Since the rise of modern pharmaceutical research and industry in the 1950s, drugs have been used to treat an increasingly wide range of diseases. From antibiotics for treating infections, to antivirals to treat HIV/AIDS, to drugs for hypertension and cancer, drug-based therapies have had enormous effects in curing or converting often fatal diseases into manageable conditions. Even pathophysiologies, such as peptic ulcers, that once required surgery are now routinely treated by drugs.
Along with the many successes, several limitations have also become evident. Many diseases, especially those that progress in severity, remain difficult to treat with drugs. The list of such disorders is long and includes aneurysms, congestive heart failure, diabetes, kidney disease and many types of cancer. Even drugs that are efficacious do not work for everybody. Effective drugs cause serious adverse events in a subset of users. As we often cannot predict who might suffer from these side effects, the drug is typically taken off the market.
These problems have generated a sense that our current approaches might have reached their limits and that we need new thinking to drive both drug discovery and usage. The extensive advances in our understanding of the basic molecular and cell biology of humans, other mammalian organisms and model organisms indicate that there are probably many more cellular components that could be targeted by drugs to fight disease. Another general insight is that cellular components interact with one another to form extensive networks. These networks have the capability to regulate and coordinate a range of subcellular functions, which gives rise to cellular phenotypes [1,2]. These cellular phenotypes underlie the tissue and organ functions that are characteristics of both health and disease.
Malfunctions at the molecular level, when propagated to a higher level of organization, give rise to disease
Genomics, molecular and cell biology and biochemistry are steadily becoming the basic elements for systems biology. As we continue to identify and characterize parts of cells and tissues, the next step in biology is to understand how these parts come together to form functional systems. The focus is not only to understand the characteristics and functions of individual entities, such as genes, proteins, lipids, sugars and so on, but also to understand how these entities interact with one another and what functions emerge from these interactions [3]. In this line of reasoning, almost all tissue and organ functions as well as organismal behaviour arise from molecular interactions. This has been explicitly demonstrated for coupled biochemical components that form positive feedback loops, which function as bistable switches. Such switches underlie, for instance, long-term depression of synaptic responses in the hippocampus [4] or hunger in mice [5].
The systems-biology view that complex networks underlie many diseases is being increasingly demonstrated for many diseases, including heart disease, kidney disease, diabetes, metabolic diseases and cancers. To cast systems of interacting entities as networks is useful because it allows the use of graph theory, a branch of mathematics that analyses how complex systems are organized and how such organization enables system-level functions. When one thinks of complex regulatory networks, we often tend to think of molecular networks, but it is important to remember that networks exist at the level of tissues and organs and between organs at the level of organisms. Tissue-level networks are best recognized in the brain, where the activity of circuits—that is, networks of neurons—can be correlated with the behaviour of animals.
The combining of drugs that act on different targets within a network could be more efficacious than treating disease with one drug
At the organismal level, current therapies for hypertension, which include multiple drugs acting at various tissues and organs—β-blockers on the heart, angiotensin-converting-enzyme inhibitors on blood vessels and diuretics on the kidney—provide compelling evidence of how blood pressure is a function of interactions between multiple tissues and organs in the body. Overall, it is reasonable to conclude that there are networks at different levels of organization: molecular networks within and between cells, cellular networks within tissues and organs, and networks of organs that functionally give rise to organismal physiology. Between each of these networks there are multiple connections, which are essential for a healthy organism (Fig 1). Malfunctions at the molecular level, when propagated to a higher level of organization, give rise to disease. Sometimes these malfunctions differ from person to person owing to variations and changes in the person's genome. These variations indicate that different malfunctions can give rise to the same disease and knowing the molecular malfunctions is essential for developing personalized therapy. The various streams of data show overall that there is reasonable evidence to support a systems-biology approach that uses a network perspective of disease genes and mechanisms [6].
