Abstract
Chromatin remodellers are ATP-dependent motor proteins that physically reposition and reorganize nucleosomes. Chd1 and Iswi-type remodellers possess a DNA-binding domain (DBD) needed for efficient nucleosome mobilization; however, it has not been clear how this domain physically contributes to remodelling. Here we show that the Chd1 DBD promotes nucleosome sliding simply by tethering the remodeller to nucleosome substrates. Nucleosome sliding activity was largely resistant to increasing length and flexibility of the linker connecting the DBD and ATPase motor, arguing that the ATPase motor does not shift DNA onto the nucleosome by pulling on the DBD.
Keywords: Chd1, chromatin remodeller, nucleosome sliding
Introduction
Chromatin packaging of eukaryotic genomes is tightly integrated into transcriptional regulation, which in turn dictates both patterns of proliferation and cellular identity [1, 2]. Central to establishing and maintaining particular chromatin states are chromatin remodellers, which reorganize the DNA wrapping around histone cores. In a manner that is poorly understood, chromatin remodellers use a helicase-like ATPase motor to disrupt histone–DNA interactions, actively directing the assembly, disassembly and repositioning of nucleosomes [3]. In addition to the ATPase motor, chromatin remodellers possess multiple non-ATPase domains that can also participate in and help guide the outcome of remodelling [4]. A central question is determining what roles the non-ATPase domains have, and whether they are integral to the core mechanism for reorganizing nucleosomes.
Chd1 and Iswi remodellers share a similarly folded DNA-binding domain (DBD) that is essential for fast and efficient nucleosome sliding [5–8]. Two contrasting models have been proposed to explain how the DBD participates in remodelling. One model has proposed that the DBD has a mechanical role, aiding buildup of tension generated by the ATPase motor for breaking histone–DNA contacts [9, 10]. This model is supported by experiments showing that weakening DNA-binding ability of the DBD still allowed the ATPase motor to engage with nucleosomes, but prevented efficient sliding [6, 11]. An alternative model has suggested that the DBD stimulates nucleosome sliding by passively tethering the remodeller to nucleosomes. In support of this model, both Chd1 and Iswi remodellers have been shown to slide nucleosomes (albeit more poorly) when their C-terminal DBDs were deleted [7, 12, 13]. Additional support for a passive, tethering role was provided by chimeric Chd1 remodellers, where the native DBD was substituted with foreign, sequence-specific DBDs or monomeric streptavidin, which promoted nucleosome sliding in a DNA sequence-specific or biotin-dependent manner, respectively [7, 14].
To distinguish between these two models and elucidate how the Chd1 DBD stimulates sliding, we altered the length and flexibility of the linker that connects the DBD to the ATPase motor. We found that the DBD linker was quite tolerant of changes in length and increased flexibility, with sliding activity maintained after deletion of 29 residues or insertion of 121 intrinsically disordered residues. Shortening the DBD linker by 45 residues, however, blocked nucleosome sliding yet allowed both the DBD and ATPase motor to interact with the nucleosome. The DBD linker might therefore permit the DBD and ATPase motor to engage with nucleosomes without physically constraining one another. These results support the model that the DBD stimulates sliding by keeping the ATPase motor close to nucleosome substrates, and that the DBD linker is not a rigid connection through which the ATPase motor transmits force to the DBD during nucleosome sliding.
