to nucleosome sliding. We monitored sliding in the context of nucleosomal arrays. Each linker DNA contained an exposed AvaI restriction site that became protected upon sliding (Fig 4A) [15].
Figure 4.
Insertion of flexible polypeptide linkers does not compromise nucleosome sliding. (A) Schematic depiction of the nucleosome sliding assay. 25-mer nucleosomal arrays containing exposed AvaI restriction sites in the linker DNA were used to follow nucleosome sliding. Accessibility to AvaI (arrows) changes upon remodelling. Nucleosomes (ovals) and AvaI sites (x) are indicated. (B) ISWIWt and the insertion mutants were incubated with nucleosomal arrays for 3, 13 and 48 min, whereas ISWIΔHSS was incubated for 6 h. Control reactions (−) were depleted of ATP with apyrase before addition of ISWI and incubated for 6 h (ISWIΔHSS) or 48 min (all other enzymes). ISWIWt, wild-type ISWI.
Surprisingly, but in accordance with the results shown above, all insertion mutants were able to slide nucleosomes over the AvaI sites (Fig 4B). In fact, time courses showed that ISWIWt and all insertion mutants moved nucleosomes with similar efficiency. As shown before [15], also ISWIΔHSS relocated nucleosomes, although higher concentrations and longer incubation times were necessary.
Discussion and Conclusions
According to recent mechanistic models, the ATPase engine of ISWI is bound to DNA well within the nucleosome and starts the remodelling process by translocating single bp of DNA in the direction of the exit side of the nucleosome. ATP hydrolysis is required for the transport of each bp. Only after the initial 7 bp of DNA have exited the nucleosome will fresh DNA enter from the opposite side of the nucleosome [10, 11]. How DNA enters the nucleosome is unclear.
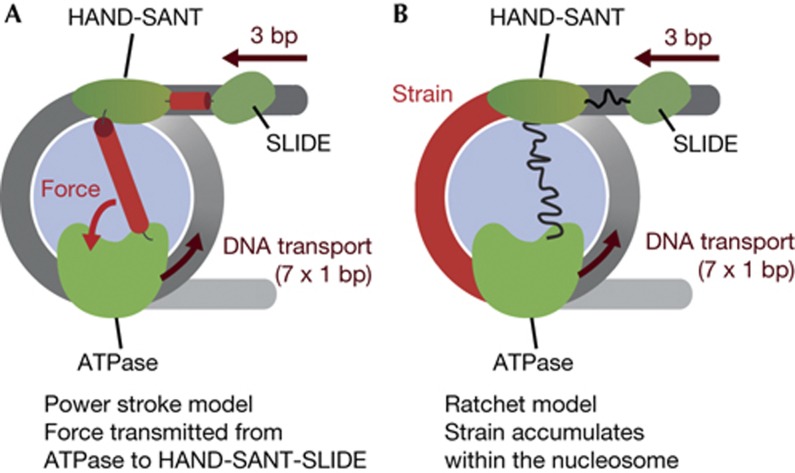
Prominent models favor a power stroke as a mechanism for how DNA enters the nucleosome [10–13]. At this stage of remodelling, hydrolysis of ATP does not fuel transport of DNA according to these models. Instead, ATP hydrolysis would be coupled to a conformational change between the HSS and ATPase modules. This conformational change exerts force onto the HSS domain and the DNA at the entry site bound by it. Three bp thereby enter the nucleosome (Fig 5A). Subsequently, the ATPase engine resumes transporting single bp toward the exit site [10, 11].
Figure 5.
Models for nucleosome remodelling by ISWI. (A) Power stroke model. First, the ATPase engine of ISWI translocates 7 bp of DNA. The ATPase and HSS domains then undergo a power stroke that exerts force on the HSS domains. The power stroke pulls 3 bp of flanking DNA into the nucleosome. Hypothetical force-transducing elements are coloured red. (B) Ratchet model. As in A, the ATPase engine translocates 7 bp. This translocation strains the structure of the nucleosome, in particular the DNA delimitated by the ATPase and HSS domains (red). When the strain on the nucleosome structure becomes too large, 3 bp of DNA ratchet into the nucleosome. No direct coordination between the HSS and ATPase domain is required for this mechanism and flexible linkage between individual domains (curved lines) does not affect remodelling. Histones are light blue; DNA is grey.