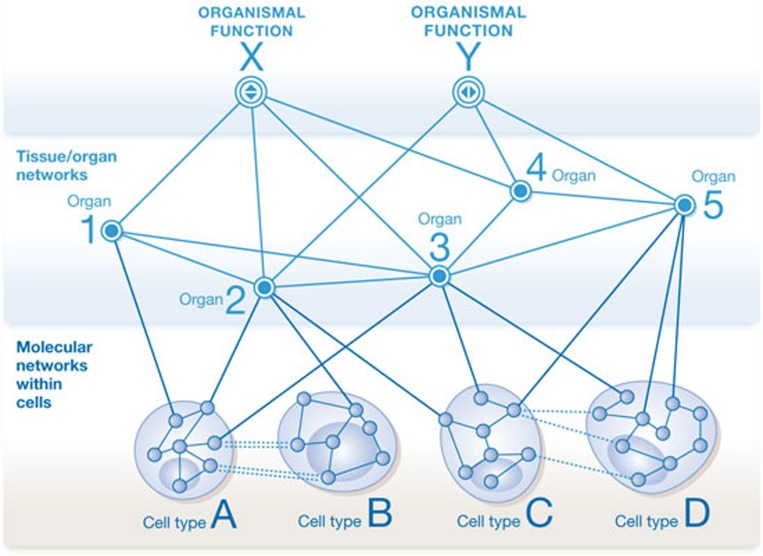
Figure 1.
A schematic representation of the layers of networks that underlie organismal function, such as control of blood pressure (hypertension) or glucose levels in the blood (type 2 diabetes). Organismal functions arise from functional interactions between multiple organs. Organ and tissue functions arise from the functions of the multiple cell types of which they are comprised. Molecular networks exist within and between cell types that give rise to cellular functions. Drugs typically change the activity of the molecular components, and this change in activity percolates up to eventually affect organismal functions or malfunctions in disease states.
Drugs, by and large, work at the molecular level, just as diseases originate from molecular malfunctions. From penicillin, which inhibits enzymes that make the bacterial cell wall, to β-blockers, such as propranolol, that inhibit β-adrenergic receptors to regulate heart function, to cancer drugs, such as imatinib, that block tyrosine kinases to inhibit the proliferation of cells, the effects of drugs start with molecular interactions. These effects are propagated across scales of organization to alter tissue or organ function to cure or relieve disease. The transmission of the drug effect is not linear. Rather, it occurs through the networks at each level of organization. This type of percolation at various scales of organization can sometimes have harmful consequences in addition to the intended good effect of treating the disease. These are called side effects, where effective treatment of one disease or its symptoms is associated with occurrence of a different type of disease in some individuals taking the drug.
Such systems-biology-based approaches are likely to be of increasing value in the treatment of cancer because most cancers undergo multiple molecular changes as they progress
Well-known examples of side effects are the occurrence of heart attacks and strokes associated with rofecoxib, which is used to treat osteoarthritis, and rosiglitazone, which is used to treat type 2 diabetes mellitus. In each case the drug is efficacious in treating the disease it is intended to treat but the risk of a serious side effect is too great and these drugs have been largely withdrawn from the market. In both cases, it appears that the side effects were a result of the networks in which the intended drug targets participate in different cell types and tissues.
Sometimes, drugs bind to unintended targets and such interactions can lead to serious side effects. Many classes of drug, for reasons that are not always clear, cause arrhythmias by binding to the HERG channel in the heart. As one of its preclinical safety checks, the US Food and Drug Administration (FDA) therefore recommends that the developers of new drugs demonstrate that their drug does not interact with the HERG channel protein. Unintended targets of drugs are also part of cellular networks and, therefore, effects on these targets can be propagated through networks.
Drug combinations can also cause unanticipated side effects. Analysis of the FDA Adverse Event Reporting System database (FAERS) by Altman and colleagues [7] showed that paroxetine, an antidepressant, and pravastatin, a cholesterol-lowering drug, raised blood glucose levels when administered in combination, whereas each drug on its own did not. Such an increase in blood glucose is an important consideration for patients with diabetes. This study showed the potential usefulness of analysing large databases, such as FAERS, to identify unanticipated biological effects associated with drug combinations and provided support for the idea that systems biology underlies combination drug therapy.
Systems pharmacology is the name that is increasingly being used for the new systems-based approach that is being used to understand drug actions and for drug discovery. Systems pharmacology will take into account genomic variations and molecular complexity in defining physiological and pathophysiological responses at the tissue, organ and organism levels. My colleagues and I have used it to understand drug actions by studying how drug targets function within cellular networks. One hypothesis we have pursued is that, in addition to networks enabling drugs to do bad things, they can also enable good effects.