Results and Discussion
A minimal DBD linker is required for sliding
Our strategy for testing how the DBD positively contributes to nucleosome sliding was to alter the length and flexibility of the segment connecting the DBD with the ATPase motor. An important consideration in the design of Chd1 variants was to limit changes to a portion of the protein chain that does not have an important functional role on its own, but is in a location that would be required for mechanical communication or force generation. The segment connecting the ATPase motor and DBD of Chd1 has been shown to consist of at least three distinct elements: a bridge, an ATPase-coupling region and a linker region (Fig 1A). A crystal structure of the chromodomain–ATPase portion of yeast Chd1 revealed an extended, ∼50, residue segment, called the carboxy-terminal bridge that packed against both lobes of the ATPase motor and thus appeared to be positioned as a regulatory element [15]. Sequence analysis and cross-linking suggested that the bridge is also present in Iswi-type remodellers [15, 16], and recent work has reported that the bridge (also called NegC) regulates the ATPase motor of Iswi [13]. We previously defined the ATPase-coupling and linker regions of Chd1 using short deletions and point substitutions [8]. Disruption of the ATPase-coupling region allowed ATPase activity to remain ∼40% that of wild type, yet abolished or severely reduced nucleosome sliding activity. In contrast, individual deletions of 11 to 18 residues in the linker region had only a minor effect, reducing sliding activity by twofold or less. As the linker did not contain an essential sequence for nucleosome sliding, we increased flexibility and varied length of this region to test models for how the DBD and ATPase motor might work together.
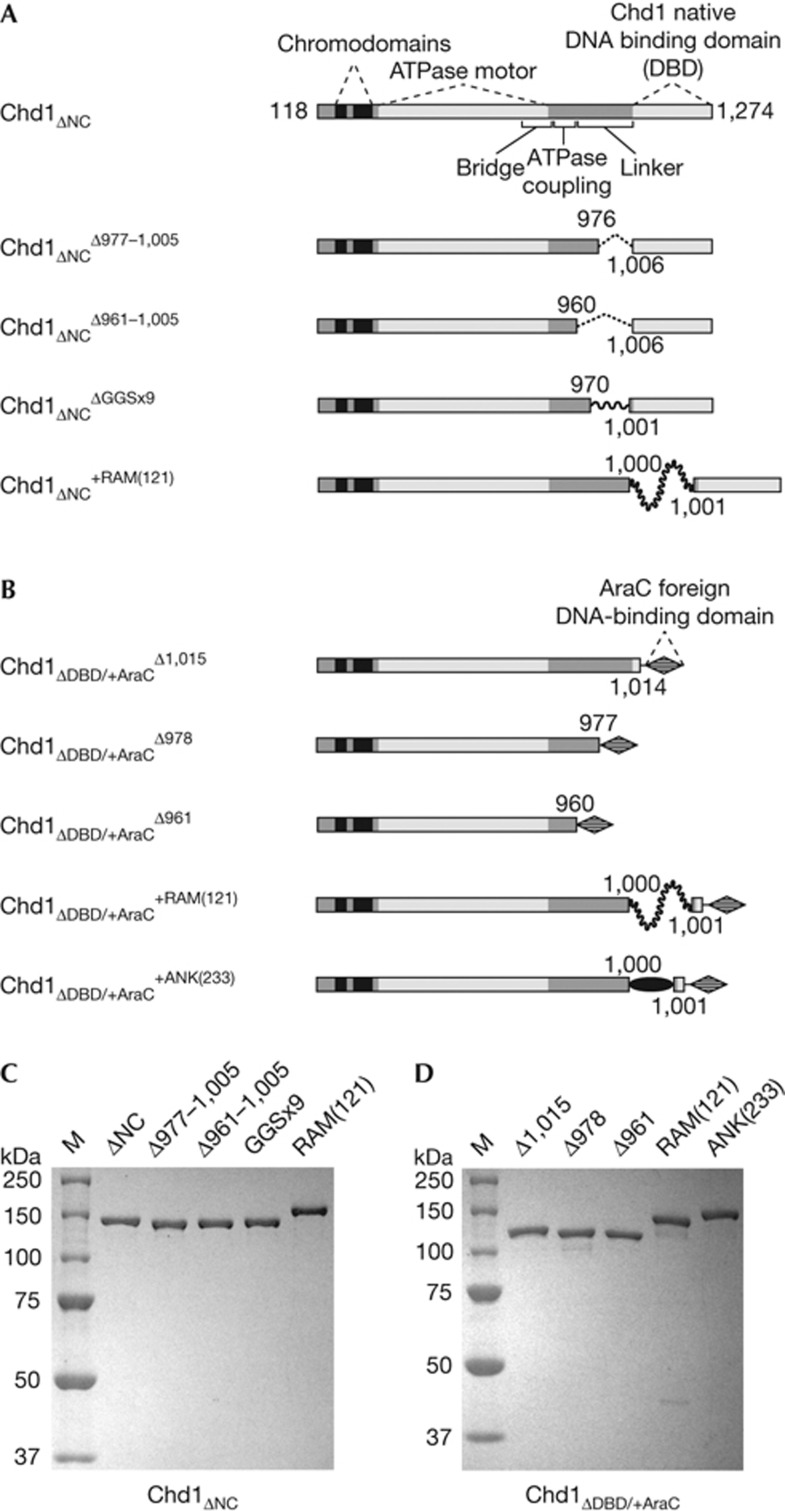
Figure 1.
