Abstract
Prim-pol is a recently identified DNA primase-polymerase belonging to the archaeao-eukaryotic primase (AEP) superfamily. Here, we characterize a previously unrecognized prim-pol in human cells, which we designate hPrimpol1 (human primase-polymerase 1). hPrimpol1 possesses primase and DNA polymerase activities in vitro, interacts directly with RPA1 and is recruited to sites of DNA damage and stalled replication forks in an RPA1-dependent manner. Cells depleted of hPrimpol1 display increased spontaneous DNA damage and defects in the restart of stalled replication forks. Both RPA1 binding and the primase activity of hPrimpol1 are required for its cellular function during DNA replication. Our results indicate that hPrimpol1 is a novel factor involved in the response to DNA replication stress.
Keywords: hPrimpol1/CCDC111, primase-polymerase, RPA1, stalled replication fork
Introduction
To maintain genetic information, human cells must accurately replicate billions of base pairs of DNA and repair a very wide range of DNA lesions. One of the proteins required for the maintenance of genetic information is the single-stranded DNA (ssDNA)-binding protein, replication protein A (RPA) [1]. Human RPA is a stable three-subunit complex composed of RPA1, RPA2 and RPA3 [1]. RPA1 contains four OB-folds, and RPA2 and RPA3 have one OB-fold each [1]. Although each OB-fold is structurally similar, the majority of the ssDNA binding occurs through two OB-folds (DBD-A and DBD-B) centrally located in RPA1, referred to as the ssDNA-binding core [2, 3]. In addition to the ssDNA-binding core, RPA1 also contains an OB-fold at each terminus. The OB-fold at the N terminus of RPA1, DBD-F, interacts with a large number of other proteins and is required for DNA repair, recombination and cell cycle regulation [3, 4]. DBD-C, at the C terminus of RPA1, is required for heterotrimeric complex formation and has been implicated in recognition of some types of DNA damage [3, 4]. As RPA is engaged in these diverse functions through its ssDNA-binding activity and its ability to interact with multiple proteins involved in these pathways, it has thus been considered as an adaptor protein that facilitates various biochemical reactions that occur at ssDNA regions during DNA replication and/or DNA repair [3–10].
DNA primases are enzymes that catalyse the synthesis of short RNA (or DNA in some organisms) sequences called primers, which serve as starting points for DNA synthesis [11–13]. Structurally, most DNA primases can be divided into two classes [11–13]. The first class contains the DnaG family enzymes found in bacteria and archaea [11, 14]. The second class comprises the heterodimeric primases of the archaeao-eukaryotic primase (AEP) superfamily found in the eukarya and archaea [11, 14]. Recently, a novel family of AEPs, called the prim-pol, which is sporadically present in crenarchaeal and Gram-positive bacterial plasmids, has been described [15, 16]. Members of the prim-pol family are able to catalyse both primase and DNA polymerase reaction in vitro, hence the name prim-pols (primase-polymerases) [15–17]. The precise physiological function of these prim-pols has remained largely unexplored.
In this study, we used an affinity purification approach to isolate RPA-containing complex and identified a novel protein CCDC111, which we refer to as hPrimpol1 (human primase-polymerase 1). hPrimpol1 belongs to the prim-pol family and possesses both primase and DNA polymerase activities in vitro. hPrimpol1 is recruited to sites of DNA damage and stalled replication forks via its direct interaction with RPA1. Cells with hPrimpol1 depletion display increased spontaneous DNA damage and defects in the restart of stalled replication forks, suggesting that hPrimpol1 normally acts to stabilize stalled replication forks. Our results provide the first evidence of the involvement of a human DNA primase-polymerase in the replication stress response.