In striking opposition to predictions derived from the power stroke model, none of the glycine-rich flexible insertions caused any detectable catalytic defects. Apparently, ISWI can tolerate considerable flexibility between individual domains. Notably, the Bowman lab came to very similar conclusions in a recent study that focused on the related remodelling enzyme Chd1 [26]. We note that inherent flexibility in the remodellers might allow the DBD and ATPase domain of one enzyme molecule to simultaneously contact two neighbouring nucleosomes, a situation that has recently been suggested to be important for remodelling by ISWI enzymes [27].
We were particularly surprised that the 10–20 amino acid long insertions on either side of the brace–bridge polypeptide did not hamper catalysis, as this polypeptide makes intimate contacts with the ATPase domain and was proposed to regulate the enzyme [15, 16, 19, 25]. Depending on whether or not the structure of the brace and bridge is disrupted by the insertions, we can either conclude that this region might be of lesser importance for remodelling than previously hypothesized [16] or that build-up of force is not necessary for proper function of the brace–bridge polypeptide.
If not by a power stroke, how else can flanking DNA enter the nucleosome? We propose that the HSS and ATPase domains work independently of each other with no need for direct coordination during catalysis (Fig 5B). The HSS domain is an important recognition module for the nucleosome [15, 20] and is expected to anchor the enzyme to the nucleosome. Anchoring increases the chance for the ATPase engine to productively engage the nucleosome and start with the translocation of DNA. After the first seven translocation steps, the structure of the nucleosome becomes highly strained, particularly around the DNA delimited by the ATPase and HSS module, such that translocation stalls. Eventually, the HSS domain loses its grip on the DNA flanking the nucleosome, allowing 3 bp to ratchet in.
Although not part of a power stroke, the HSS module clearly evolved to carry out important functions that collectively optimize remodelling by an order of magnitude [10, 15]. Besides established functions such as anchoring the remodeller to the nucleosome and increasing the processivity [10, 15], we hypothesize that the HSS module improves catalysis by changing the structure of the nucleosome around the DNA entry site, perhaps by locally separating the DNA from the histone surface [13, 28]. Other remodelling subfamilies that do not interact with flanking DNA and therefore cannot engage in a power stroke in the first place might in fact use a similar mechanism [29], pointing to an unified remodelling strategy shared between several remodeller subfamilies.
Methods
Protein expression and purification. pPROEX-HTb-based expression plasmids with genes encoding Drosophila melanogaster ISWIWt and ISWIΔHSS were kindly provided by C. Müller (EMBL, Heidelberg, Germany). All genes were fused amino-terminally to a His6-TEV tag. Flexible linkers were introduced into ISWIWt by polymerase incomplete primer extension at the appropriate positions [30]. All ISWI derivatives were fully sequenced. Expression and purification were performed as described [25]. The His6-TEV tag was cleaved off by TEV protease for all ISWI constructs except for ISWIΔHSS. Catalytic parameters of ISWIΔHSS are unaffected by the presence of the tag [15].
Enzyme assays and enzyme ligands. All assays were performed in 25 mM HEPES-KOH, pH 7.6, 50 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.2 g/l BSA and 1 mM DTT at 26 °C in the presence of an ATP-regenerating system as described [15]. Nucleosomes were reconstituted with recombinant Drosophila melanogaster histones by salt-gradient dialysis [31]. The concentration of nucleosomal DNA was determined by measuring its UV absorption at 260 nm. For nucleosomal arrays, concentrations refer to the concentration of individual nucleosomes.
ATP hydrolysis assays. ATP hydrolysis was monitored using an NADH-coupled assay as described [25]. Saturating concentrations of ATP-Mg2+ (1 mM), linearized plasmid DNA (pT7blue derivative; 0.2 mg/ml) and nucleosomes reconstituted on the same plasmid DNA (0.1 mg/ml) were used. Saturation was controlled in all cases by titration of the ligand at least over a 16-fold range.