Overview of S. cerevisiae Chd1 constructs used in this study. (A) Chd1 variants possessing the native Chd1 DBD. All constructs were on the basis of an N- and C-terminally truncated variant spanning residues 118–1,274, which includes the double chromodomains, ATPase motor and DBD, and are indicated with the subscript ΔNC. All changes to the DBD linker are denoted with superscripts, with the locations of the deletions, replacement and insertions labelled on the schematics. GGSx9 stands for nine Gly-Gly-Ser repeats, which replaced Chd1 residues 971–1,000. The +RAM(121) construct contains a 121-residue insertion of the RAM segment (residues 1,763 to 1,883) from the Drosophila melanogaster Notch receptor. (B) Chd1 variants possessing the foreign AraC DBD. All constructs contained the DBD of the AraC transcriptional regulator (residues 175–281) in place of the native Chd1 DBD, denoted with the subscript ΔDBD/+AraC. As in (A), superscripts refer to changes in the DBD linker, with Δ1,015, Δ978 and Δ961 referring to the beginning of the deletion replaced by the AraC DBD. The +ANK(233) construct contained the ankyrin domain (residues 1,907–2,139) of D. melanogaster Notch. Note that domains in A and B are not drawn to scale. (C) and (D) Purified proteins containing the native Chd1 DBD (C) or with the foreign AraC DBD (D) on SDS–PAGE gels stained with GelCode Blue. ANK, Ankyrin; DBD, DNA-binding domain; SDS–PAGE, SDS–polyacrylamide gel electrophoresis
To determine how shortening the DBD linker length affected nucleosome sliding ability, we generated two deletion variants that lacked either 29 or 45 residues before the DBD, called Chd1ΔNCΔ977–1,005 and Chd1ΔNCΔ961–1,005, respectively (Fig 1A,C). These and other linker variations were introduced in a previously characterized Saccharomyces cerevisiae Chd1 variant called Chd1ΔNC that possessed the Chd1 family-specific amino-terminal double chromodomains, central helicase-like ATPase motor and C-terminal DBD and was active in nucleosome sliding and centering [7, 8]. Nucleosome sliding activities were determined by monitoring migration of fluorescently labelled, end-positioned (0-N-80) mononucleosomes by native polyacrylamide gel electrophoresis (PAGE). As previously demonstrated for Chd1 and similar to many Iswi-type remodellers [17, 18], Chd1ΔNC preferentially shifts mononucleosomes away from DNA ends and thus more towards central locations on short DNA fragments.