Combining drugs that act on different targets within a network could be more efficacious than treating disease with one drug. Sometimes, complex diseases cannot be treated effectively by modulating a single target. Asthma is a good example: long-acting stimulators of the β-adrenergic receptors and corticosteroids together are effective and are widely used in combination. The combined effects are through drug action at varying timescales in cellular and tissue networks: the long-acting β2-adrenergic activator acutely relaxes the airways while the corticosteroids suppress inflammation with a slower time course.
The combination of long-acting β2-adrenergic activators with muscarinic-receptor blockers is going through the approval process for treatment of chronic obstructive pulmonary disease [8]. These drug combinations are based on knowledge of how the targets of these drugs work in the context of cellular regulatory networks, and represent good examples of how systems-level thinking can lead to useful therapies. Such systems-biology-based approaches are likely to be of increasing value in the treatment of cancer because most cancers undergo multiple molecular changes as they progress. The combination of drugs that block the effects of multiple activators and inhibitors of cell growth are likely to become efficacious targeted therapy as we start to obtain detailed knowledge of the molecular networks underlying many cancers.
Not all drug combinations are based on network logic. The commonly used antibacterial, Augmentin, combines the antibiotic amoxicillin with clavulanic acid, an inhibitor of the β-lactamase that breaks down the antibiotic. Here, the second drug extends the life of the first drug thus making it more efficacious.
A novel systems approach in cancer has been described for treatment of some types of leukaemia and involves the use of genetically engineered T cells, which produce a cytokine storm that can kill off cancerous cells [9]. However, there are serious life-threatening side effects. An article in the New York Times describes how physicians have combined the genetically engineered T cells with antibodies against interleukin-6 by using tocilizumab to keep the effects of the T cells within a therapeutic range [10]. Although the news report suggests that this combination was developed empirically for a medical emergency, post hoc it is clear that the physicians have used an implicit systems approach to select a second drug to manage the risk–benefit ratio of the first drug by considering the source and target cells as part of a multicellular response network.
A study from my laboratory [11] also shows that combination therapy can substantially reduce the serious adverse effects associated with a useful drug. We analysed FAERS and found many cases in which a drug B was given for a different reason and reduced a serious adverse event associated with drug A. We studied the combination of rosiglitazone and exenatide in some depth. Patients who were prescribed rosiglitazone and exenatide had a greatly reduced risk of heart attack than did patients prescribed rosiglitazone in combination with other drugs. This finding suggests that exenatide selectively reduces the risk of heart attacks and stroke associated with rosiglitazone. We were able to build molecular networks to show how signals from the targets of these two drugs might intersect and found that the blood protein PAI1 might be involved. PAI1 regulates the protease that breaks down blood clots. Increases in PAI1 levels lead to an increased risk of clots. We validated the network-based molecular mechanisms underlying the drug combination effects in a mouse model of diabetes.
…the studies described here and many others are starting to show that systems-level analysis can be a powerful driver for understanding drug action
This case is not unique. We identified nearly 19,000 other drug combinations in FAERS in which a second drug mitigated a serious adverse event associated with a first drug. Some of these combinations and effects are surprising. H2 antagonists, typically given for acid reflux diseases, were associated with a decreased number of suicides associated with selective serotonin reuptake inhibitors, and the blood-pressure medication lisinopril reduces statin-associated muscle wasting. We have been able to build plausible molecular networks for several of these drug combinations, suggesting that current molecular and cell biological knowledge could be used to develop a network-based understanding of the beneficial effects of drug combinations. Of note, the second drug is often given to treat an entirely different disease and the decreased side effects are unanticipated benefits of drug combination.
