Nature (2013) doi: ; DOI: 10.1038/nature12542
Nature (2013) doi: ; DOI: 10.1038/nature12653
Cell Host Microbe (2013) doi: ; DOI: 10.1016/j.chom.2013.08.015
Our innate immune system constitutes a first line of defence against invading microorganisms, long before pathogen-specific adaptive immune responses become effective. Intense global research efforts over the past 30 years have deepened our understanding of the molecular principles that underlie the early innate recognition of evolutionarily conserved structures on pathogens, the signalling cascades this elicits, and the proinflammatory mediators and host cell effectors that inhibit pathogen replication. On the other side of the coin, microbes have evolved strategies to circumvent these processes, an understanding of which could enable new therapeutic approaches. Innate pathogen recognition by the host typically results in the expression of proinflammatory cytokines, which orchestrate early defence mechanisms to infection. A characteristic of this response is the secretion of interferons (IFNs), named after their capacity to ‘interfere’ with viral replication in host cells through the induction of IFN-stimulated genes (ISGs). Three reports from the Malim, Bieniasz and Liang labs, published in Nature and Cell Host & Microbe, now describe human MxB/Mx2 as a potent IFN-induced inhibitor of the early phase of human immunodeficiency virus 1 (HIV-1) infection in target cells [1,2,3].
Landmark studies of influenza viruses in the 1980s identified the murine myxovirus resistance protein 1 (Mx1) as the first cellular ISG with potent antiviral activity, which is conserved in the human orthologue MxA [4]. MxA is a GTPase of the dynamin superfamily, the expression of which is potently induced by type I and type III IFNs. MxA multimers apparently adopt ring-like structures that allow them to function as restriction factors for influenza viruses by binding to incoming viral capsids [5]. In addition to MxA, humans encode for a second Mx protein, MxB, also referred to as Mx2. Despite having 63% sequence identity with MxA, MxB was reported not to have antiviral activity against influenza or vesicular stomatitis virus [6], and its biological role has remained largely elusive.
In contrast to the influenza virus field, studies on HIV for many years focused on strategies to induce broadly effective adaptive responses and to interfere with cellular cofactors of infection. The interest in innate immunity against HIV was spurred by the characterization of potent anti-HIV activity by IFNs in tissue culture models, the detection of strong proinflammatory cytokine/IFN responses in patients during the acute phase of HIV-1 infection, and the identification of several IFN-inducible restriction factors—including APOBEC3 proteins, TRIM5α, SAMHD1 and BST2/CD317/tetherin— that target HIV-1 at early and late steps of the replication cycle (Fig 1) [7]. However, these restriction factors cannot fully account for the anti-HIV state elicited in HIV target cells by IFN. As a consequence, systematic efforts were launched to identify additional ISGs with anti-HIV activity. Overexpression screening approaches to test the antiviral activity of more than 380 human ISGs initially reported that MxB has some antiviral activity against HIV-1 [8], as well as against vesicular stomatitis virus and mouse herpes virus type 68 [9].