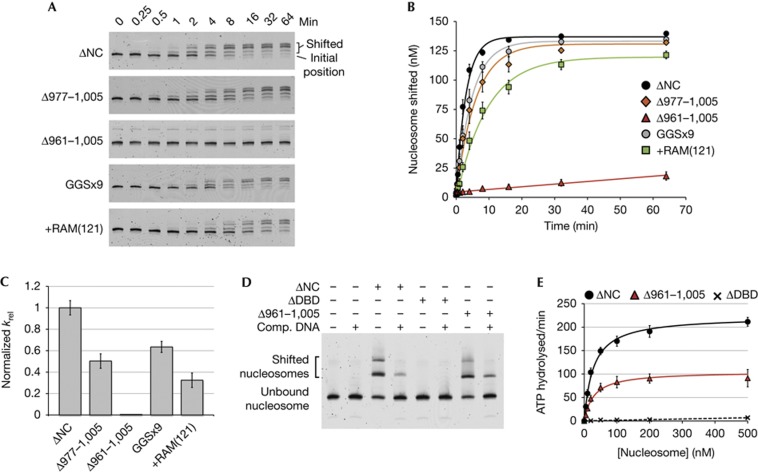
Whereas deletion of 29 residues from the linker only modestly slowed down the sliding rate of Chd1ΔNCΔ977–1,005 compared with Chd1ΔNC, the larger deletion of 45 residues significantly disrupted sliding activity of Chd1ΔNCΔ961–1,005, estimated to be ∼200-fold slower than Chd1ΔNC (Fig 2A–C). This larger deletion might have interfered with nucleosome sliding by preventing either the DBD or ATPase motor from interacting with the nucleosome, or by disrupting coordination between the remodeller domains. It was previously shown that in the absence of ATP, robust nucleosome binding requires the Chd1 DBD [6]. We therefore investigated whether the 961–1,005 deletion, which left the DBD intact, perturbed nucleosome binding. As assessed by native PAGE, binding of Chd1ΔDBD (lacking the DBD) to nucleosomes was not observed, yet Chd1ΔNCΔ961–1,005 associated with nucleosomes to a similar extent as Chd1ΔNC (Fig 2D).
Figure 2.
Chd1 tolerates increased length and flexibility in the DBD linker, but requires a minimal length linker for coordinated action of DBD and ATPase domains. (A) Nucleosome sliding as assessed by native PAGE. Data shown are representative experiments (n≥4). (B) Quantification of nucleosome sliding reactions shown in (A). Error bars represent standard deviations (s.d.) (n≥4). (C) Relative rate constants for nucleosome sliding. Rate constants were obtained from single exponential fits to the data shown in (B), with the relative rates shown as krel=kobs, Chd1 variant/kobs, Chd1ΔNC. For Chd1ΔNC, kobs was 0.33±0.02 min−1. Error bars represent the calculated errors of the fits. (D) Deletion of the DBD linker does not disrupt nucleosome binding, as shown by native PAGE. Super-shifted bands indicate Chd1–nucleosome complexes. (E) Deletion of the DBD linker still allows the Chd1 ATPase motor to be stimulated by nucleosomes. Shown are the averages and s.d. (n≥3) of nucleosome-stimulated ATPase activities with increasing concentrations of nucleosome substrate. Nucleosome-free rates were subtracted from all values, and the resultant activities were fit to the Michaelis–Menton equation (Chd1ΔNC and Chd1ΔNCΔ961–1,005, solid lines) or linearly (Chd1ΔDBD, broken line). Chd1ΔDBD contained residues 118–1,005. DBD, DNA-binding domain; PAGE, polyacrylamide gel electrophoresis.
As these results suggested that the DBD of Chd1ΔNCΔ961–1,005 was able to interact with nucleosomes, we measured ATP hydrolysis rates to see whether nucleosomes stimulated the ATPase motor (Fig 2E). In agreement with previous work, deletion of the entire Chd1 DBD essentially abolished nucleosome-stimulated ATPase activity, consistent with low nucleosome-binding affinity [7, 8]. For Chd1ΔNCΔ961–1,005, in contrast, the maximal nucleosome-stimulated ATPase activity was ∼50% that of Chd1ΔNC, with a similar apparent KM for (28-N-35) nucleosomes (29±6 nM for Chd1ΔNCΔ961–1,005 and 27±3 nM for Chd1ΔNC). These results indicate that although the reduced length of the DBD linker in Chd1ΔNCΔ961–1,005 blocked nucleosome sliding, both the DBD and ATPase motor could engage with the nucleosome. One likely explanation for this defect is that the linker deletion disrupted neighbouring ATPase-coupling residues, which in turn led to an ATPase-coupling defect. As this linker deletion would be expected to reduce the distance that the DBD could extend from the ATPase motor, an alternative possibility is that shortening the linker prevented the DBD from playing a mechanical role in pulling DNA onto the nucleosome.