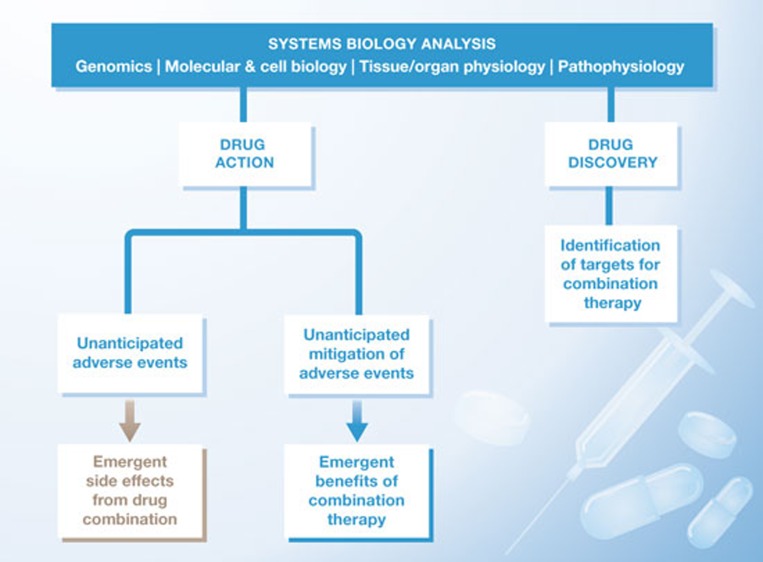
At a general level, the studies described here and many others show that systems-level analysis can be a powerful driver for understanding drug action. One can envisage three kinds of new knowledge coming from such analyses (Fig 2). First is the identification of unanticipated adverse events that each drug might not produce on its own. Identification and prediction of such adverse effects could prove useful to guide physicians regarding which medicines can be co-prescribed. The second kind of knowledge is the opposite of the first: identification of unanticipated beneficial effects by drug combinations, such as mitigation of side effects. This type of knowledge might lead to repurposing of approved drugs if their efficacy in suppressing adverse events could be established in rigorous clinical trials. The third kind of knowledge, which is the most forward-looking, is that network biology can be used for the discovery of new drugs. Network analysis can provide a rational basis for identifying targets, which, when modulated together by drug combinations, might be distinctively efficacious in treating complex diseases.
Figure 2.
A flow chart of how systems biology can affect various facets of pharmacology and therapeutics.
Combination therapy based on network biology could become efficacious for the treatment of progressive diseases, such as type 2 diabetes, kidney disease, congestive heart failure and, of course, many cancers. While the necessary knowledge is not yet available, the path forward can be readily seen. Large databases, such as FAERs, can provide empirical knowledge of good and bad outcomes associated with combination therapies in humans. As large amounts of genomic and molecular data are integrated with clinical data when electronic medical records become more widely used and molecular characterization of patients becomes more standardized, it will probably generate a wealth of systems-level information to analyse and generate hypotheses. These hypotheses might help with the design of studies to better understand the progression of diseases, and design new drugs or repurpose existing drugs that, in combination, are more effective for treating complex diseases.

Ravi Iyengar
Footnotes
R.I. receives support for one postdoctoral position from GlaxoSmithKline (GSK). The person supported by GSK has had no role in this paper.
References
- Jordan JD, Landau EM, Iyengar R (2000) Signaling networks: the origins of cellular multitasking. Cell 103: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A, Blitzer RD, Iyengar R (2005) Toward predictive models of mammalian cells. Annu Rev Biophys Biomol Struct 34: 319–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Iyengar R (1999) Emergent properties of networks of biological signaling pathways. Science 283: 381–387 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Augustine GJ (2008) A positive feedback signal transduction loop determines timing of cerebellar long-term depression. Neuron 59: 608–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y et al. (2011) Hunger states switch a flip-flop memory circuit via a synaptic AMPK-dependent positive feedback loop. Cell 146: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási AL (2011) Interactome networks and human disease. Cell 144: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatonetti NP et al. (2011) Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 90: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson H (2013) Pipeline for COPD drugs flows with combination candidates. Nat Med 19: 1079. [DOI] [PubMed] [Google Scholar]
- Kalos M et al. (2011) T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med 3: 95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady D (2012) In Girl's Last Hope, Altered Immune Cells Beat Leukemia. New York Times 9 Dec http://www.nytimes.com/2012/12/10/health/a-breakthrough-against-leukemia-using-altered-t-cells.html?_r=0 [Google Scholar]
- Zhao S et al. (2013) Systems pharmacology of adverse event mitigation by drug combinations. Sci Transl Med 5: 206ra140. [DOI] [PMC free article] [PubMed] [Google Scholar]