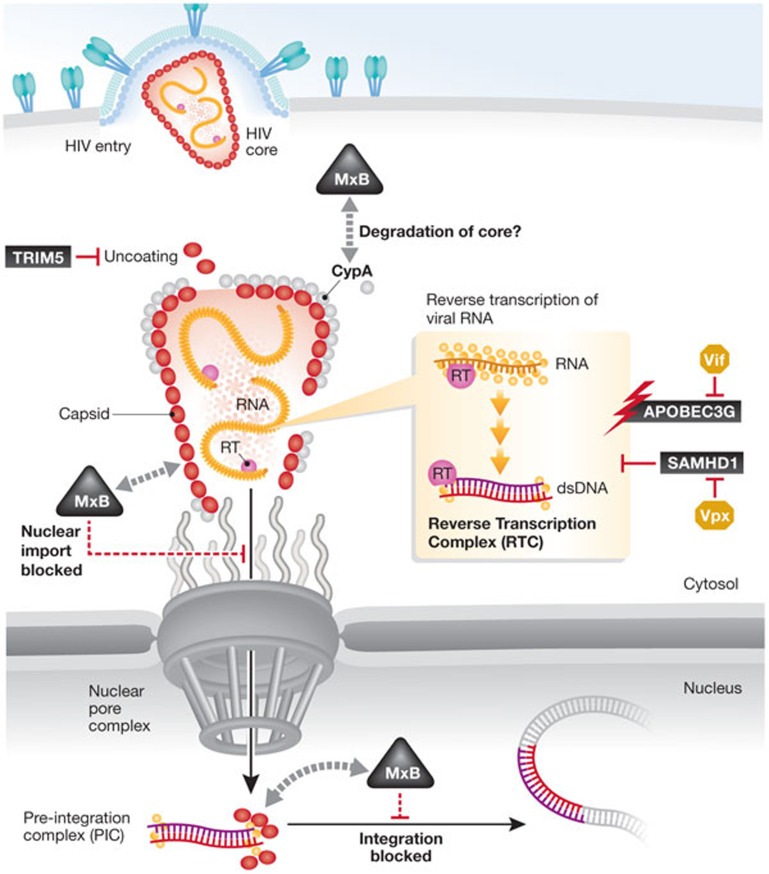
Figure 1.
Interferon-induced anti-HIV restriction factors. Several proteins induced by the interferon response target various early steps of the lentiviral life cycle, and the virus encodes proteins that counteract some of them. MxB/Mx2 is the latest addition to the cell's arsenal against infection by HIV-1. Three proposed models for the action of MxB/Mx2 are depicted. See text for details. HIV, human immunodeficiency virus.
The Malim and Bieniasz labs used comparative gene expression profiling in cell lines that differed in their capacity to restrict the early post-entry phase of HIV infection after type I IFN treatment as an unbiased screening approach for ISGs with anti-HIV activity [1,3]. Both of these studies, as well as the one from the Liang lab [2], showed that ectopic overexpression of MxB potently reduces the permissiveness of the cells in a single-cycle HIV-1 infection assay and results in a pronounced reduction of virus spread over multiple rounds of replication. Moreover, silencing of MxB expression by RNA interference in type I-IFN-treated cells markedly reduced the inhibitory potency of the cytokine, pinpointing MxB as an important IFN effector with anti-HIV-1 activity.
Next, the three groups addressed which specific post-entry step of the HIV replication cycle is affected by MxB expression by using established quantitative PCR methodology. Reverse transcription of the HIV-1 RNA within incoming particles to cDNA occurred normally, whereas integration of HIV-1 proviruses into the host cell genome was markedly diminished in MxB-expressing cells. An MxB-dependent reduction of HIV-1 2-long terminal repeat circles was also observed by Goujon et al and Kane et al [1,3], but not by Liu et al [2]. This HIV-1 cDNA species acts as a quantitative marker for successful nuclear import of the reserved transcribed viral genome in infected cells. These results place the anti-HIV activity of MxB in the post-entry phase of HIV-1 infection after cDNA synthesis, possibly involving nuclear import, nuclear stability and/or integration of the viral cDNA (Fig 1). Analysis of a broad range of primate lentiviruses and retroviruses revealed that human MxB potently inhibits HIV-1, HIV-2 and simian immunodeficiency virus, has a lower effect on the infection by feline immunodeficiency virus and equine infectious anaemia virus, and only marginal or no activity against murine leukaemia virus.
Two experimental approaches were taken to define the viral component targeted—directly or indirectly—by MxB and gain a first insight into its mode of action. HIV-1 capsid protein is known to play a central role in mediating physical interactions with several host proteins involved in the post-entry phase of infection, so the Malim and Bieniasz laboratories [1,3] focused their attention on this component of the viral reverse transcription complex (RTC)/pre-integration complex (PIC). They identified residues in capsid that are involved in binding to cyclophilin A (CypA), TRIM5α, TNP03, CPSF6, NUP153 and NUP358/RanBP2, as also critical for the sensitivity of HIV-1 to the antiviral action of MxB. Consistently, the study by the Liang laboratory [2] observed that passaging HIV-1 in SupT1 T cells, which constitutively express high levels of MxB, resulted in the rapid emergence of HIV-1 variants that had similar mutations in capsid. Thus, the viral capsid governs the sensitivity of HIV-1 to MxB. Furthermore, the results obtained by Liu and colleagues indicate that the action of MxB might be associated with that of CypA, as silencing of CypA expression or disruption of the capsid–CypA interaction by addition of the immunosuppressive drug cyclosporine A abrogated the anti-HIV activity of MxB. Moreover, expression of a recombinant CypA–MxB fusion protein significantly increased the antiviral potency of MxB, presumably by facilitating its tethering to the RTC/PIC.