Increased linker flexibility modestly impacts sliding
To determine whether remodelling required a rigid connection between the DBD and ATPase domains, we next altered the flexibility and length of the DBD linker. To increase flexibility, we designed a Chd1 variant with residues 971–1,000 replaced by a 29 amino-acid segment containing nine Gly-Gly-Ser repeats (GGSx9; Fig 1A). As shown by nucleosome sliding assays, Chd1 activity was only modestly (<2-fold) reduced on substitution of this intrinsically flexible GGSx9 segment (Fig 2A–C), arguing against a rigid connection being required for mobilizing nucleosomes. Although flexible, however, it could be argued that the DBD might mechanically participate in the nucleosome sliding process if the length of the DBD linker was limiting. If, for example, translocation of the ATPase motor on nucleosomal DNA resulted in pulling the DBD, bound at the entry site, a flexible but taut connection could aid the buildup of torque between these two points of contact. To investigate this possibility, we lengthened the linker connecting the DBD to the rest of the remodeller by inserting a 121-residue segment, known as RAM from the Drosophila Notch receptor, that is intrinsically disordered [19]. Sliding activity of this protein, called Chd1ΔNC+RAM(121), was approximately one-third as fast as Chd1ΔNC (Fig 2A–C). The activity exhibited by Chd1ΔNC+RAM(121) demonstrates that although long and flexible, this linker still allowed the DBD to participate in nucleosome sliding. These results indicate that the DBD does not need to be connected to the rest of the remodeller via a rigid linker to support nucleosome sliding, and suggest that a key role of the DBD is to tether the remodeller to nucleosome substrates.
Lengthening the DBD linker allows for a farther reach
To further explore the idea that the DBD positively stimulates nucleosome sliding simply by tethering, we turned to fusion Chd1 remodellers with a foreign, sequence-specific DBD. We previously showed that replacement of the native DBD of Chd1 with the DBD of the Escherichia coli transcriptional regulator AraC produced a hybrid remodeller that shifted nucleosomes towards the AraC binding site (araI1) [7]. Although the DBD of Chd1 presumably binds to extranucleosomal DNA, the sequence-nonspecific nature of the DBD makes it unclear where exactly the DBD binds, or if interactions between the DBD and nucleosome must change during the remodelling cycle. With the sequence-specific binding of the AraC DBD, the Chd1–AraC remodellers offered the opportunity to test how the flexibility and length of the DBD linker affect remodeller activity with the DBD targeted to specific locations.
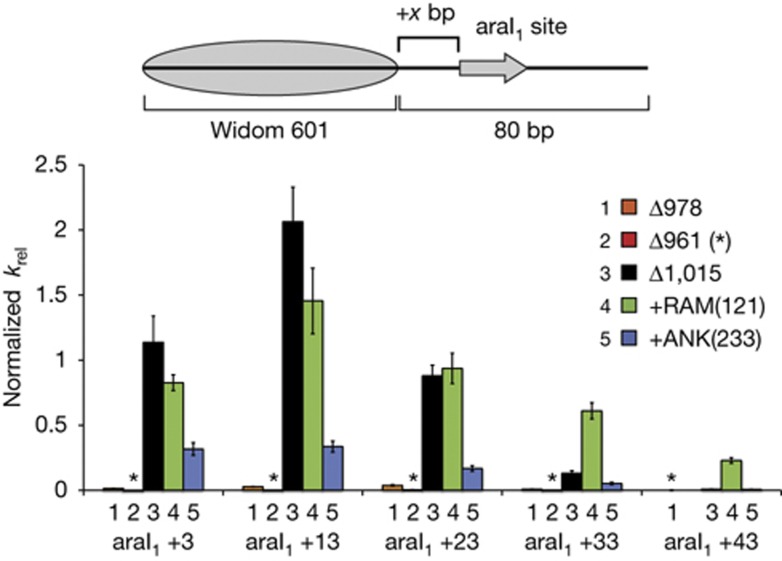
Using a Chd1–AraC background, deletions and insertions in the DBD linker were generated that paralleled the linker variants studied with the native Chd1 DBD (Fig 1B,D). Five different nucleosomes were generated, each having the araI1 different distances from the edge of the nucleosome. Similar to previous observations [7], a Chd1–AraC fusion remodeller with the DBD of AraC replacing the C-terminal portion of the protein beginning at residue 1,015 (Chd1ΔDBD/+AraCΔ1,015) moved nucleosomes as fast or faster than Chd1ΔNC when the edge of the araI1 was