Results and discussion
Identification of hPrimpol1/CCDC111
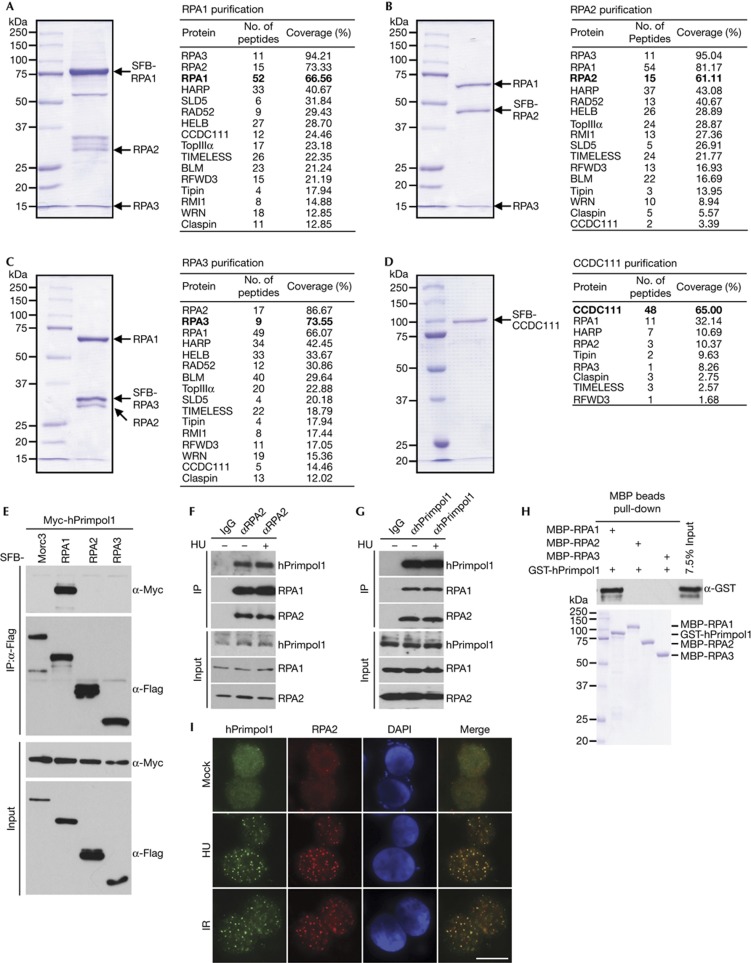
To search for previously undetected proteins present in RPA-containing complex, we performed tandem affinity purification using HEK293T cells stably expressing SFB-tagged wild-type RPA1, RPA2 or RPA3 for the identification of RPA-interacting proteins. Mass spectrometry analysis revealed several known RPA-associated proteins, including SMARCAL1/HARP, RAD52 and BLM (Fig 1A–C). Interestingly, we also repeatedly identified a previously uncharacterized RPA-binding protein as CCDC111 (Fig 1A–C). To ensure that CCDC111 indeed associates with RPA, we performed reverse tandem affinity purification using a cell line stably expressing tagged CCDC111, and identified RPA1, RPA2 and RPA3 as major CCDC111-associated proteins (Fig 1D). These data strongly suggest that CCDC111 is a bone fide RPA-binding protein. The CCDC111 gene encodes a deduced polypeptide of 560 amino acids with a predicted molecular mass of 65 kDa. Structure and sequence analysis revealed that CCDC111 belongs to the prim-pol family and contains two conserved domains: an AEP domain (residues 101–240) and a Zn ribbon-like domain (residues 392–470) [15]. We have now designated this protein as hPrimpol1 (for ‘human primase-polymerase 1’).
Figure 1.
Identification of hPrimpol1/CCDC111 as a novel RPA-binding partner. (A–D) 293T cells stably expressing SFB-tagged-RPA or hPrimpol1/CCDC111 were used for TAP of protein complexes specifically from chromatin fractions. Tables are summaries of proteins identified by mass spectrometry analysis. Letters in bold indicate the bait proteins. (E) hPrimpol1 interacts with RPA1. 293T cells were transfected with indicated plasmids. Cell lysates were immunoprecipitated with anti-Flag antibody and western blot analysis was performed as indicated. (F,G) Association of endogenous hPrimpol1 with the RPA complex in HeLa cells was performed by co-immunoprecipitation using anti-RPA2 or anti-hPrimpol1 antibody. HeLa cells were lysed in the presence of benzonase, cell lysates were then incubated with protein A agarose beads conjugated with indicated antibodies and western blot analysis was carried out as indicated. (H) Direct binding between recombinant GST-tagged hPrimpol1 and MBP-tagged-RPA. Upper panel: hPrimpol1 was detected by immunoblotting. Lower panel: Purified proteins visualized by Coomassie staining. (I) hPrimpol1 colocalizes with RPA2. SFB-tagged hPrimpol1 was expressed in 293T cells. Foci assembled by this fusion protein and by RPA2 following exposure to HU (2 mM) for 16 h or IR (10 Gy) followed by recovery for 3 h were detected by immunofluorescence using anti-Flag and anti-RPA2 antibodies, respectively. A merged image shows colocalization. Scale bar, 10 μm. GST, glutathione S-transferase; hPrimpol1 human primase-polymerase 1; MBP, maltose binding protein; RPA, replication protein A; TAP, tandem affinity purification.
hPrimpol1 interacts with RPA
To validate our tandem affinity purification results, we first performed co-immunoprecipitation experiments using epitope-tagged hPrimpol1 and RPA. We found that Myc-tagged hPrimpol1 interacts strongly with SFB-tagged RPA1 but not with the Morc3 control protein (Fig 1E). In addition, the interaction between hPrimpol1 and RPA was confirmed by reciprocal immunoprecipitations using antibodies against endogenous RPA2 and hPrimpol1, respectively (Fig 1F,G; supplementary Fig S1A,B online). Moreover, the hPrimpol1–RPA complex formation was DNA damage-independent (Fig 1F,G). To determine whether the interaction between RPA and hPrimpol1 is direct, we expressed and purified recombinant MBP-tagged RPA1, RPA2, RPA3 and GST-tagged hPrimpol1. Pull-down experiments revealed that hPrimpol1 binds strongly with RPA1 but not with RPA2 or RPA3 (Fig 1H), indicating that hPrimpol1 associates with RPA complex through RPA1.