The first results from mutational analyses of MxB provided more questions than answers. In analogy to MxA, the GTPase activity of MxB was expected to be required for its antiviral effect. Surprisingly, this enzymatic activity seems in fact to be dispensable, as mutating residues essential for GTP binding or hydrolysis only mildly affects the anti-HIV-1 activity of MxB [1,3]. Also, the functional importance of the subcellular localization of MxB is unclear: although the amino-terminal 25 amino acids—which contain the nuclear localization signal of MxB—were strictly required, the primarily cytoplasmic MxB (Tyr131Ala) mutant retained antiviral activity.
The authors propose several models for MxB action (Fig 1), which are not mutually exclusive. MxB might directly bind to capsid within the RTC/PIC in a CypA-dependent manner and consequently inhibit nuclear import. In MxA-sensitive RNA viruses, oligomeric rings of MxA presumably target tubular nucleocapsid structures [4]. Conceivably, analogous ordered MxB complexes could recognize the highly structured capsid lattice of incoming HIV-1 particles to exert antiviral activity. Alternatively, MxB might block specific nuclear import pathways that are typically used by HIV-1. Mutations in capsid might simply allow the virus to exploit alternative, MxB-insensitive, transport machineries as ways into the nucleus. MxB might also act after nuclear entry and primarily affect proviral stability and integration. The precise mechanism of MxB action remains elusive and will undoubtedly become the focus of intense investigations.
In addition to ascribing a biological function to MxB and identifying it as a major anti-HIV effector of the IFN response, these three landmark studies raise several important questions: How potent is MxB in primary non-cycling and proliferating cells that are natural targets of HIV? Are there variations in the constitutive expression and IFN responsiveness of MxB at sites of transmission within the human population? Why does HIV not encode a bona fide MxB antagonist, as it does encode such antagonists for other restriction factors (Fig 1)? Do transmission and/or founder viruses have (transient) variations in capsid to escape MxB? What is the functional role of the different isoforms of MxB?
From a more general perspective, the post-entry phase of HIV-1 appears to be particularly vulnerable to the actions of IFN-inducible restriction factors: TRIM5α, APOBEC3 proteins, SAMHD1 and now MxB use distinct mechanisms to ultimately prevent chromosomal integration of this pathogenic lentivirus. The innate immune system undoubtedly plays a central role in the evolutionary arms race between virus and host. Deciphering the regulation of the growing number of IFN-induced anti-HIV restriction factors, their potential to cooperate and means of interfering with viral countermeasures could instruct innovative antiviral strategies.
Footnotes
The authors declare that they have no conflict of interest.
References
- Goujon C et al. (2013) Nature doi:; DOI: 10.1038/nature12542 [DOI] [Google Scholar]
- Liu Z et al. (2013) Cell Host Microbe doi:; DOI: 10.1016/j.chom.2013.08.015 [DOI] [PubMed] [Google Scholar]
- Kane M et al. (2013) Nature doi:; DOI: 10.1038/nature12653 [DOI] [Google Scholar]
- Haller O, Kochs G (2011) J Interferon Cytokine Res 31: 79–87 [DOI] [PubMed] [Google Scholar]
- Gao S et al. (2011) Immunity 35: 514–525 [DOI] [PubMed] [Google Scholar]
- Pavlovic J et al. (1990) J Virol 64: 3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD (2012) Cold Spring Harbour Persp Med 2: a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW et al. (2011) Nature 472: 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY et al. (2012) Proc Natl Acad Sci USA 109: 4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]