within 23 bp of the nucleosome. However, sliding rates were considerably slower with more distant binding sites, being only ∼10% and ∼1% as fast with the araI1 site at +33 and +43, respectively (Fig 3). For Chd1ΔDBD/+AraC, deletions of the DBD linker similar to those used with the native DBD dramatically reduced sliding at all distances. A Chd1–AraC remodeller with the AraC DBD fused after residue 977 (called Chd1ΔDBD/+AraCΔ978) showed <5% the sliding rate as Chd1ΔDBD/+AraCΔ1,015, and the shorter fusion at residue 960 (called Chd1ΔDBD/+AraCΔ961) was <1% as active. Using the same nucleosome substrates, Chd1ΔDBD/+AraCΔ978 was considerably slower (<5% activity) than Chd1ΔNCΔ977–1,005 (∼50% activity, data not shown), which had the analogous DBD linker deletion in the presence of the native Chd1 DBD. Thus, the combination of this linker deletion and the site-specific binding of the AraC DBD appeared to limit the action of the ATPase motor on the nucleosome.
Figure 3.
Insertion of a long flexible segment into the DBD linker allows the Chd1–AraC remodeller to reach farther away from the nucleosome. End-positioned nucleosomes were generated containing the 17 bp araI1 site located +3, +13, +23, +33 or +43 bp away from the nucleosome edge. For each remodeller variant, a rate constant was calculated for each nucleosome substrate from single exponential fits to the data (n≥3). The bar graph shows rate constants, along with calculated errors of fitting, relative to Chd1ΔNC as in Fig 2. For Chd1ΔNC, the average kobs for nucleosomes with araI1 at +3 and +13 was 0.32±0.02 min−1. Asterisks denote relative rate constants≤0.004. araI1, AraC binding site; DBD, DNA-binding domain.
Whereas shortening the DBD linker coupled with site-specific binding by AraC appeared more constraining, one prediction of tethering is that a longer linker should allow the remodeller to effectively reach binding sites farther away from the nucleosome. To test this idea, sliding rates of the five araI1-containing nucleosomes were measured for a Chd1–AraC remodeller containing a flexible RAM insertion in the DBD linker (Chd1ΔDBD/+AraC+RAM(121)). Compared with Chd1ΔDBD/+AraCΔ1,015, Chd1ΔDBD/+AraC+RAM(121) showed higher sliding activity for araI1 +33 and +43 nucleosomes (Fig 3). In addition to a farther reach, the RAM insertion also gave a more gradual decrease in activity with more distant araI1 sites. These traits are consistent with the AraC DBD increasing the local concentration of the ATPase motor around the nucleosome, with a longer, flexible connection providing a wider range for the ATPase to the sample.
As an alternative means of lengthening the DBD linker, we also generated a Chd1–AraC fusion with the folded protein domain from Ankyrin, also from the Notch receptor, inserted between the ATPase motor and the foreign AraC DBD. Ankyrin has N- and C-termini on the opposite sides of the domain, separated by ∼60 Å [20], and yet this insertion failed to increase sliding activity for +33 and +43 araI1 nucleosomes, and decreased sliding of nucleosomes with +23 and closer araI1 sites (Fig 3). In conjunction with the higher activity and longer reach of Chd1ΔDBD/+AraC+RAM(121), these data suggest that flexibility might not only be tolerated in the DBD linker, but in fact preferable.