Upon the occurrence of DNA damage or inhibition of DNA replication, RPA could form large nuclear foci. A physical interaction between hPrimpol1 and RPA as demonstrated above raises the possibility that hPrimpol1 might colocalize with RPA at sites of DNA damage or other barriers in the cell. Indeed, discrete foci of hPrimpol1 were readily detected in cells following hydroxyurea (HU) or IR treatment (Fig 1I). Moreover, these foci colocalize with RPA2 foci, indicating that the localization of hPrimpol1, like that of RPA, is regulated in response to DNA damage and DNA replication stress (Fig 1I).
RPA-binding is required for hPrimpol1 foci formation
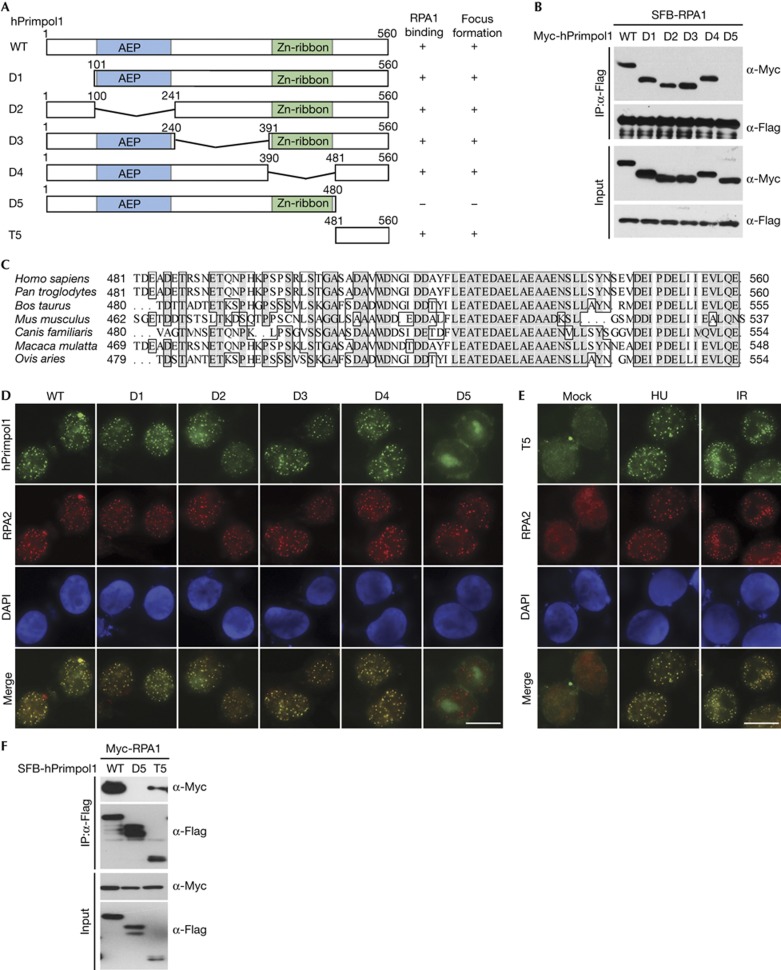
To further define the binding between hPrimpol1 and RPA1, we sought to identify the region(s) within hPrimpol1 responsible for its interaction with RPA1 (Fig 2A). Co-immunoprecipitation experiments revealed that hPrimpol1 associated with RPA1 through its very C terminus, as deletion mutant lacking the C-terminal 80 amino acids (D5) failed to co-precipitate with RPA1 (Fig 2B). Interestingly, the C terminus of hPrimpol1 has been highly conserved throughout evolution, suggesting that it might carry out an important function of hPrimpol1 (Fig 2C). Indeed, although distinct nuclear foci of full-length hPrimpol1 and the other mutants were readily detected in HU- or IR-treated cells, the D5 mutant, which does not bind to RPA1, failed to form nuclear foci after HU or IR treatment (Fig 2D). Thus, the conserved C-terminal region of hPrimpol1 is important not only for its interaction with RPA1, but also for its proper localization in response to DNA damage and replication stress.
Figure 2.