Nucleosome remodelling assay. Remodelling activity was probed as previously described [15] by incubating 13-mer nucleosomal arrays (100 nM) with ISWI derivatives at the indicated concentrations, ATP-Mg2+ (1 mM) and KpnI (2 U/μl). Reactions were quenched with SDS (0.4%) and EDTA (20 mM) before the samples were deproteinized and analysed as described [15].
Nucleosome sliding assay. Nucleosome sliding was performed as described [15] by incubation of 25-mer nucleosomal arrays (30 nM) with ATP-Mg2+ (0.2 mM) and the respective ISWI derivative (ISWIΔHSS: 300 nM; all other enzymes: 5 nM). After quenching the reaction with apyrase (2.5 U/μl), arrays were digested with AvaI (1.1 U/μl) at 26 °C for 3–3.5 h. The AvaI digest was terminated with EDTA (40 mM) and SDS (0.4%) before the samples were deproteinized and analysed as described [15].
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to Nadine Harrer for collecting preliminary data, Katharina Braunger and Franziska Henze for help with protein purification, Nicola Hepp for preparation of 13-mer nucleosomal DNA and Gregory D. Bowman and Peter B. Becker for discussions and comments. J.L. thanks the Ernst Schering foundation for granting a predoctoral fellowship. H.K. was supported by a grant of the 'Center for Integrated Protein Science Munich' available to Peter B. Becker. This research was supported by the Deutsche Forschungsgemeinschaft by grant MU3613/1-1 available to F.M.-P.
Author contributions: J.L. and H.K. performed all experiments, interpreted the results and contributed to the manuscript. F.M.-P. conceived the study, interpreted the results and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2002) Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev 16: 2120–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T (2003) Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol 23: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13: 339–346 [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2005) Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol 12: 747–755 [DOI] [PubMed] [Google Scholar]
- Schwanbeck R, Xiao H, Wu C (2004) Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem 279: 39933–39941 [DOI] [PubMed] [Google Scholar]
- Dang W, Kagalwala MN, Bartholomew B (2006) Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol 26: 7388–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B (2008) Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol 28: 6010–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Bhardwaj SK, Deindl S, Lin YC, Zhuang X, Bartholomew B (2013) Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat Struct Mol Biol 20: 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X (2013) ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 152: 442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001) ISWI induces nucleosome sliding on nicked DNA. Mol Cell 8: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Langst G (2005) A 'loop recapture' mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol 12: 683–690 [DOI] [PubMed] [Google Scholar]
- Dang W, Bartholomew B (2007) Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol 27: 8306–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Planitz F, Klinker H, Ludwigsen J, Becker PB (2013) The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat Struct Mol Biol 20: 82–89 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2012) Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T (2011) The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J 30: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD (2011) Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol 31: 4746–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, McKnight JN, Nodelman IM, Bowman GD (2010) The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW (2003) Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell 12: 449–460 [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD, Stock MF, Moore J, Patterson RA (2003) Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry 42: 12011–12018 [DOI] [PubMed] [Google Scholar]
- Evers TH, van Dongen EM, Faesen AC, Meijer EW, Merkx M (2006) Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry 45: 13183–13192 [DOI] [PubMed] [Google Scholar]
- Sahoo H, Roccatano D, Zacharias M, Nau WM (2006) Distance distributions of short polypeptides recovered by fluorescence resonance energy transfer in the 10 A domain. J Am Chem Soc 128: 8118–8119 [DOI] [PubMed] [Google Scholar]
- Forne I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F (2012) Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics 11: M111 012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Bowman GD (2013) Nucleosome sliding by Chd1 does not require rigid coupling between DNA-binding and ATPase domains. EMBO Rep (e-pub ahead of print; doi:10.1038/embor.2013.158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ (2011) Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 472: 448–453 [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B (2009) Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell 35: 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD (2010) Mechanism of chromatin remodeling. Proc Natl Acad Sci USA 107: 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock HE, Lesley SA (2009) The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol 498: 91–103 [DOI] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K (2004) Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375: 23–44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.