In conclusion, these experiments indicate that the DBD is not physically pulled by the ATPase motor as part of the nucleosome sliding mechanism. Instead, the data presented here support the idea that the linker connecting the DBD with the rest of the remodeller must be long enough and sufficiently flexible to accommodate conformational changes that occur during remodelling. Shortening the DBD linker by 45 residues blocked nucleosome sliding. This sliding defect may have been due to disrupting residues in the neighbouring ATPase-coupling region or premature dissociation of the DBD during the remodelling cycle. Lengthening the linker with the RAM insertion, in contrast, had only a modest impact on sliding activity and allowed the remodeller to reach DNA farther from the nucleosome. The experiments in this study highlight how flexibility might be a natural property of the native DBD linker. These conclusions are consistent with recent work with the related Iswi remodeller from Drosophila, where flexible insertions between the ATPase motor and DBD were found to be well tolerated [21]. Nucleosome sliding has been shown to consist of multiple ATP-dependent translocation events that must occur in succession to reposition nucleosomes [22]. During the multistep remodelling cycle, the DBD likely has a critical role in maintaining the ATPase motor close to the nucleosome substrate. By tethering the remodeller to nucleosomes, these and previous experiments suggest that the Chd1 DBD increases processivity of the ATPase motor during remodelling [6, 7, 14], with the DBD linker allowing for conformational changes in the remodeller and/or nucleosome during the remodelling reaction.
Methods
Chd1 constructs and purification. A pDEST17 expression vector containing S. cerevisiae Chd1ΔNC (residues 118–1,274), flanked by a cleavable N-terminal 6xHis tag, served as a template for generating all other constructs using PCR mutagenesis. The Chd1ΔNCGGSx9 variant was made in two stages, first PCR amplifying the GGS template (100 bp complementary oligonucleotides) using overlapping primers to add flanking sequence complementary to Chd1, and second using this purified PCR product for insertional mutagenesis into the Chd1ΔNC template. The resulting construct contained a 29 amino-acid substitution for residues 971–1,000, which consisted of two arginines (substituting for Lys972 and Lys973) followed by nine Gly-Gly-Ser repeats. All Chd1 variants were expressed in E. coli BL21(DE3)star cells containing the Trigger chaperone expression plasmid (a kind gift of Li Ma and Guy Montelione) and pRIL as previously described [15]. All Chd1 proteins were purified essentially as described [8], with the exception of Chd1ΔDBD/+AraC variants, which were purified using a SourceQ instead of an SP-FF column. The final step of purification was size exclusion chromatography on a Superdex 200 column, which allowed for removal of the aggregates and contaminating proteins (supplementary Fig S1 online and Fig 1C,D).
Nucleosome preparation. Xenopus laevis and S. cerevisiae histone proteins were recombinantly expressed and purified as previously described ([23], and web protocol from the Tsukiyama laboratory: http://www.research.fhcrc.org/tsukiyama/en/protocols.html). Using PCR-amplified DNA containing the Widom 601 positioning sequence [24], nucleosomes were reconstituted and purified on a Bio-Rad Mini-Prep Cell. DNA containing the araI1 sequence (5′-TAGCATTTTTATCCATA-3′) was amplified from modified pGEM-601 templates containing araI1 sites introduced by insertional mutagenesis.
Nucleosome binding and sliding. Nucleosome sliding was measured by following migration of end-positioned 0-N-80 nucleosomes by native PAGE as described [8]. Reactions were carried out in 1 × slide buffer (50 mM KCl, 20 mM Hepes pH 7.8, 5 mM MgCl2, 5% sucrose, 0.1 mg/ml bovine serum albumin, 1 mM dithiothreitol) with 2.5 mM ATP at room temperature. Nucleosome sliding experiments for Chd1 variants containing the native Chd1 DBD were carried out using 50 nM remodeller and 150 nM nucleosomes made with X. laevis histones and FAM-labelled DNA, whereas those for Chd1–AraC variants were carried out with 10 nM remodeller and 100 nM nucleosomes made with S. cerevisiae histones and Cy5-labelled DNA. Activities of Chd1ΔNC and Chd1ΔNCΔ977–1,005 were also assessed on S. cerevisiae nucleosomes. Gels were quantified in ImageJ and the rates determined by fitting progress curves to single exponentials in Kaleidagraph. For nucleosome-binding experiments, Chd1 and nucleosomes were mixed in equimolar ratios (150 nM each) in sliding buffer minus ATP, and incubated for 1 h at room temperature before being resolved on a 3.5% native acrylamide gel at 4 °C. Where indicated, plasmid DNA as competitor was added to a final concentration of 125 ng/μl.