The conserved C terminus of hPrimpol1 is required for RPA1 binding and its foci formation. (A) Schematic representation of wild-type and deletion mutants of hPrimpol1 used in this study. (B) The 80 amino acids at the C terminus of hPrimpol1 are required for RPA1 binding. Co-immunoprecipitation experiments were carried out as indicated. (C) Alignment of C-terminal sequences of hPrimpol1 from different species. (D) The conserved C terminus of hPrimpol1 is responsible for its foci formation. Immunostaining experiments were performed 16 h after HU treatment using indicated antibodies. Scale bar, 10 μm. (E) The conserved C terminus of hPrimpol1 is sufficient for its foci formation. 293T cells were transfected with SFB-tagged T5 mutant of hPrimpol1. Immunostaining experiments were performed using indicated antibodies. Scale bar, 10 μm. (F) The C-terminal fragment (T5) of hPrimpol1 is sufficient for RPA1 binding. Co-immunoprecipitation experiments were carried out as indicated. hPrimpol1 human primase-polymerase 1; HU, hydroxyurea; RPA, replication protein A.
To test whether this C-terminal region of hPrimpol1 is also sufficient to bind RPA1 and localize hPrimpol1 to sites of DNA damage and stalled replication forks, we generated a construct encoding the C-terminal region alone (T5) (Fig 2A). As shown in Fig 2E, the T5 protein localized to sites of DNA damage and stalled replication forks as efficiently as full-length hPrimpol1. Moreover, T5 retained the ability to interact with RPA1, although this ability was reduced in comparison with the wild-type hPrimpol1 (Fig 2F). These results suggest that hPrimpol1 is likely to be recruited to sites of DNA damage and stalled replication forks via an interaction between its conserved C-terminal 80 amino acids and RPA1. Consistent with this hypothesis, CtIP depletion abolished IR-induced recruitment of RPA and the downstream hPrimpol1 to DNA damage sites (supplementary Fig S1C,D online).
The DBD-C domain of RPA1 interacts with hPrimpol1
To identify the hPrimpol1-binding domain on RPA1, several RPA1 truncation and internal deletion mutants were generated (supplementary Fig S2A online). Although wild-type and other mutants of RPA1 could be co-immunoprecipitated with hPrimpol1, the D5 mutant, which is deleted of DBD-C domain, failed to bind to hPrimpol1 (supplementary Fig S2B online). These results suggest that the DBD-C domain of RPA1 is required for its binding to hPrimpol1.
We next examined whether the physical interaction between hPrimpol1 and RPA1 might be important for RPA1 localization. As shown in supplementary Fig S2C online, the D5 mutant was still able to form discrete foci following HU treatment. By contrast, the DBC-A domain deletion mutant and the DBC-B domain deletion mutant totally lost their foci formation ability. These observations suggest that the interaction between hPrimpol1 and RPA1 is critical for hPrimpol1 focus formation, but not vice versa.
hPrimpol1 has primase and DNA polymerase activities
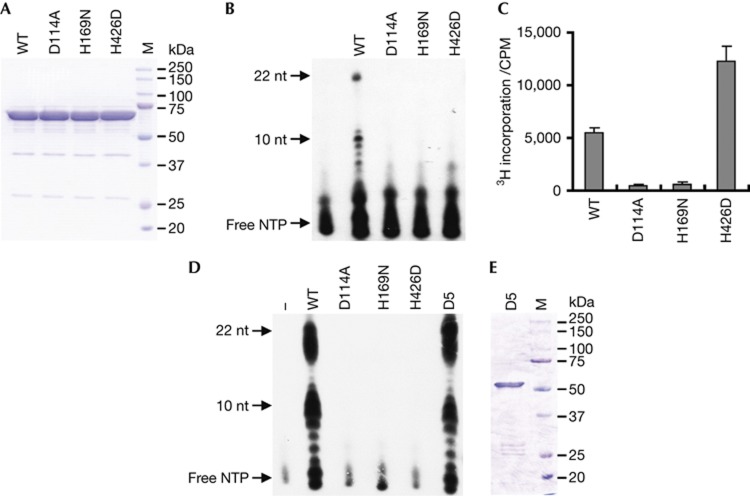
As hPrimpol1 contains a highly conserved AEP-primase domain, we tested whether hPrimpol1 is a bona fide primase. We purified wild-type hPrimpol1 protein (Fig 3A) and examined its primase activity using an assay described previously [18, 19]. As shown in Fig 3B, after incubation with various ssDNA templates in the presence of NTPs containing [α-32P] ATP, hPrimpol1 shows strong primase activity in vitro. Primers of variable size (mostly 7–11 nucleotides) were synthesized according to different template sequences, with the longest RNA primers (about 22 nucleotides) being produced on T20CTGT20 template (Fig 3B). hPrimpol1 showed profoundly robust activity with a triplet (5′-Pyrimidine-Pyrimidine-G-3′) in the template as herpes simplex virus primase, whereas no primers longer than 3 nucleotides in length could be detected on poly(dC)40 template (unpublished data). To confirm that the primase activity we observed is intrinsic to hPrimpol1, we generated hPrimpol1 mutations at three highly conserved residues within its primase domain (D114A and H169N) and Zn ribbon-like domain (H426D), respectively (Fig 3A). All three mutations abolished the primase activity of hPrimpol1 (Fig 3B). Meanwhile, hPrimpol1 was capable to incorporate 3H-dTTP into poly(dA)500/oligo(dT)18 substrate (Fig 3C), indicating that it also has DNA polymerase activity. Interestingly, H426D mutant showed even stronger DNA polymerase activity than wild type, whereas mutations in the primase domain (D114A and H169N) destroyed the DNA polymerase activity (Fig 3C). These results suggest that the Zn ribbon-like domain is indispensable for de novo RNA primer synthesis but not necessary for primer extension activity of hPrimpol1.