ATPase activity. ATPase activity was measured using an NADH-coupled assay, similarly to previously described protocols [8, 25]. Each reaction was carried out in 1 × slide buffer, and contained 4 μl of a 10 mg/ml NADH solution, 5 μl of 50 mM phosphoenolpyruvate, 4 μl of a pyruvate kinase (PK)/lactate dehydrogenase (LDH) solution (P0294-Sigma) or 6 μl of PK/LDH mixture (0.01 units/μl PK (10128155001-Roche) and 0.00275, units/μl LDH (10128155001-Roche)), varying amounts of unlabelled (28-N-35) nucleosomes reconstituted with X. laevis histones, and 5 μl of 1 μM Chd1 protein, in a total volume of 95 μl. After addition of 5 μl of 50 mM ATP to initiate the reactions, the solutions were rapidly mixed and transferred to a 96 well microtiter plate, where the absorbances were monitored at 340 nm every 10 s for 15 min at 24 °C.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Karolin Luger and Toshio Tsukiyama for histone expression plasmids, Jonathan Widom for the pGEM-601 plasmid, Guy Montelione and Li Ma for the Trigger chaperone coexpression plasmid, Jeff McKnight for Chd1ΔDBD/+AraCΔ978 and Chd1ΔDBD/+AraCΔ1,015 constructs, Doug Barrick for RAM and Ankyrin templates, and Ashok Patel for Chd1ΔDBD protein and nucleosome reagents for ATPase assays. We appreciate Herschel Wade, Sarah Woodson and members of the Bowman lab for stimulating discussions. This work was supported by a grant from the National Institutes of Health (R01-GM084192).
Author contributions: I.M.N. designed and carried out all experiments, analysed data, and contributed to the manuscript. G.D.B. designed experiments, analysed data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Cairns BR (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Ryan DP, Owen-Hughes T (2011) Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol 15: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Bartholomew B (2011) Diversity of operation in ATP-dependent chromatin remodelers. Biochim Biophys Acta 1809: 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüne T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Müller CW (2003) Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell 12: 449–460 [DOI] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T (2011) The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J 30: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD (2011) Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol 31: 4746–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, McKnight JN, Genzor P, Bowman GD (2011) Identification of residues in chromo-helicase-DNA-binding protein 1 (Chd1) required for coupling ATP hydrolysis to nucleosome sliding. J Biol Chem 286: 43984–43993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Cairns BR (2007) Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol 14: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Bhardwaj SK, Deindl S, Lin YC, Zhuang X, Bartholomew B (2013) Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat Struct Mol Biol 20: 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Planitz F, Klinker H, Ludwigsen J, Becker PB (2013) The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat Struct Mol Biol 20: 82–89 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2012) Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Chakravarthy S, Morrone S, Nodelman IM, McKnight JN, Bowman GD (2013) Decoupling nucleosome recognition from DNA binding dramatically alters the properties of the Chd1 chromatin remodeler. Nucleic Acids Res 41: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, McKnight JN, Nodelman IM, Bowman GD (2010) The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forne I, Ludwigsen J, Imhof A, Becker PB, Müller-Planitz F (2012) Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive cross-linkers and mass spectrometry. Mol Cell Proteomics 11: M111.012088.1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T (2006) Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem 281: 16279–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ (2006) The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol 13: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Bertagna A, Toptygin D, Brand L, Barrick D (2008) The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem Soc Trans 36: 157–166 [DOI] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Hughson FM, Barrick D (2003) Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci 12: 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwigsen J, Klinker H, Müller-Planitz F (2013) No need for a power stroke in ISWI-mediated nucleosome sliding. EMBO Rep (e-pub ahead of print 11 October 2013; doi:10.1038/embor.2013.160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X (2013) ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 152: 442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J (1998) New DNA sequence rules for high-affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD (2003) NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high-specific activities. Anal Biochem 321: 266–271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.