Figure 3.
hPrimpol1 possesses primase and DNA polymerase activities. (A) SDS–PAGE profile of purified wild-type and mutants of hPrimpol1. (B) hPrimpol1 is a bona fide primase. Wild type and mutants of hPrimpol1 were incubated with ssDNA template (T20GTCT20) and NTPs containing [α-32P]-ATP as substrates. Reaction products were separated by 8 M Urea-SDS–PAGE and detected by autoradiography. (C) hPrimpol1 displays DNA polymerase activity. The ability of wild-type and mutant proteins to synthesize DNA was analysed by classical tritium incorporation assay. Incorporation of [3H] dTTP into poly(dA)500/oligo(dT)18 was quantified by scintillation counting. Error bars are s.d.; n=3. (D) Both wide-type and the D5 mutant of hPrimpol1 can elongate the primers synthesized by its own primase activity efficiently. Reactions were carried out as standard primase assays in Fig. 3B except that 100 μM dNTPs were added together with 100 μM NTPs containing [α-32P]-ATP. (E) SDS–PAGE profile of purified D5 mutant of hPrimpol1. hPrimpol1 human primase-polymerase 1; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; ssDNA, single-stranded DNA.
As hPrimpol1 exhibits primase and DNA polymerase activities, the primase-polymerase coupled assays were conducted by addition of dNTPs together with NTPs into standard primase reactions. As shown in Fig 3D, compared with NTPs only reactions, much more abundant products (ranging from 7–22 nucleotides) could be detected in the presence of wild-type hPrimpol1, but not the mutants including H426D. These results indicate that hPrimpol1 can elongate the primers synthesized by themselves. Moreover, the D5 mutant can extend the primers as efficiently as wild-type hPrimpol1, suggesting that the C-terminal-conserved RPA1-binding domain might not be required for the in vitro biochemical activities of hPrimpol1 (Fig 3D,E).
hPrimpol1 is involved in replication stress response
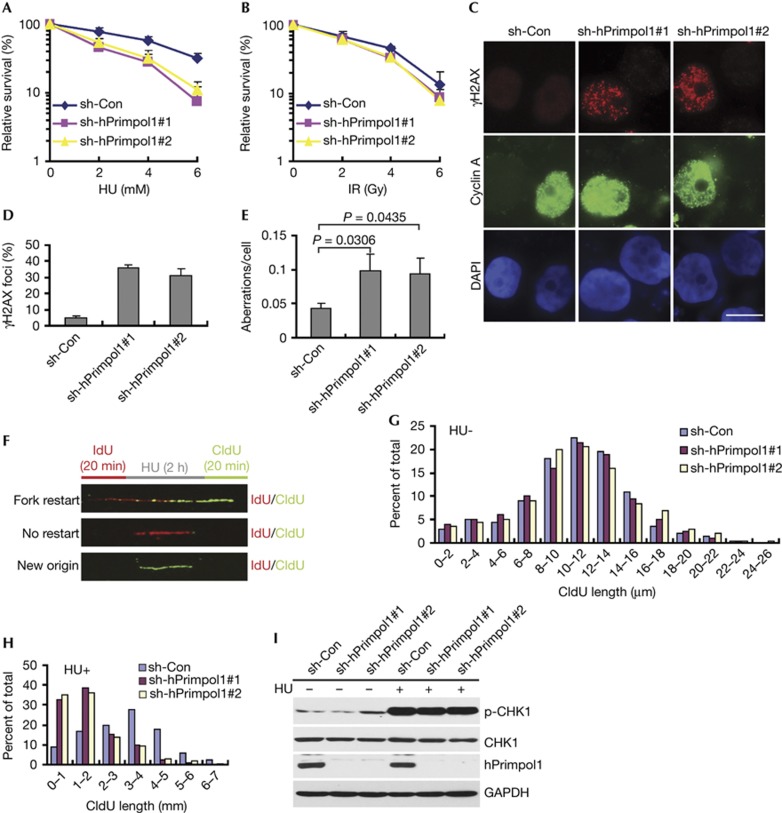
In order to investigate the cellular function of hPrimpol1, we knocked down its expression in human HeLa cells using two independent small hairpin RNAs targeting hPrimpol1. Interestingly, hPrimpol1 knockdown cells showed significantly elevated sensitivity to HU as compared with cells treated with control shRNA, suggesting an involvement of hPrimpol1 in the response to HU-induced replication fork stalling (Fig 4A). hPrimpol1 knockdown cells, however, showed little or no hypersensitivity to IR, indicating that hPrimpol1 might only have a mild role in the repair of IR-induced DNA damage (Fig 4B).
Figure 4.
hPrimpol1 functions at stalled replication forks. (A,B) Clonogenic survival assays in hPrimpol1-depleted HeLa cells following HU or IR treatment. Error bars are s.d.; n=3. (C,D) γH2AX foci are greatly increased in hPrimpol1-depleted HeLa cells. HeLa cells were infected with lentiviruses carrying non-target control or hPrimpol1 shRNAs. Seventy-two hours later, cells were subjected to immunostaining using indicated antibodies (C). Scale bar, 10 μm. The quantification of foci-positive cells was performed by counting a total of 200 cells per sample (D). Error bars are s.d.; n=3. (E) Quantification of chromosomal aberrations in control and hPrimpol1-depleted HeLa cells. At least 50 cells were counted in each experiment. Error bars are s.d.; n=3; significances of differences by a two-tailed unpaired t-test. (F) Schematic representation of the labelling protocol for DNA fibre analysis of replication forks. (G,H) CIdU tract length distributions from DNA fibres from control and hPrimpol1-depleted HeLa cells in the presence (H) or absence of HU (G). (I) hPrimpol1 is not required for CHK1 activation. HeLa cells were introduced with indicated shRNA and then treated with HU (2 mM) for 2 h. Cells were lysed and immunoblotting was performed using indicated antibodies. hPrimpol1 human primase-polymerase 1; HU, hydroxyurea; shRNA, small hairpin RNA.
The absence of proteins that function in DNA replication and/or repair often results in the accumulation of intrinsic DNA damage. Indeed, hPrimpol1-depleted cells accumulated spontaneous γH2AX, 53BP1 and pATM foci and displayed a slight spontaneous phosphorylation of CHK2 (Fig 4C,D; supplementary Fig S3A–C online). In addition, more than 90% of cells with γH2AX foci were cyclin A positive, suggesting that hPrimpol1 prevents DSBs from arising during DNA replication (Fig 4C). Moreover, hPrimpol1-depleted HeLa cells displayed a significant increase in chromosomal aberrations (Fig 4E). These results demonstrate that hPrimpol1 is important for maintaining genomic stability.
hPrimpol1 promotes restart of replication forks
The localization of hPrimpol1 to stalled replication forks, appearance of γH2AX foci in hPrimpol1-depleted HeLa cells and its ability to act as a primase-polymerase in vitro indicate that hPrimpol1 might normally be required during the restart of stalled replication forks. To test this possibility, we analysed replication fork progression on single DNA fibres. HeLa cells were pulse labelled with the modified thymidine analogue iododeoxyuridine (IdU) for 20 min, and then treated with HU for 2 h to arrest replication fork progression. Following removal of HU, cells were labelled with another thymidine analogue, chlorodeoxyuridine (CIdU), for an additional 20 min, and DNA fibres were then isolated and stained for IdU and CIdU. Replication forks that were able to restart following HU treatment were visualized as a stretch of IdU incorporation (Fig 4F, labelled in red) followed by a stretch of CIdU incorporation (Fig 4F, labelled in green). Tracts showing only IdU incorporation (Fig 4F, red only) indicate stalled forks that were unable to restart replication following the removal of HU. Replication origins that fired after removal of HU show only CIdU incorporation (Fig 4F, green only). As shown in Fig 4G and supplementary Fig S3D online, depletion of hPrimpol1 showed a modest effect on fork progression in the absence of replication stress. However, the restart of replication forks following HU treatment was notably reduced in the hPrimpol1-depleted cells (Fig 4H; supplementary Table S1 online). By contrast, new origin firing after HU treatment was not significantly affected upon hPrimpol1 depletion (supplementary Fig S3E online). These results indicate that hPrimpol1 is likely involved in stabilizing stalled replication forks following replication stress by directly mediating the reinitiating of DNA replication.
As cells depleted of hPrimpol1 were sensitive to replication stress, we also examined whether hPrimpol1 is involved in replication checkpoint control. As shown in Fig 4I, there was no detectable change in phospho-CHK1 level in hPrimpol1-depleted cells following HU treatment. Our results suggest that although hPrimpol1 depletion results in destabilization of stalled replication forks, it does not participate in replication checkpoint control.
Cellular functions of hPrimpol1 are regulated by RPA
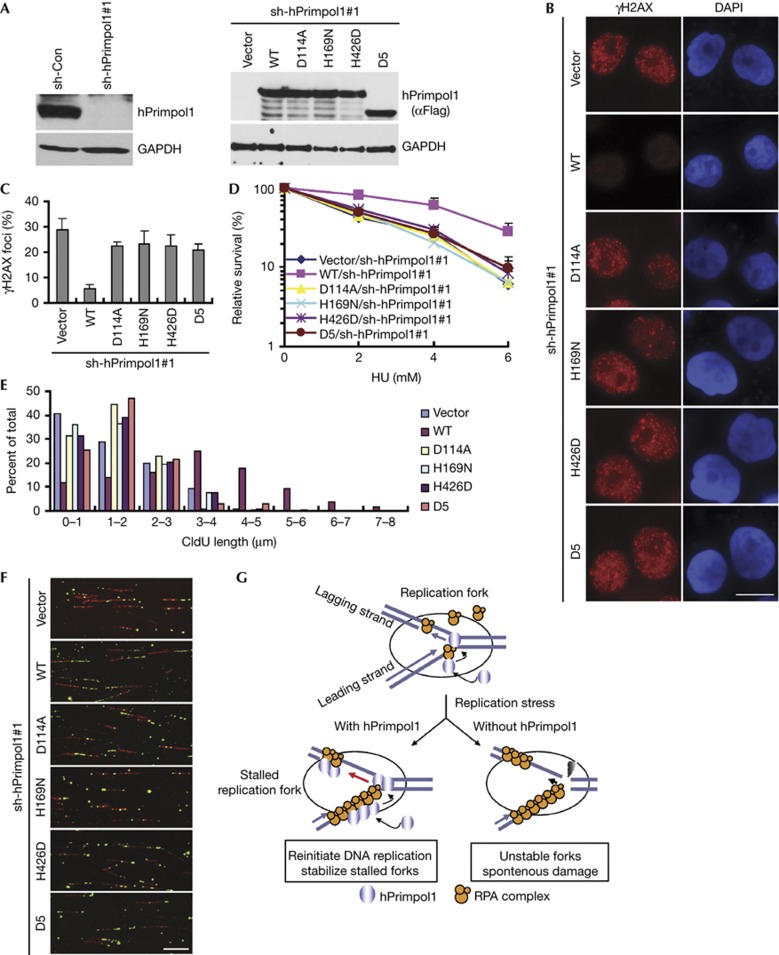
To explore the physiological relevance of the enzymatic activities and the RPA1-binding ability of hPrimpol1 in replication fork stabilization, we took advantage of the inducible expression system to express the shRNA#1-resistant full-length hPrimpol1, D5, or the primase-inactivating mutants (D114A, H169N and H426D) of hPrimpol1 in hPrimpol1-depleted HeLa cells. The expression of wild-type and mutated hPrimpol1 was induced in hPrimpol1 knockdown cells when the cells were treated with doxycycline (Fig 5A). Notably, neither expression of the D5 mutant nor the primase-inactivating mutants was capable of preventing the appearance of γH2AX foci and suppressing the elevated HU sensitivity when endogenous hPrimpol1 was silenced (Fig 5B–D). Likewise, the defects in fork restart in hPrimpol1-depleted cells after replication arrest could be reversed by the expression of wild-type hPrimpol1, but not the D5 and the primase-inactivating mutants of hPrimpol1 (Fig 5E,F; supplementary Table S1 online). These results suggested that both the RPA1-binding domain and the primase activity of hPrimpol1 are critical for its in vivo function.
Figure 5.
The RPA1 binding and the primase activity of hPrimpol1 are required for its cellular functions. (A) shRNA-resistant hPrimpol1 and its indicated mutants were transduced into hPrimpol1-deplected HeLa cells. The indicated proteins were analysed in HeLa cells cultured with doxycycline (Dox) for 24 h. (B,C) hPrimpol1 depletion-induced increase of γH2AX foci was rescued by the expression of wild-type hPrimpol1 but not the D5 and the enzyme-inactivating mutants of hPrimpol1. shRNA-resistant hPrimpol1 and its indicated mutants were transduced into hPrimpol1-depleted HeLa cells. Cells were cultured with Dox for 24 h and then subjected to immunostaining using indicated antibodies (B). Scale bar, 10 μm. The quantification of foci-positive cells was performed by counting a total of 200 cells per sample (C). Error bars are s.d.; n=3. (D) Both RPA1 binding and the primase activity of hPrimpol1 are required for restoring cellular resistance to HU. Error bars are s.d.; n=3. (E,F) Defects in fork restart in hPrimpol1-deplected HeLa cells were rescued by wild-type hPrimpol1 but not by its mutants. Quantification of CIdU track lengths was shown (E). Images of DNA fibres isolated from cells treated with hPrimpol1 shRNA following the expression of shRNA-resistant wild-type, the D5, or the enzyme-inactivating mutants of hPrimpol1 (F). Scale bar, 10 μm. (G) A proposed model of hPrimpol1 function at replication forks. Please refer the text for details. hPrimpol1 human primase-polymerase 1; HU, hydroxyurea; shRNA, small hairpin RNA.
Re-priming is a mechanism to allow resumption of DNA synthesis following replication fork stalling [20, 21]. How the re-priming occurs is largely unexplored. In this study, we report the identification of a unique protein designated hPrimpol1, which might serve as a primase-polymerase that helps to stabilize stalled replication forks. We propose that, in response to replication stress, replication forks would be stalled and allow the formation of ssDNA regions that become coated with RPA. Through the interaction with RPA1, hPrimpol1, SMARCAL1/HARP [5–9] and/or other RPA-binding proteins are recruited to these stalled replication forks and exerts their enzymatic activities to reinitiate DNA replication and thus to limit replication-associated DNA damage (Fig 5G).
Methods
Primase assays. Primase activity was analysed using ssDNA templates as described [18, 19]. Reaction mixtures (20 μl) contained 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 1 mM dithiothreitol, 50 μg ml−1 bovine serum albumin, 5 μM ssDNA template, 100 μM NTP containing [α-32P] ATP or GTP (800 ci mmol−1, PerkinElmer Life Sciences), and 15 μg hPrimpol1 protein. After incubation at 37 °C for 30 min, reactions were quenched by adding equal volume of gel-loading buffer (100% formamide, 0.05% bromophenol blue). The sequences of ssDNA template are 5′-T20CTGT20-3′ (T20CTGT20) and 5′-C40-3′ (poly(dC)40). Products were separated by denaturing polyacrylamide gel electrophoresis (20% polyacrylamide, 8 M urea) and visualized by autoradiography.
Polymerase assays. Incorporation of 3H-dTTP into poly(dA)500/oligo(dT)18 DNA substrate was carried out as described [19]. The poly(dA)500/oligo(dT)18 (1:20, template to primer chains) was annealed in 25 mM Hepes (pH 7.1), 60 mM KCl. Reaction mixtures (60 μl) consisted of 50 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 120 mM NaCl, 2 mM dithiothreitol, 17 μg ml−1 poly(dA)500/oligo(dT)18, 10% glycerol, 100 μg ml−1 bovine serum albumin, and 50 μM [3H] dTTP (100 cpm pmol−1). The reactions were conducted at 37 °C for 30 min. The mixtures were directly spotted on Whatman DE81 filter paper. The filters were washed five times with 0.5 M Na2HPO4, two times with water and then rinsed with 95% ethanol. Radioactivity was monitored by scintillation counter Triathler.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank all our colleagues in the Huang laboratory for insightful discussions. This work was supported by National Basic Research Program of China Grants 2012CB944402 and 2013CB911003, National Natural Science Foundation of China 31071095 and 31271331, and the China’s Fundamental Research Funds for the Central Universities.
Author contributions: L.W., J.L., Y.X., B.S., T.L., J.C. and Y.S. performed the experiments; J.H. and H.L. designed the experiments, analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Wold MS (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92 [DOI] [PubMed] [Google Scholar]
- Arunkumar AI, Stauffer ME, Bochkareva E, Bochkarev A, Chazin WJ (2003) Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J Biol Chem 278: 41077–41082 [DOI] [PubMed] [Google Scholar]
- Haring SJ, Mason AC, Binz SK, Wold MS (2008) Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J Biol Chem 283: 19095–19111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT (2009) The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 23: 2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J (2009) The annealing helicase HARP protects stalled replication forks. Genes Dev 23: 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D (2009) The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ (2009) The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FB et al. (2013) ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D (2008) The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol 28: 7345–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Richardson CC (2001) DNA primases. Annu Rev Biochem 70: 39–80 [DOI] [PubMed] [Google Scholar]
- Kuchta RD, Stengel G (2010) Mechanism and evolution of DNA primases. Biochim Biophys Acta 1804: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao-Sirieix SH, Pellegrini L, Bell SD (2005) The promiscuous primase. Trends Genet 21: 568–572 [DOI] [PubMed] [Google Scholar]
- Swiatek A, Macneill SA (2010) The archaeo-eukaryotic GINS proteins and the archaeal primase catalytic subunit PriS share a common domain. Biol Direct 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L (2005) Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 33: 3875–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps G, Rother S, Hart C, Krauss G (2003) A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J 22: 2516–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P (2004) Structure of a bifunctional DNA primase-polymerase. Nat Struct Mol Biol 11: 157–162 [DOI] [PubMed] [Google Scholar]
- Ramirez-Aguilar KA, Low-Nam NA, Kuchta RD (2002) Key role of template sequence for primer synthesis by the herpes simplex virus 1 helicase-primase. Biochemistry 41: 14569–14579 [DOI] [PubMed] [Google Scholar]
- Lou H, Duan Z, Huo X, Huang L (2004) Modulation of hyperthermophilic DNA polymerase activity by archaeal chromatin proteins. J Biol Chem 279: 127–132 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Elvers I, Johansson F, Groth P, Erixon K, Helleday T (2011) UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res 39: 7049–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.